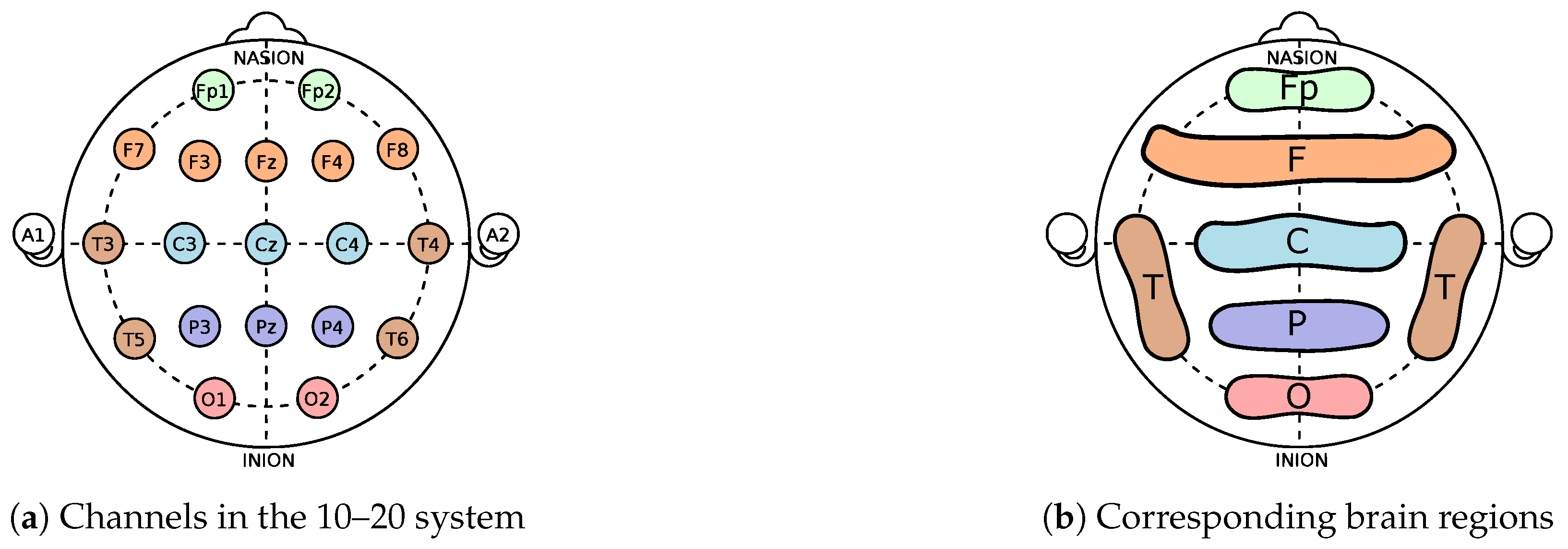1. Introduction
There is a myriad of physiological processes in biological systems [
1] that enable an organism to perform multiple activities. The modelling of these phenomena is a challenging task wherein different mathematical techniques capable of adequately describing such models have been employed [
2,
3,
4,
5,
6]. The brain is composed of a complex network of actions and reactions working in a coordinated effort to control several processes in the whole body. These networks incorporate a mixture of integration, differentiation, feedback loops, and other regulatory mechanisms that enable an organism to perform multiple activities, a quality typical of complex systems with time-adaptable features. Considering their non-stationary behaviour, modelling these phenomena is a very challenging task. Electrical activity can provide significant information about the dynamics pertaining to the behaviour of brain systems [
7].
A record of the brain electrical activity can be obtained by means of an electroencephalogram (EEG). EEG signals are the essence of the synchronous electrical activity of neuronal cells, which change their membrane potentials according to mental, motor, and sensory activities. These signals contain relevant information that can contribute to a better understanding of brain activity and to the diagnosis and treatment of several different pathologies, especially those with psychiatric and neurophysiological origins. In this context, EEG signals comprise valuable information for better understanding brain activity since they are influenced by both physiological and exogenous factors and have random characteristics. Indeed, there is an increasing demand for novel medical therapies with lower costs and higher efficiencies, motivating several researchers from different fields of knowledge, including physicians, physicists, and engineers, towards the same goal.
EEG signals can be obtained through the use of invasive and noninvasive electrodes. Noninvasive electrodes are widely used for their convenience and allow for capturing information with less risk to the patient. Invasive electrodes are mainly used when one needs to capture information from an internal region of the brain where noninvasive electrodes are insufficient. In general, typical EEG equipment uses noninvasive electrodes normally placed in caps or attached to the scalp [
8]. Usually, EEG data collection follows standards such as the International 10/20 scheme, which is detailed in
Section 3.
EEG analyses can be employed in cases of epilepsy [
9,
10] or clinical suspicion of this disease [
11]; in patients with altered consciousness; and for diagnostic evaluations of patients with other neurological (i.e., infectious [
12], degenerative [
13]), and psychiatric diseases [
14]. Furthermore, EEG can also be employed to investigate behavioural aspects (disease-related or not). It has been used to assess sleep quality, to characterise sleep stages [
15], and even to identify underlying disorders [
16,
17]. Recent fields of study concerning EEG conduct predictions and analyses of the performance pertaining to mobile cognition including sports competition [
18], stress and emotional regulation [
19], the identification of drowsiness/alertness patterns [
20], and even the exploration of the effect of music on the brain [
21].
Still concerning performance and cognition, EEG has also been used as a source for exploring and classifying signals during complex mental activities [
22], including arithmetic tasks, which usually demand several simultaneous cognitive processes and strategies. Different approaches have been employed for EEG analysis on that note, mostly related to machine learning, such as neural networks based on particle swarm optimisation [
23] and other approaches trying to avail real-time performance recognition [
24]. In general, several studies have focused on identifying EEG patterns using different methods that can detect and quantify both linear and nonlinear mechanisms and, therefore, somehow reflect patients’ specific characteristics (pertaining to diseases or cognitive aspects) [
25,
26,
27,
28,
29,
30,
31,
32,
33].
Recently, the implementation of computational intelligence towards the analysis of EEG signals in light of brain–computer interfaces has become an increasingly expanding and promising field concerning health and behavioural applications [
34]. While this vast field broadens several different tools that have been used to classify and recognise EEG patterns, such as autoregressive models [
35], mode decomposition [
36], and pattern recognition [
37], there seems to be a significant advantage of classifications employing a particular machine learning subfield: deep learning models [
38].
