No Clear Clustering Dysbiosis from Salivary Microbiota Analysis by Long Sequencing Reads in Patients Affected by Oral Squamous Cell Carcinoma: A Single Center Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Population Recruitment
- i.
- Age older than 18 years;
- ii.
- Patients affected by OSCC confirmed by pathological findings;
- iii.
- Ability to provide informed consent.
2.3. Outcome Measures
2.4. Sample Collection
2.5. DNA Extraction
2.6. Samples Sequencing
2.7. Bioinformatics and Statistical Analysis
3. Results
3.1. Characteristics of Study Participants
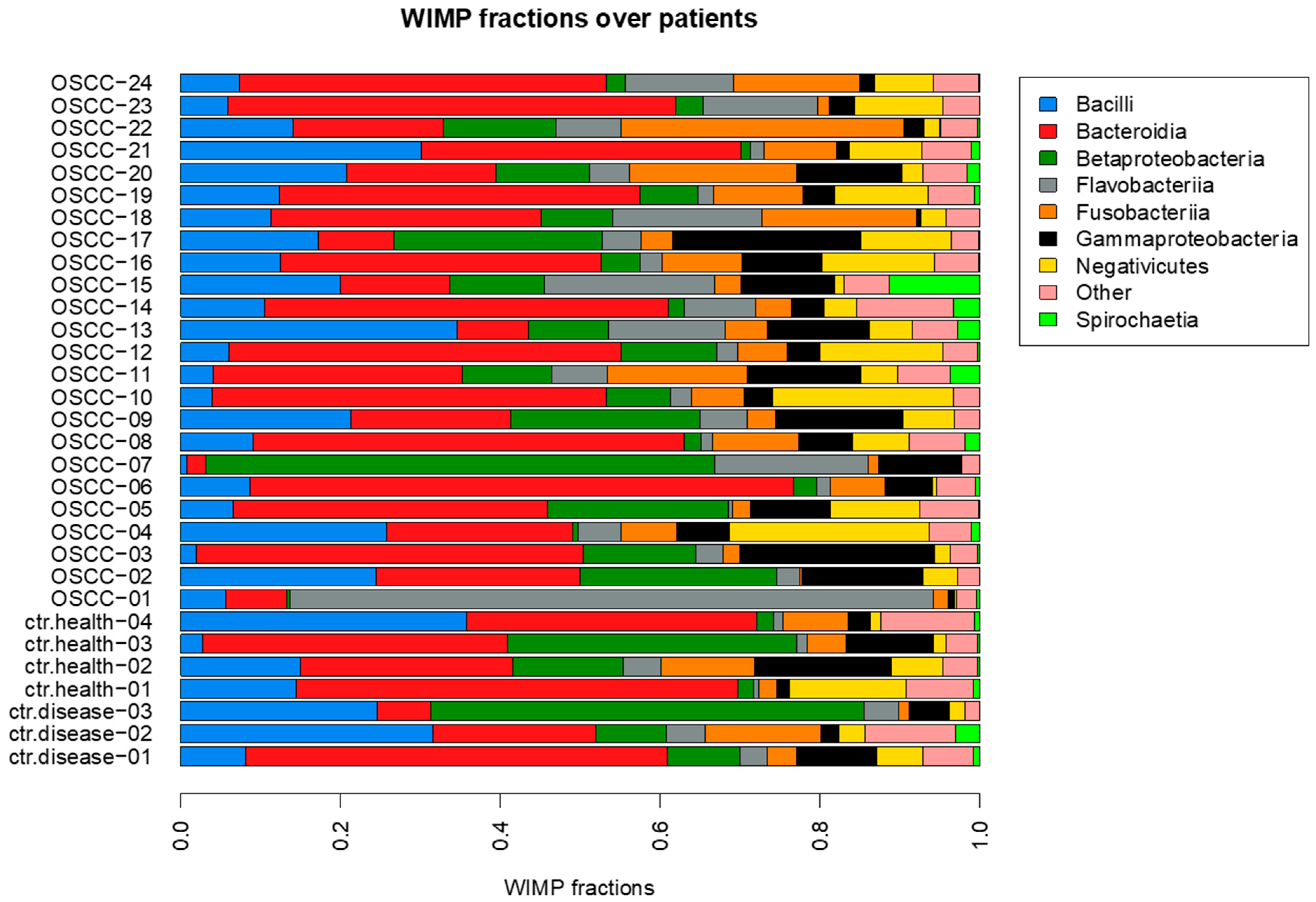
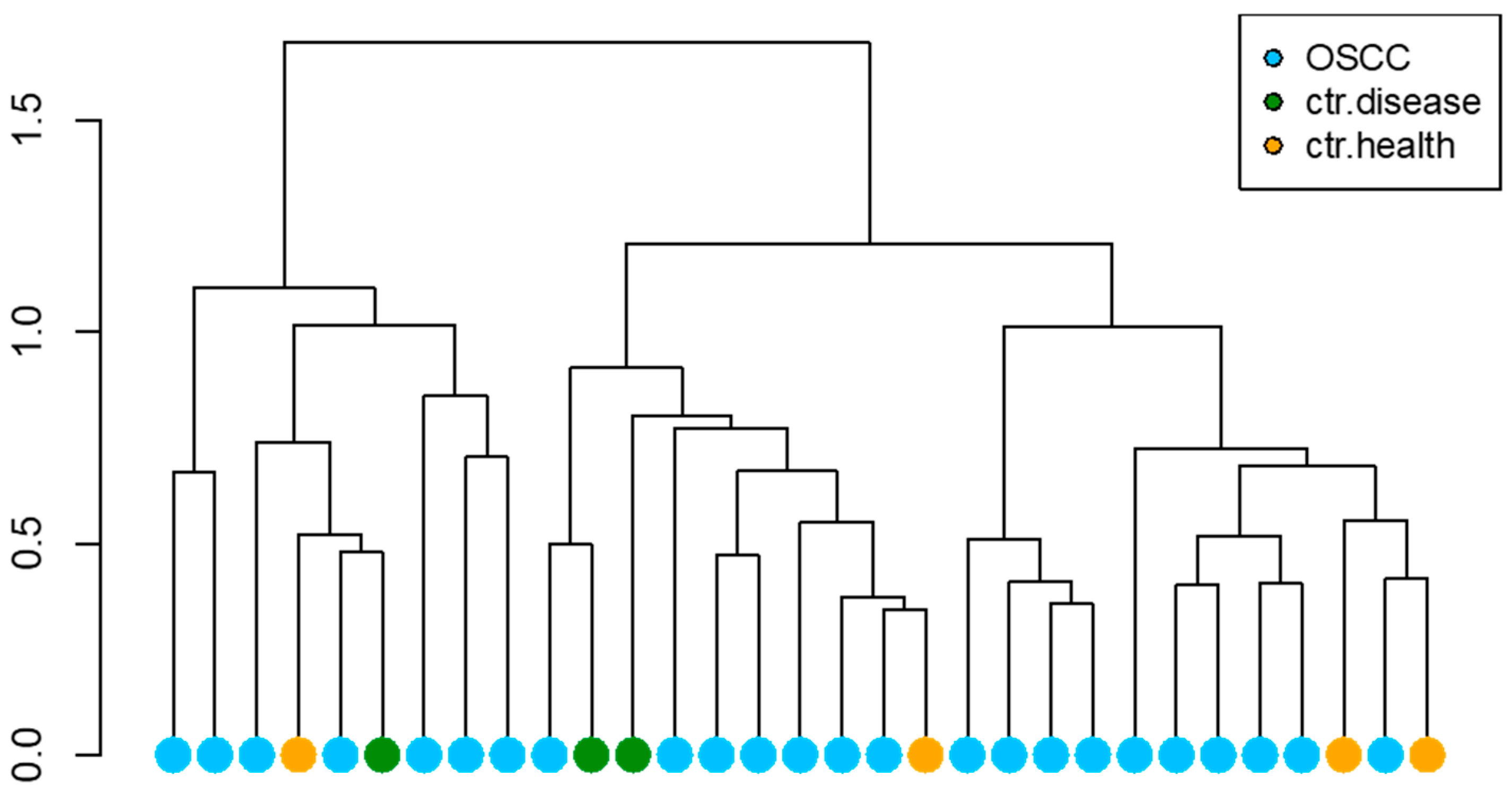
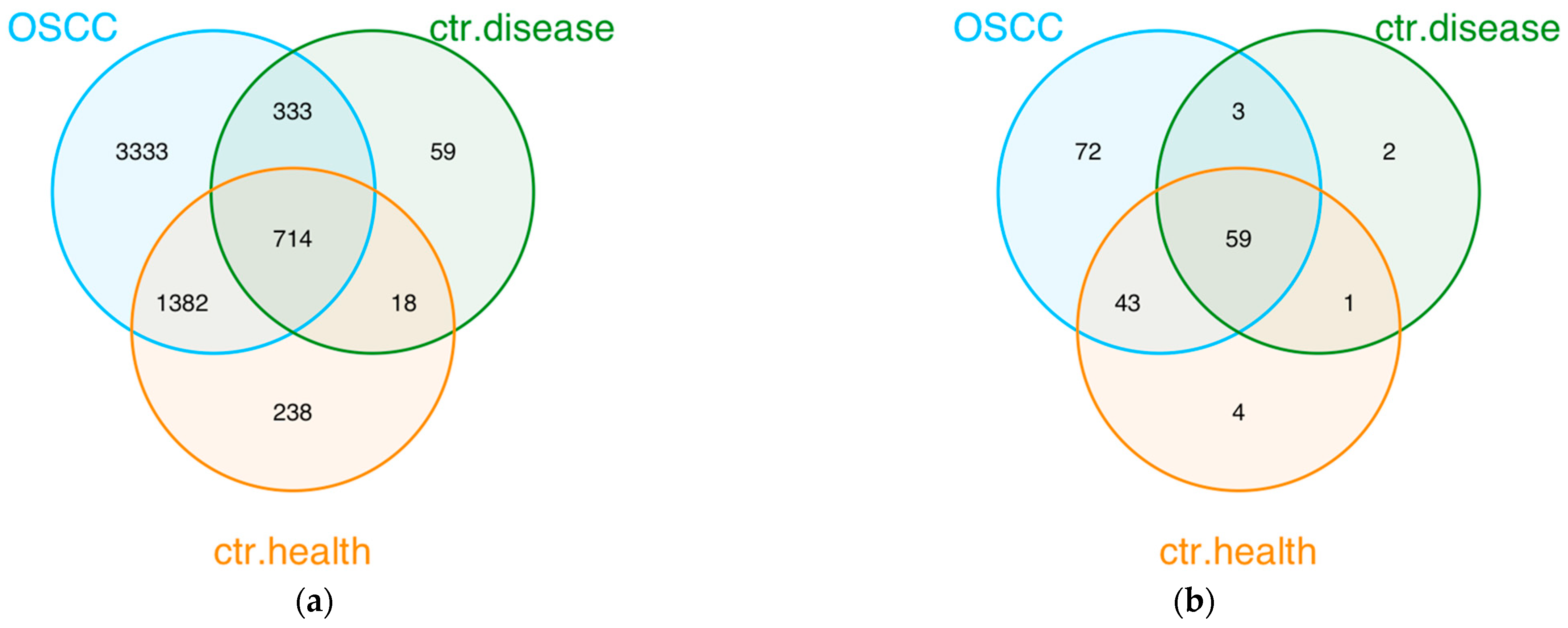
3.2. Microbiota Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The Oral Microbiota: Dynamic Communities and Host Interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Ravel, J. The Vocabulary of Microbiome Research: A Proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Costalonga, M.; Herzberg, M.C. The Oral Microbiome and the Immunobiology of Periodontal Disease and Caries. Immunol. Lett. 2014, 162, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Blaser, M.J. The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd_Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9, 2868. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Eslami, H.; Yousefi, M.; Asgharzadeh, M.; Aghazadeh, M.; Kafil, H.S. Role of Oral Microbiome on Oral Cancers, a Review. Biomed. Pharmacother. 2016, 84, 552–558. [Google Scholar] [CrossRef]
- Khatoon, J. Role of Helicobacter Pylori in Gastric Cancer: Updates. World J. Gastrointest. Oncol. 2016, 8, 147. [Google Scholar] [CrossRef]
- Koshiol, J.; Wozniak, A.; Cook, P.; Adaniel, C.; Acevedo, J.; Azócar, L.; Hsing, A.W.; Roa, J.C.; Pasetti, M.F.; Miquel, J.F.; et al. Salmonella Enterica Serovar Typhi and Gallbladder Cancer: A Case–Control Study and Meta-analysis. Cancer Med. 2016, 5, 3310–3235. [Google Scholar] [CrossRef]