Based on multilayer neural networks, deep learning models are a family of supervised-learning algorithms usually tailored very carefully for a specific application or use case. Such design specificity along with an efficient use of computational power enables this category of models to achieve remarkable accuracy rates [
39]. Deep learning has been studied to improve the diagnosis of brain diseases, such as Parkinson’s [
40]. Some models also proved reliable for classifying epilepsy cases beyond simple binary diagnoses [
41] while still fast-responsive and not excessively memory-consuming, thus making them capable of being implemented for real-time clinical settings. Based on EEG time series, deep learning models have also been used to successfully detect fatigue status of pilots [
42], to classify driver mental states [
43], to identify alcoholic patients, and to recognise emotions [
44,
45].
In this context, the main advantage of deep learning models seems to be their ability to exploit hidden or unknown particularities in the structure of data, extracting from low-level to high-level features that can be objectively compared and explored [
46]. Those features are highly dependent on the application and can be related to several different aspects of the original time series, ranging from power, auto-regressive model coefficients, statistical parameters, fractal coefficients, variance, energy, entropy, and others. Particularly for the automated classification of mental arithmetic tasks, nonlinear entropy features from each multi-channel EEG signal have been used [
47]. Overall, the model features can be explored either in time or frequency domains depending on the approach chosen, and it may be complicated to compare them with so-called engineered or hand-designed features from other model approaches, since deep learning models often employ features that cannot be immediately identified or extracted from data using other techniques.
Nonetheless, while machine learning methods have showed high success rates, extracting relevant information from EEG signals is quite a complex challenge and other methods are also important—particularly when there are not a lot of original data to train algorithms. Accordingly, in order to analyse EEG data, it is common to transform the signal to the frequency domain. The power spectral density is a powerful tool that applies Fourier transforms to analyse the amount of power of a signal for determined frequencies, and it can be estimated through different techniques [
48,
49,
50]. In the context of EEG and biomedical systems, it has been applied in several different situations including to analyse the effects of age and gender [
51], disruptions caused by Alzheimer’s [
52], cognitive alterations when patients are under mental stress [
53], and sleep classification [
54].
On the other hand, time series analysis [
55,
56,
57] is an important technique that can be applied to a range of physiological measures including respiratory signals [
58,
59,
60], cardiac evaluation [
61,
62], anaesthesia dosage monitoring [
63,
64], and electrophysiological signals from the brain [
65,
66]. Dynamic time series analysis of EEG signals may reveal complex phenomena associated with long-range correlation and distinct classes of nonlinear interactions [
65], improving the understanding about brain activity. In this context and based on the inherent complex nature of the brain electrical, EEG signals can also be explored under nonlinear features, such as ARIMA (autoregressive integrated moving average) and ARFIMA (autoregressive fractionally integrated moving average) models [
67], the Hurst exponent [
68], and fractal dimensions, that might be able to extract hidden complex information within the signals.
In this context, this paper explore an EEG dataset from patients undertaking arithmetic subtractions aiming to obtain insights regarding possible trends and the relation between changes in the randomness pattern of the signal. In this sense, we compare the EEG signal for the subjects at rest and during activity. We also try to identify if the data reveal any hints on the performances of the subjects. In order to approach the analysis from both the frequency and time domains, different tools are employed. First, we apply Welch’s power spectral density [
69] to analyse the spectrum of EEG data and how the signal behaves according to each frequency band. Next, we compare the Hurst exponent (H) [
70] obtained via the detrented fluctuation analysis (DFA) method [
71], and fractional dimensions obtained by means of the Hall–Wood (HW) and Robust–Genton (RG) estimators [
72]. By applying the aforementioned methods, we therefore resort to a two-fold approach considering both time (H, DFA, HW, and RG parameters) and frequency (PSD)-domain analyses. We then interpret the results obtained in order to investigate if one or more approaches seems to be more suitable (i) for showing significant differences between the electrical activities for subjects at rest and during activity, (ii) for categorising subjects’ performance groups (good, average, and poor) based solely on EEG data, and (iii) for checking for differences in the activity of different brain areas.
Therefore, this paper is organised as follows:
Section 2 describes the EEG data assessed in the study, and
Section 3 briefly presents the applied methodology;
Section 4 explores and discusses the main results obtained via power spectral density and time-domain analysis; finally,
Section 5 provides the authors’ main thoughts and highlights the major trends identified.
2. Data Characteristics
The data herein assessed consists of EEG time series (TS) from 36 healthy patients while they were conducting mental subtractions and originates from an extant study [
73], obtained through the PhysioNet database [
74]. All patients are equivalently aged and come from the same educational background. They were eligible to enroll in the study if they had normal or corrected-to-normal visual acuity; normal color vision; and no clinical manifestations of mental or cognitive impairment, or verbal or non-verbal learning disabilities. Exclusion criteria were the use of psychoactive medication, drug or alcohol addiction, and psychiatric or neurological complaints.
According to the authors, the EEG collection was conducted following the International 10/20 scheme, with all electrodes referenced to the interconnected ear reference electrodes. The sample rate was 500 Hz per channel. Regarding filters and artifact removal, a high-pass filter with a 0.5 Hz cut-off frequency and low-pass filter with 45 Hz were used. All recordings employed are artifact-free EEG segments of 60 s duration. At the stage of data pre-processing, the Independent Component Analysis (ICA) was used to eliminate artifacts (eyes, muscle, and cardiac overlapping of the cardiac pulsation).
The arithmetic task the continuous subtraction of two numbers (different each time). Each trial started with the verbal communication of four-digit (minuend) and two-digit (subtrahend) numbers (e.g., 2040 and 20). The performance of each subject was calculated based on the number of subtractions and the accuracy of the results during the whole duration of the test (4 min). The EEG signals for each subject were collected during the first 60 s of the activity, with a second same-sized dataset collected while the subjects were resting before the task. It is important to register that the original rest dataset was much larger than the corresponding dataset for activity, since they were collected for a longer duration of time. Regardless, we sliced the data so both datasets were equivalent (60 s, corresponding to 30,000 samples). The two datasets were called “Activity dataset” and “Rest dataset”, respectively, and were compared. Based on their performance during the mental task, the participants were categorised into three groups: good, average, or poor performers. Each group consisted of an equal number of participants (N = 12). Please refer to
Table A1 for more information on this classification.