- Nosho, K. Association of Fusobacterium Nucleatum with Immunity and Molecular Alterations in Colorectal Cancer. World J. Gastroenterol. 2016, 22, 557. [Google Scholar] [CrossRef]
- Lee, W.-H.; Chen, H.-M.; Yang, S.-F.; Liang, C.; Peng, C.-Y.; Lin, F.-M.; Tsai, L.-L.; Wu, B.-C.; Hsin, C.-H.; Chuang, C.-Y.; et al. Bacterial Alterations in Salivary Microbiota and Their Association in Oral Cancer. Sci. Rep. 2017, 7, 16540. [Google Scholar] [CrossRef]
- Chamoli, A.; Gosavi, A.S.; Shirwadkar, U.P.; Wangdale, K.V.; Behera, S.K.; Kurrey, N.K.; Kalia, K.; Mandoli, A. Overview of Oral Cavity Squamous Cell Carcinoma: Risk Factors, Mechanisms, and Diagnostics. Oral Oncol. 2021, 121, 105451. [Google Scholar] [CrossRef]
- Panzarella, V.; Campisi, G.; Giardina, Y.; Maniscalco, L.; Capra, G.; Rodolico, V.; Di Fede, O.; Mauceri, R. Low Frequency of Human Papillomavirus in Strictly Site-Coded Oral Squamous Cell Carcinomas, Using the Latest NHI/SEER-ICD Systems: A Pilot Observational Study and Critical Review. Cancers 2021, 13, 4595. [Google Scholar] [CrossRef]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef]
- Chen, T.; Yu, W.-H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The Human Oral Microbiome Database: A Web Accessible Resource for Investigating Oral Microbe Taxonomic and Genomic Information. Database J. Biol. Database Curation 2010, 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Mechan Llontop, M.E.; Sharma, P.; Aguilera Flores, M.; Yang, S.; Pollok, J.; Tian, L.; Huang, C.; Rideout, S.; Heath, L.S.; Li, S.; et al. Strain-Level Identification of Bacterial Tomato Pathogens Directly from Metagenomic Sequences. Phytopathology 2020, 110, 768–779. [Google Scholar] [CrossRef]
- Chamberlain, S.A.; Szöcs, E. taxize: Taxonomic search and retrieval in R. F1000Research 2013, 2, 191. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R. An Introduction to the Bootstrap; Monographs on Statistics and Applied Probability; Chapman & Hall: New York, NY, USA, 1993; ISBN 978-0-412-04231-7. [Google Scholar]
- Qian, Y.; Kehr, B.; Halldórsson, B.V. PopAlu: Population-Scale Detection of Alu Polymorphisms. PeerJ 2015, 3, e1269. [Google Scholar] [CrossRef][Green Version]
- Mauceri, R.; Coppini, M.; Vacca, D.; Bertolazzi, G.; Panzarella, V.; Fede, O.D.; Tripodo, C.; Campisi, G. Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review. Cancers 2022, 14, 5441. [Google Scholar] [CrossRef] [PubMed]
- Efron, B. Bootstrap Methods: Another Look at the Jackknife. Ann. Stat. 1979, 7, 1–26. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving Pictures of the Human Microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.; Holmes, N.; Rakyan, V.; Loose, M. BulkVis: A Graphical Viewer for Oxford Nanopore Bulk FAST5 Files. Bioinformatics 2019, 35, 2193–2198. [Google Scholar] [CrossRef]
- Depledge, D.P.; Wilson, A.C. Using Direct RNA Nanopore Sequencing to Deconvolute Viral Transcriptomes. Curr. Protoc. Microbiol. 2020, 57. [Google Scholar] [CrossRef]
- Juul, S.; Izquierdo, F.; Hurst, A.; Dai, X.; Wright, A.; Kulesha, E.; Pettett, R.; Turner, D.J. What’s in My Pot? Real-Time Species Identification on the MinION. BioRxiv 2015. [Google Scholar] [CrossRef]
- Brandariz-Fontes, C.; Camacho-Sanchez, M.; Vilà, C.; Vega-Pla, J.L.; Rico, C.; Leonard, J.A. Effect of the Enzyme and PCR Conditions on the Quality of High-Throughput DNA Sequencing Results. Sci. Rep. 2015, 5, 8056. [Google Scholar] [CrossRef]
- Tett, A.; Pasolli, E.; Masetti, G.; Ercolini, D.; Segata, N. Prevotella Diversity, Niches and Interactions with the Human Host. Nat. Rev. Microbiol. 2021, 19, 585–599. [Google Scholar] [CrossRef]
- Granato, D.C.; Neves, L.X.; Trino, L.D.; Carnielli, C.M.; Lopes, A.F.B.; Yokoo, S.; Pauletti, B.A.; Domingues, R.R.; Sá, J.O.; Persinoti, G.; et al. Meta-Omics Analysis Indicates the Saliva Microbiome and Its Proteins Associated with the Prognosis of Oral Cancer Patients. Biochim. Biophys. Acta BBA—Proteins Proteom. 2021, 1869, 140659. [Google Scholar] [CrossRef]
- Chocolatewala, N.; Chaturvedi, P.; Desale, R. The Role of Bacteria in Oral Cancer. Indian J. Med. Paediatr. Oncol. 2010, 31, 126–131. [Google Scholar] [CrossRef]