As data collection in the aforementioned study followed the International 10/20 scheme, 19 time series were obtained for every subject, each pertaining a different channel and thus capturing electrical activity in a distinct brain part. Auditory channels were disregarded as they were used for referencing purposes. Instead of using all available signals, in this paper, we use only six TSs for one patient, each one representing the average electric tension for each brain region: central C (C3, Cz, and C4), frontal F (F7, F3, Fz, F4, and F8), pre-frontal Fp (Fp1 and Fp2), occipital O (O1 and O2), parietal P (P3, P4, and Pz), and temporal T (T3, T4, T5, and T6). Please refer to
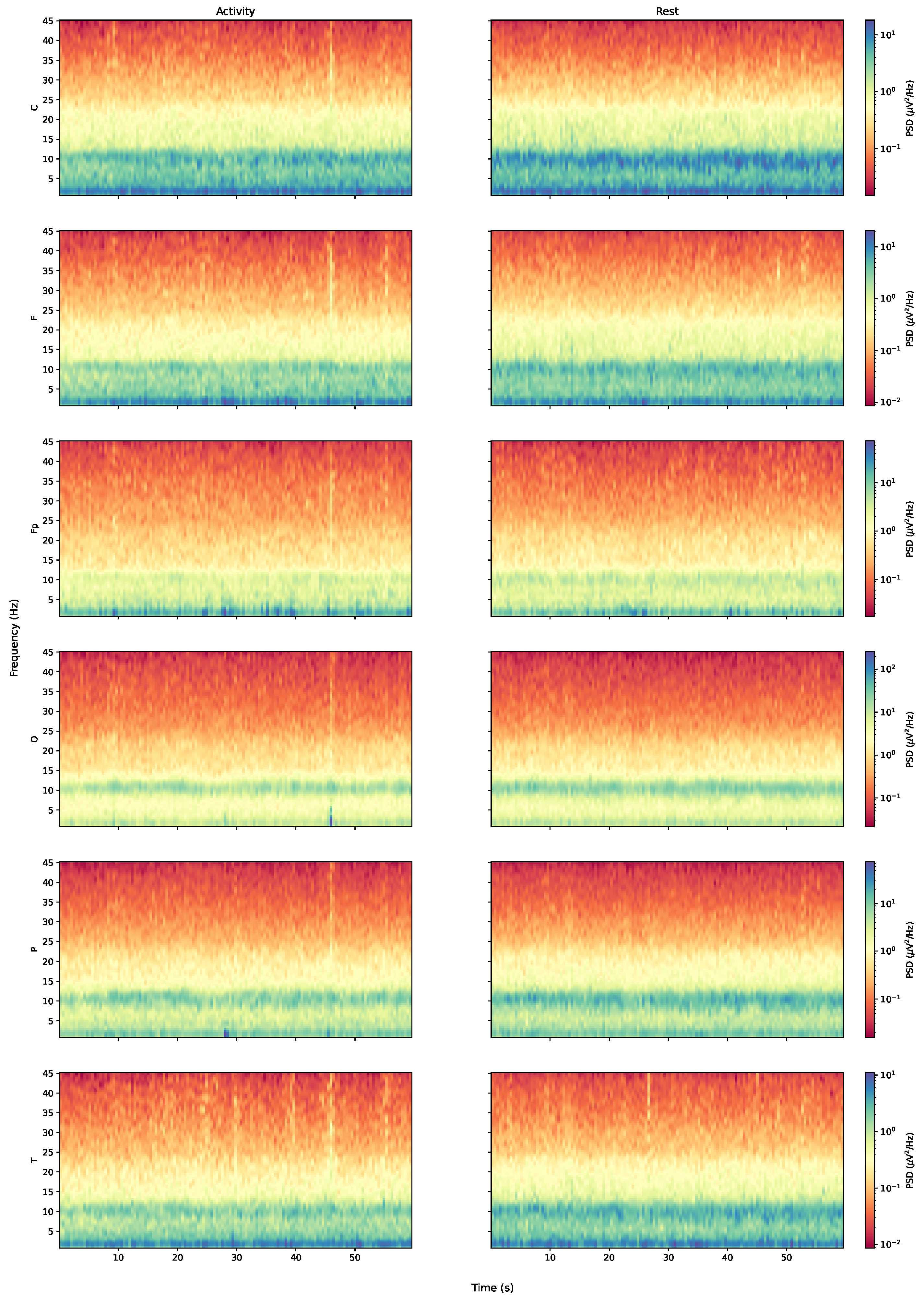
Figure 1 for a graphical representation of this organisation.
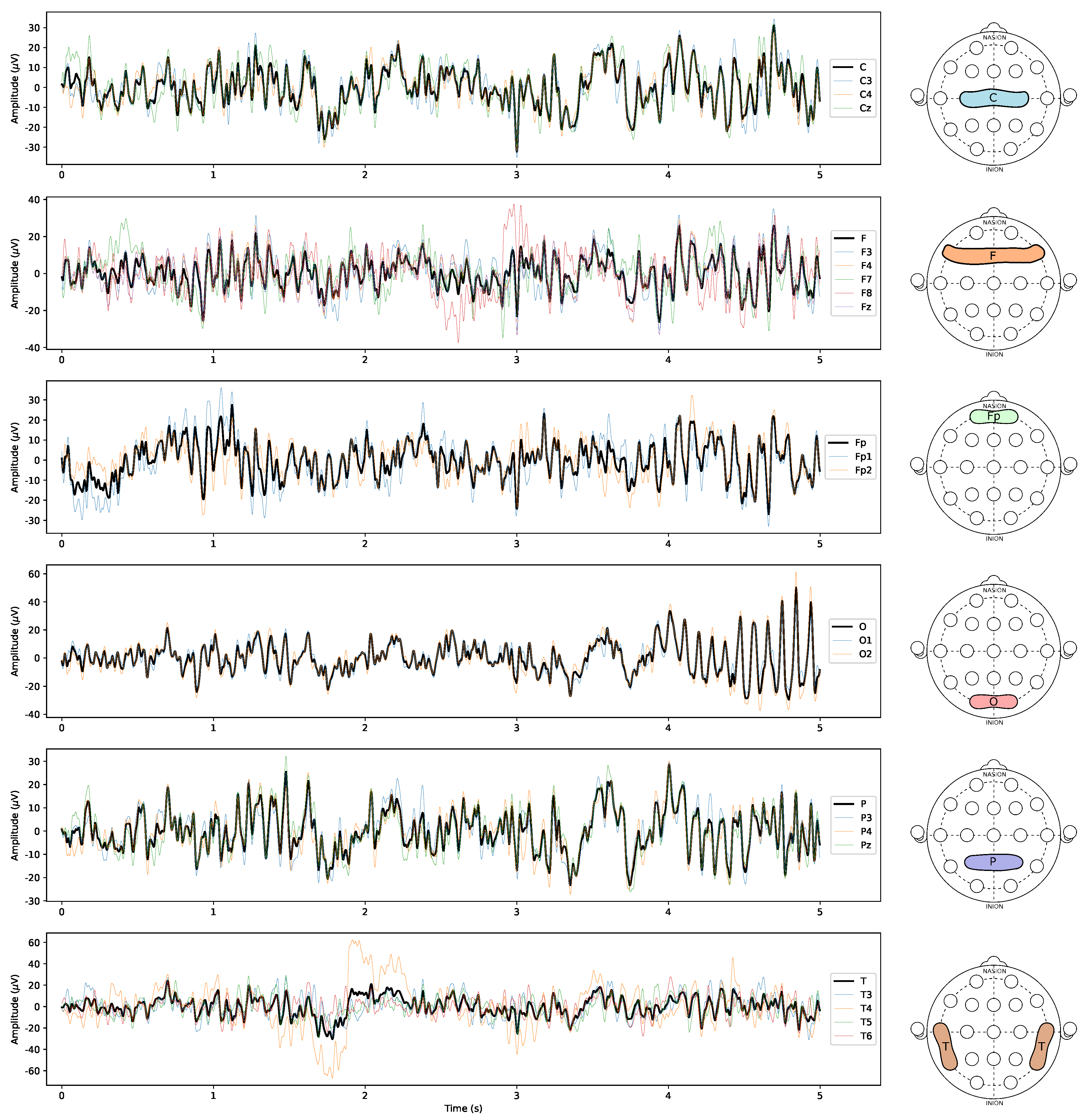
In summary, each of the 36 subjects is represented by 6 time series (each pertaining to an average of the multichannel signals, as illustrated in
Figure 1) for each dataset (rest and activity), totalling 432 time series analysed in the paper.The TS dynamics of the aforesaid averages of each channel are presented for a representative subject in
Figure 2. More details regarding the EEG records can be found in the original study in which they were collected [
73].