- Pignatelli, P.; Iezzi, L.; Pennese, M.; Raimondi, P.; Cichella, A.; Bondi, D.; Grande, R.; Cotellese, R.; Di Bartolomeo, N.; Innocenti, P.; et al. The Potential of Colonic Tumor Tissue Fusobacterium Nucleatum to Predict Staging and Its Interplay with Oral Abundance in Colon Cancer Patients. Cancers 2021, 13, 1032. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.; Haffajee, A.; Devlin, P.; Norris, C.; Posner, M.; Goodson, J. The Salivary Microbiota as a Diagnostic Indicator of Oral Cancer: A Descriptive, Non-Randomized Study of Cancer-Free and Oral Squamous Cell Carcinoma Subjects. J. Transl. Med. 2005, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Pietrobon, G.; Tagliabue, M.; Stringa, L.M.; De Berardinis, R.; Chu, F.; Zocchi, J.; Carlotto, E.; Chiocca, S.; Ansarin, M. Leukoplakia in the Oral Cavity and Oral Microbiota: A Comprehensive Review. Cancers 2021, 13, 4439. [Google Scholar] [CrossRef] [PubMed]
- Maitimu, F.C.; Soeroso, Y.; Sunarto, H.; Bachtiar, B.M. Association between Volatile Sulfur Compounds Prevotella Intermedia and Matrix Metalloproteinase-8 Expression. Pesq. Bras. Em Odontopediatria E Clínica Integr. 2022, 22, e210002. [Google Scholar] [CrossRef]
- Bhuvanendran Pillai, A.; Wong, C.M.; Zainal Abidin, N.D.I.; Syed Nor, S.F.; Mohamed Hanan, M.F.; Abd Ghani, S.R.; Aminuddin, N.A.; Safian, N. Chlamydia Infection as a Risk Factor for Cervical Cancer: A Systematic Review and Meta-Analysis. Iran. J. Public Health 2022, 51, 508–517. [Google Scholar] [CrossRef]
- Reed, S.G.; Lopatin, D.E.; Foxman, B.; Burt, B.A. Oral Chlamydia Trachomatis in Patients with Established Periodontitis. Clin. Oral Investig. 2000, 4, 226–232. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota Disbiosis Is Associated with Colorectal Cancer. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Li, Y.; Tan, X.; Zhao, X.; Xu, Z.; Dai, W.; Duan, W.; Huang, S.; Zhang, E.; Liu, J.; Zhang, S.; et al. Composition and Function of Oral Microbiota between Gingival Squamous Cell Carcinoma and Periodontitis. Oral Oncol. 2020, 107, 104710. [Google Scholar] [CrossRef]