5. Conclusions
This study employed frequency- and time-based methods to explore extant EEG data in search of patterns that could differentiate signals of subjects undertaking a mental task (i) while at rest and during activity, (ii) in respect of how well the subjects performed during the arithmetic tests, and (iii) with spatial differences in the electrical activity in the brain. We chose to organise the EEG data for every patient into six time series, each corresponding to a brain region, which are averages according to the EEG channels in that area of the brain.
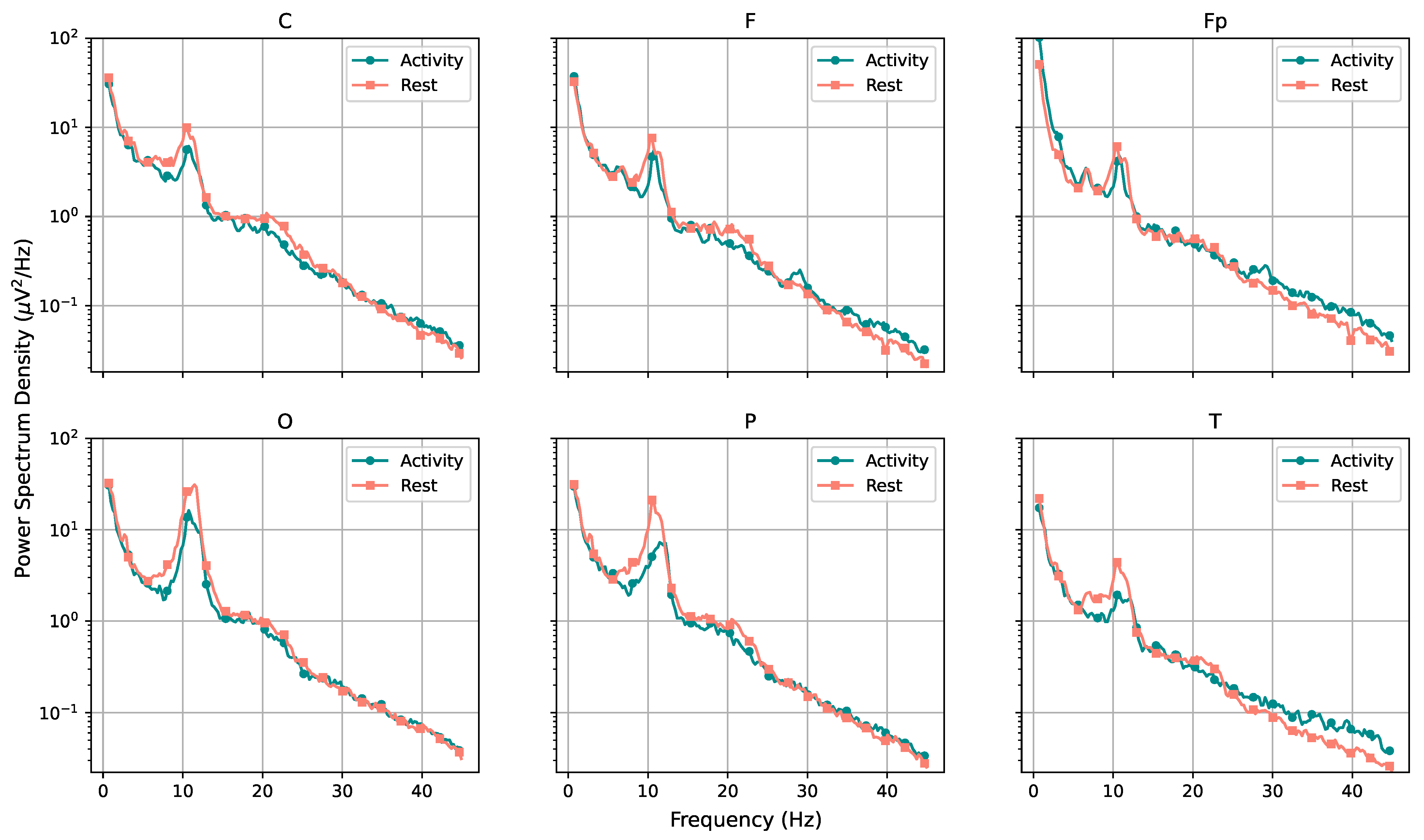
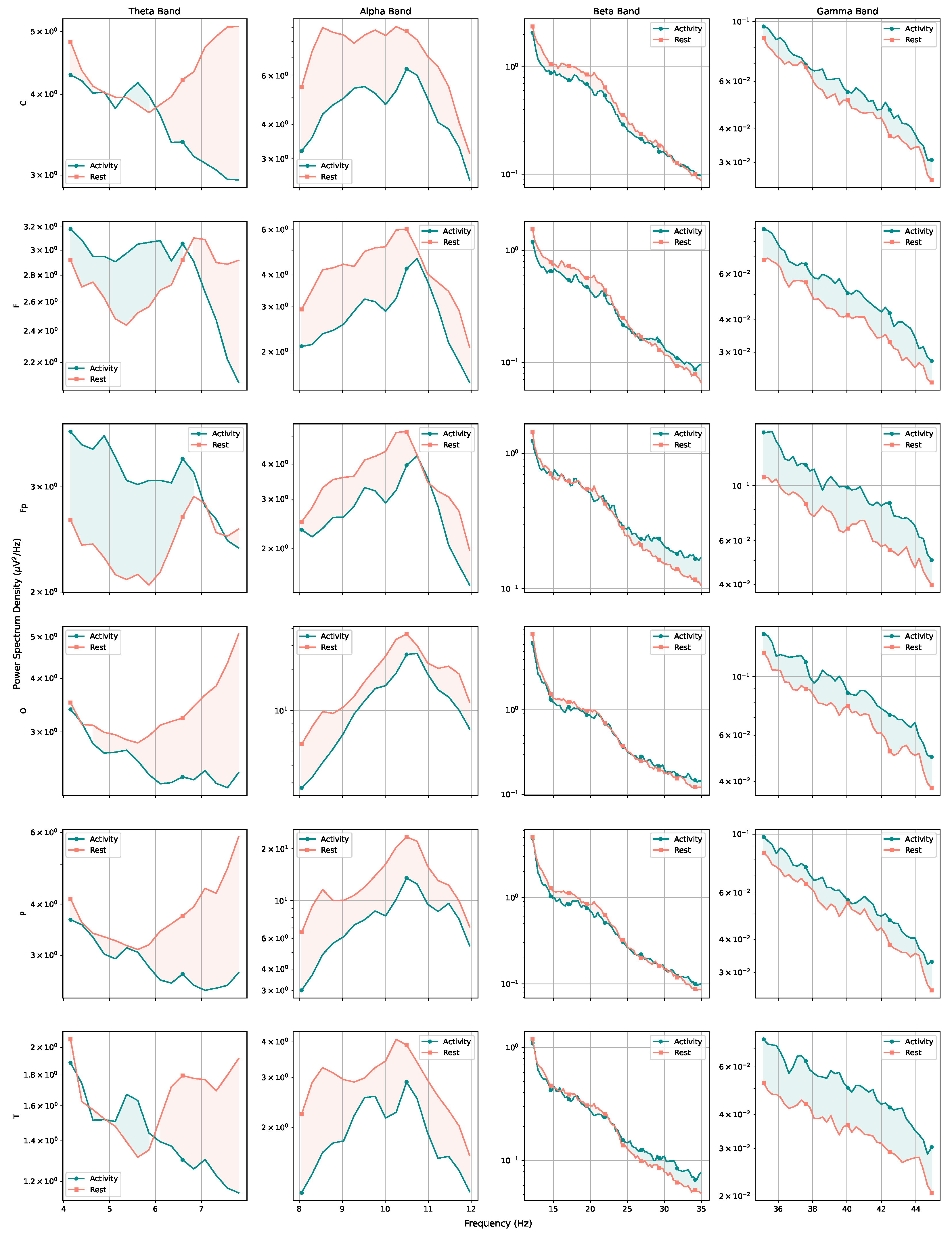
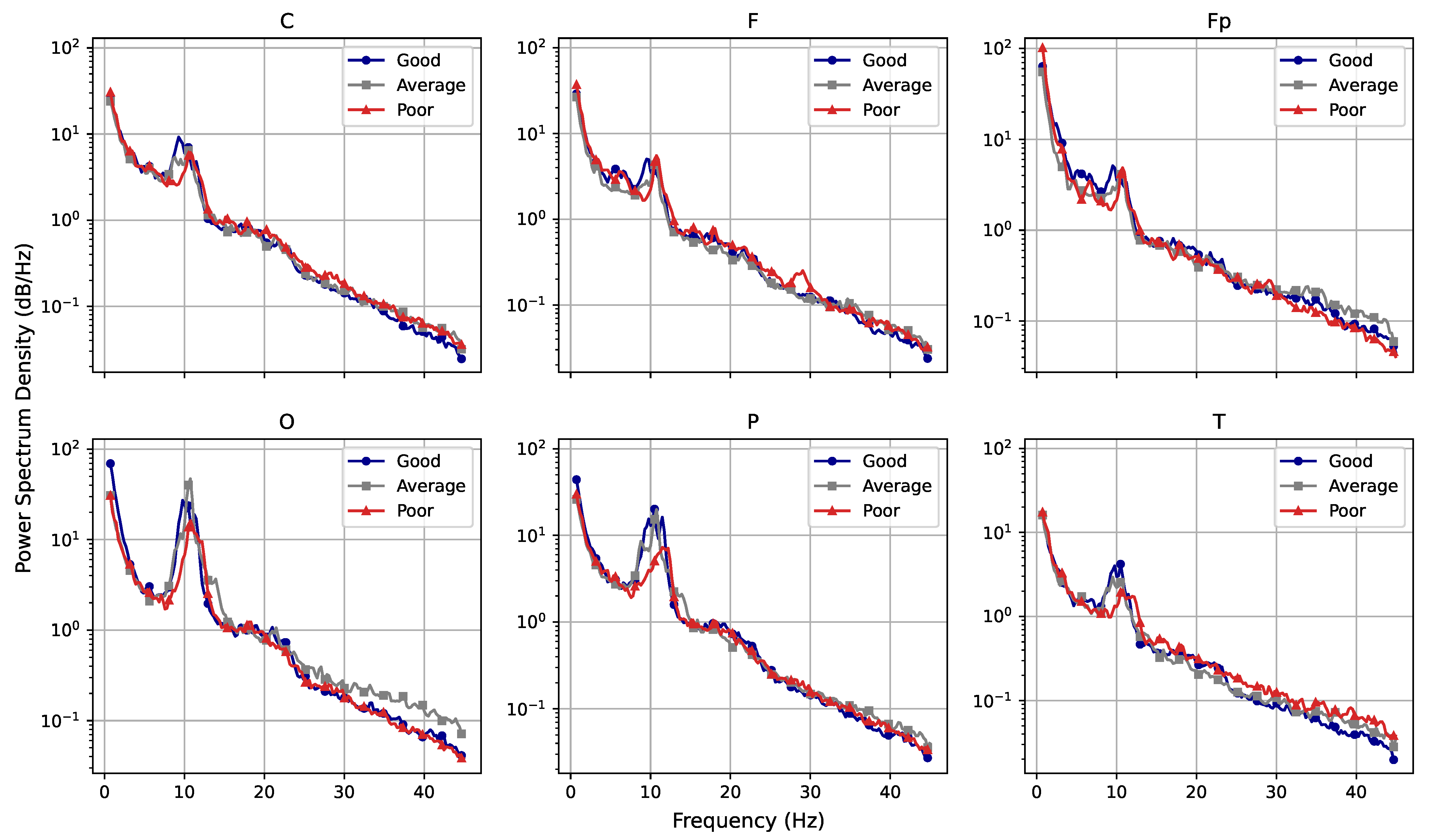