- Zhou, X.; Hao, Y.; Peng, X.; Li, B.; Han, Q.; Ren, B.; Li, M.; Li, L.; Li, Y.; Cheng, G.; et al. The Clinical Potential of Oral Microbiota as a Screening Tool for Oral Squamous Cell Carcinomas. Front. Cell. Infect. Microbiol. 2021, 11, 728933. [Google Scholar] [CrossRef]
- Sawant, S.; Dugad, J.; Parikh, D.; Srinivasan, S.; Singh, H. Oral Microbial Signatures of Tobacco Chewers and Oral Cancer Patients in India. Pathogens 2023, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Qamar, H.; Hussain, K.; Soni, A.; Khan, A.; Hussain, T.; Chénais, B. Cyanobacteria as Natural Therapeutics and Pharmaceutical Potential: Role in Antitumor Activity and as Nanovectors. Molecules 2021, 26, 247. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, S.; Menakha, M. Pharmaceutical Applications of Cyanobacteria—A Review. J. Acute Med. 2015, 5, 15–23. [Google Scholar] [CrossRef]
- Hernandez, B.Y.; Zhu, X.; Risch, H.A.; Lu, L.; Ma, X.; Irwin, M.L.; Lim, J.K.; Taddei, T.H.; Pawlish, K.S.; Stroup, A.M.; et al. Oral Cyanobacteria and Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2022, 31, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Y.; Li, C.; Lei, G.; Zhou, L.; Chen, X.; Jia, X.; Lu, Y. Intestinal Candida Albicans Promotes Hepatocarcinogenesis by Up-Regulating NLRP6. Front. Microbiol. 2022, 13, 812771. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Sufiawati, I.; Piliang, A.; Ramamoorthy, V.R. Oral Microbiota in Oral Cancer Patients and Healthy Individuals: A Scoping Review. Dent. J. Maj. Kedokt. Gigi 2022, 55, 186–193. [Google Scholar] [CrossRef]
- Chen, J.-W.; Wu, J.-H.; Chiang, W.-F.; Chen, Y.-L.; Wu, W.-S.; Wu, L.-W. Taxonomic and Functional Dysregulation in Salivary Microbiomes During Oral Carcinogenesis. Front. Cell. Infect. Microbiol. 2021, 11, 663068. [Google Scholar] [CrossRef]
- Chipashvili, O.; Utter, D.R.; Bedree, J.K.; Ma, Y.; Schulte, F.; Mascarin, G.; Alayyoubi, Y.; Chouhan, D.; Hardt, M.; Bidlack, F.; et al. Episymbiotic Saccharibacteria Suppresses Gingival Inflammation and Bone Loss in Mice through Host Bacterial Modulation. Cell Host Microbe 2021, 29, 1649–1662. [Google Scholar] [CrossRef]