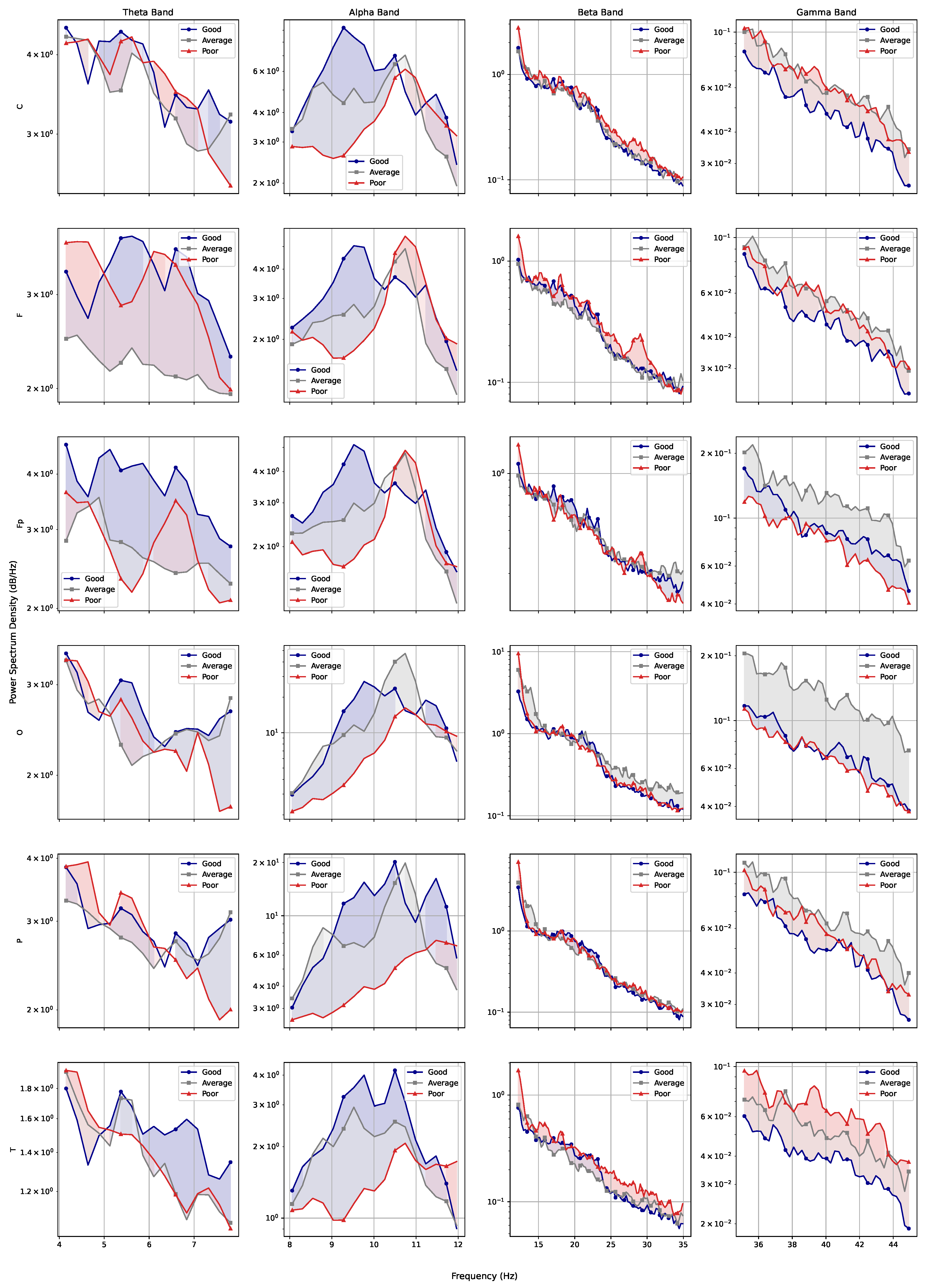
Considering the frequency domain, we estimated the power spectral density of the signals and found that, while the rest and activity datasets seem very close on a first analysis, the full picture changes when the amount of power pertaining each type of brain waves (theta, alpha, beta, and gamma) is considered. The results obtained through the estimations indicate that the subjects seem to have greater alpha-wave activity while at rest and increased gamma-wave activity while performing mental tasks. The spectrogram also reveals that the T region presents a larger number of synchronous frequency peaks when compared with other regions. The PSD curves for each region also indicated a few trends for performance curves, with average and poor performers apparently showing increased brain effort when compared with good performers (particularly when considering high-frequency ranges).
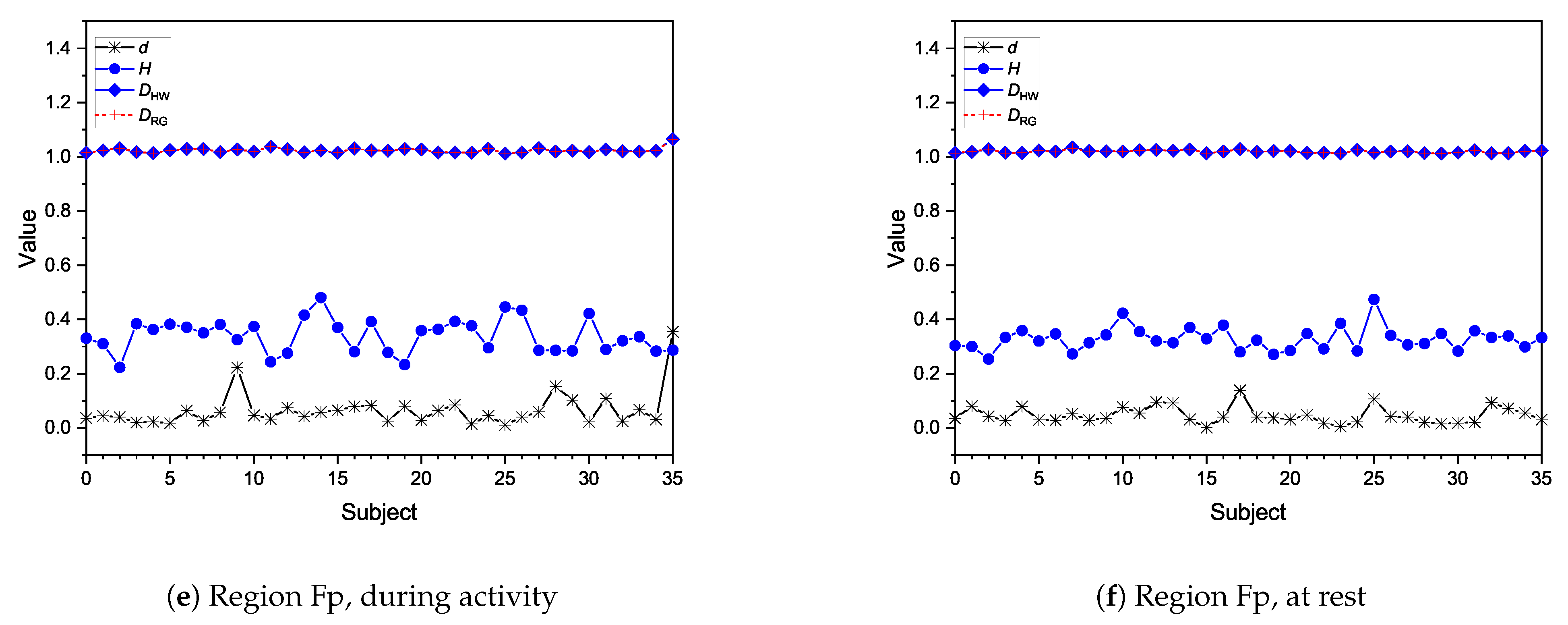
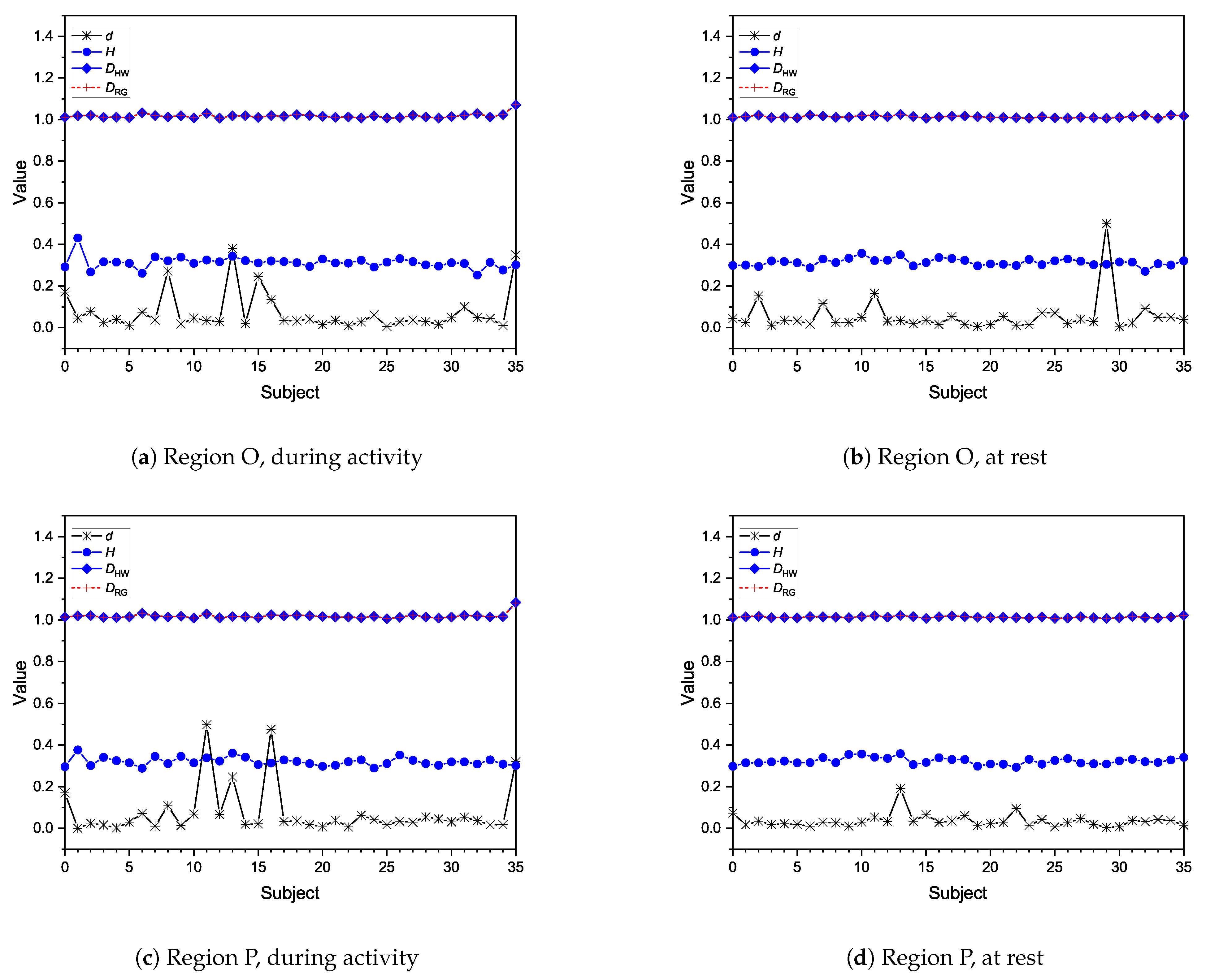
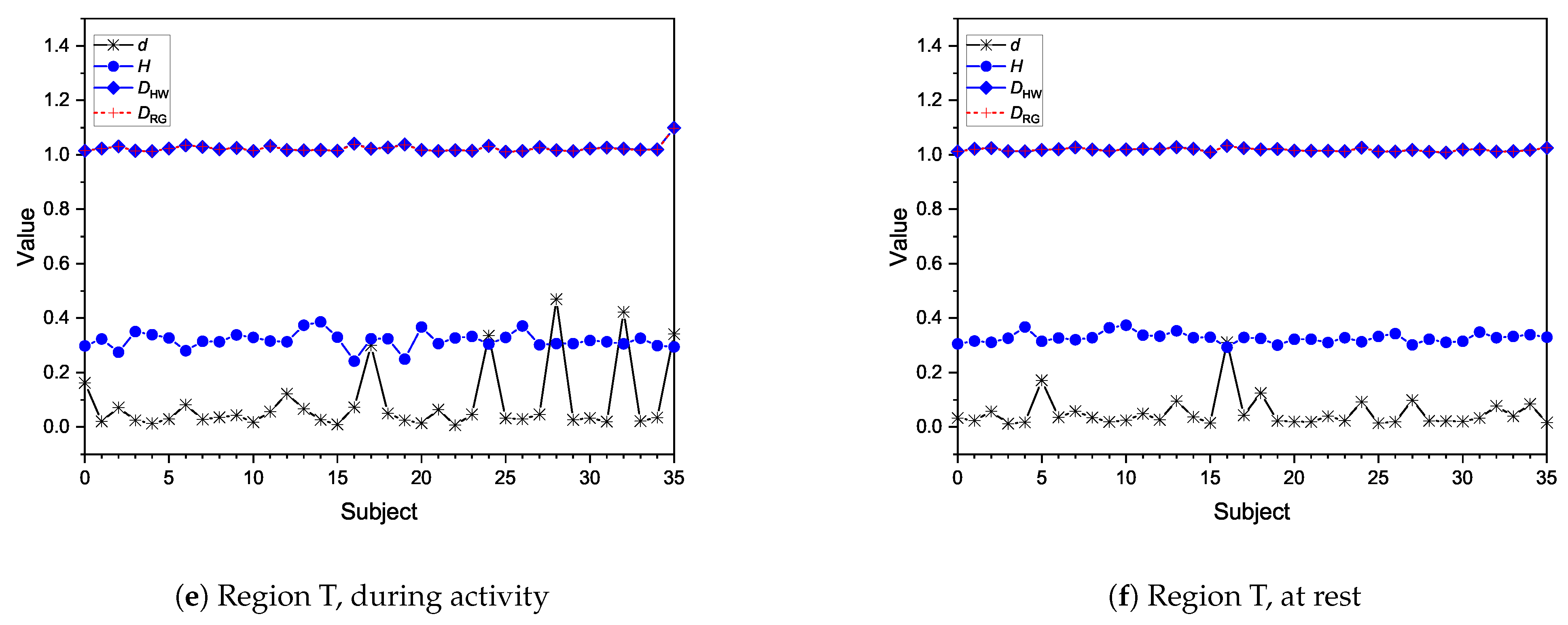
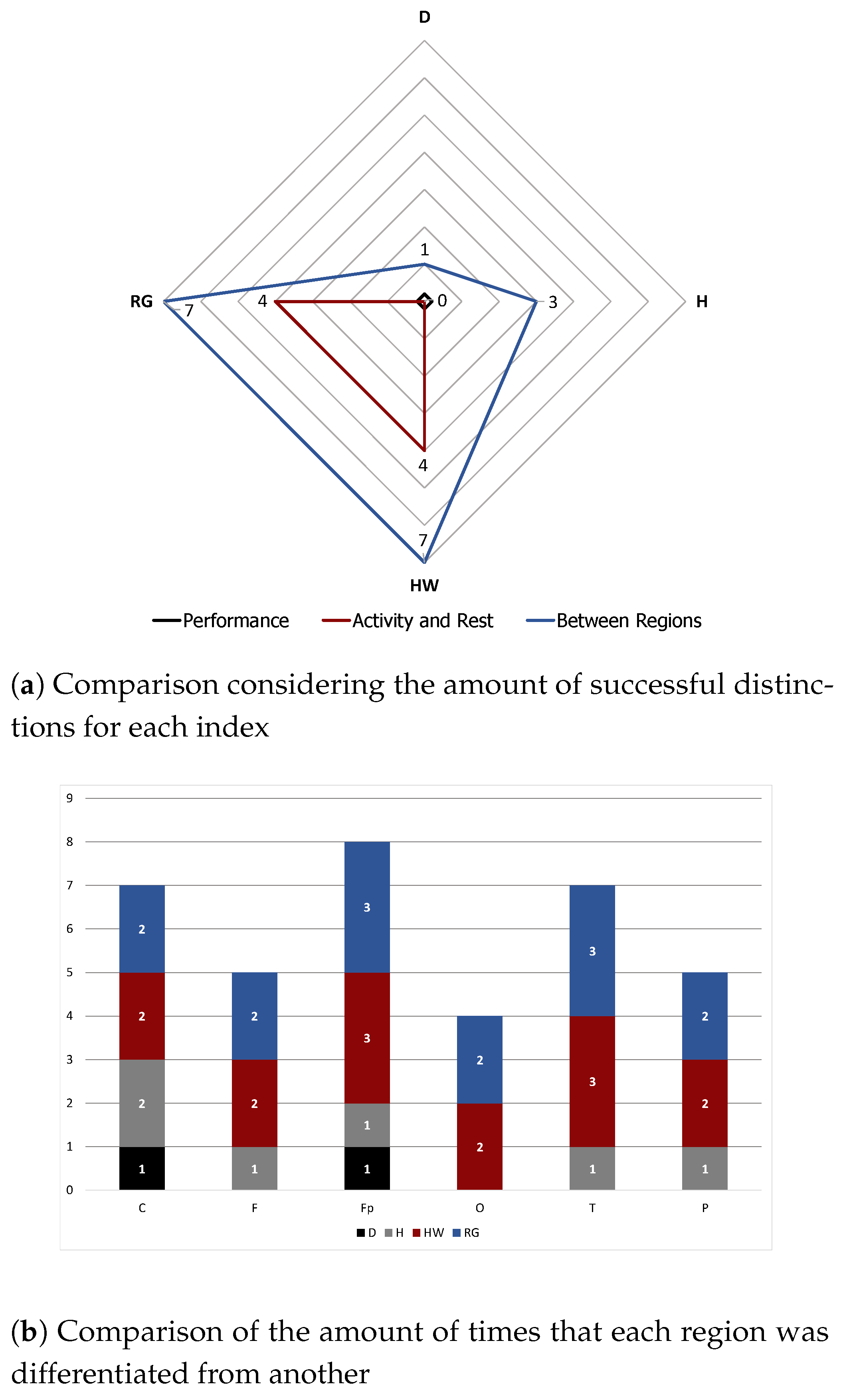
Moreover, fractional dimensions using the HW and RG estimators in addition to the H exponent by means of the DFA method were also explored. We adopted the Shapiro–Wilk normality test and variance analysis to assess if these indexes could offer any assertive statistical information regarding the three analyses in question. The aforementioned methods showed that the results achieved were very similar when comparing performance groups, with no statistical difference identified. As for differences between performance and brain regions, HW and RG estimators seemed to be better indicators. We also found that Fp seems to be the most differentiable brain region while O is the least differentiable, indicating that the former might be the most appropriate for analysing EEG data of patients undertaking mental tasks.
In conclusion, we believe that both frequency- and time-based methods were useful in the analysis and recommend that they should be used together in order to gain insights towards building a classifier of EEG data regarding mental tasks. In order to further investigate the matter and to exhaust related possibilities, in future work, we intend to explore some of the following options: (i) an analysis of the signals from other kinds of mental tasks; (ii) a consideration of the EEG channels individually (i.e., not using averages for each brain region); (iii) an extention of the number of mathematical tools, both within the scope of fractal dimensions as the frequency domain; (iv) the design of distinct analyses involving Lyapunov exponents applied to time series; and (v) the employment of machine learning approaches, such as deep learning and parametric models to extract features and to compare it with the results herein obtained.