- Bor, B.; Bedree, J.K.; Shi, W.; McLean, J.S.; He, X. Saccharibacteria (TM7) in the Human Oral Microbiome. J. Dent. Res. 2019, 98, 500–509. [Google Scholar] [CrossRef]
- Gao, L. Oral Microbiomes: More and More Importance in Oral Cavity and Whole Body. Protein Cell 2018, 13, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.-R.; Chang, C.-C.; Lee, W.-T.; Huang, C.-C.; Ou, C.-Y.; Tsai, S.-T.; Chen, K.-C.; Huang, J.-S.; Wong, T.-Y.; Lai, Y.-H.; et al. The Interplay between Oral Microbiome, Lifestyle Factors and Genetic Polymorphisms in the Risk of Oral Squamous Cell Carcinoma. Carcinogenesis 2018, 39, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Park, J.; Hosomi, K.; Yamada, T.; Kobayashi, A.; Yamaguchi, Y.; Iketani, S.; Kunisawa, J.; Mizuguchi, K.; Maeda, N.; et al. Analysis of Oral Microbiota in Japanese Oral Cancer Patients Using 16S RRNA Sequencing. J. Oral Biosci. 2019, 61, 120–128. [Google Scholar] [CrossRef]
- Ganly, I.; Yang, L.; Giese, R.A.; Hao, Y.; Nossa, C.W.; Morris, L.G.T.; Rosenthal, M.; Migliacci, J.; Kelly, D.; Tseng, W.; et al. Periodontal Pathogens Are a Risk Factor of Oral Cavity Squamous Cell Carcinoma, Independent of Tobacco and Alcohol and Human Papillomavirus: Periodontal Pathogens Are a Risk Factor for Oral Cancer. Int. J. Cancer 2019, 145, 775–784. [Google Scholar] [CrossRef]
- Feller, L.; Lemmer, J. Oral Squamous Cell Carcinoma: Epidemiology, Clinical Presentation and Treatment. J. Cancer Ther. 2012, 3, 263–268. [Google Scholar] [CrossRef]
- Pałasz, P.; Adamski, Ł.; Górska-Chrząstek, M.; Starzyńska, A.; Studniarek, M. Contemporary Diagnostic Imaging of Oral Squamous Cell Carcinoma—A Review of Literature. Pol. J. Radiol. 2017, 82, 193–202. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Patients Affected by OSCC | Patients OSCC Free |
|---|---|---|
| 24 (77.4) | 7 (22.6) | |
| Gender | ||
| Male | 13 (54.2) | 5 (71.4) |
| Female | 11 (45.8) | 2 (28.6) |
| Demographic data | ||
| Median age | 64.5 | 55 |
| Q1–Q3 | 53.5–79.7 | 30–76 |
| Mean age ± SD | 65.5 ± 13.9 | 51.4 ± 19.2 |
| Risk factors | ||
| Tobacco smoking | 4 (16.6) | 0 |
| Tobacco smoking and alcohol consumption | 3 (12.5) | 0 |
| Former smoking | 3 (12.5) | 0 |
| Characteristics of patients affected by OSCC (n. 24) | ||
| OSCC anatomical site | ||
| Border of the tongue | 10 (41.7) | |
| Buccal mucosa | 6 (25) | |
| Floor of the mouth | 3 (12.5) | |
| Lower gingiva | 3 (12.5) | |
| Upper gingiva | 1 (4.1) | |
| Hard palate | 1 (4.1) | |
| TNM staging | ||
| Stage I | 7 (29.2) | |
| Stage II | 6 (25) | |
| Stage III | 5 (20.8) | |
| Stage IV | 6 (25) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mauceri, R.; Coppini, M.; Vacca, D.; Bertolazzi, G.; Cancila, V.; Tripodo, C.; Campisi, G. No Clear Clustering Dysbiosis from Salivary Microbiota Analysis by Long Sequencing Reads in Patients Affected by Oral Squamous Cell Carcinoma: A Single Center Study. Cancers 2023, 15, 4211. https://doi.org/10.3390/cancers15174211
Mauceri R, Coppini M, Vacca D, Bertolazzi G, Cancila V, Tripodo C, Campisi G. No Clear Clustering Dysbiosis from Salivary Microbiota Analysis by Long Sequencing Reads in Patients Affected by Oral Squamous Cell Carcinoma: A Single Center Study. Cancers. 2023; 15(17):4211. https://doi.org/10.3390/cancers15174211
Chicago/Turabian StyleMauceri, Rodolfo, Martina Coppini, Davide Vacca, Giorgio Bertolazzi, Valeria Cancila, Claudio Tripodo, and Giuseppina Campisi. 2023. "No Clear Clustering Dysbiosis from Salivary Microbiota Analysis by Long Sequencing Reads in Patients Affected by Oral Squamous Cell Carcinoma: A Single Center Study" Cancers 15, no. 17: 4211. https://doi.org/10.3390/cancers15174211
APA StyleMauceri, R., Coppini, M., Vacca, D., Bertolazzi, G., Cancila, V., Tripodo, C., & Campisi, G. (2023). No Clear Clustering Dysbiosis from Salivary Microbiota Analysis by Long Sequencing Reads in Patients Affected by Oral Squamous Cell Carcinoma: A Single Center Study. Cancers, 15(17), 4211. https://doi.org/10.3390/cancers15174211








