Pre-Clinical Models to Study Human Prostate Cancer
Abstract
Simple Summary
Abstract
1. Pre-Clinical Models for Human Prostate Cancer
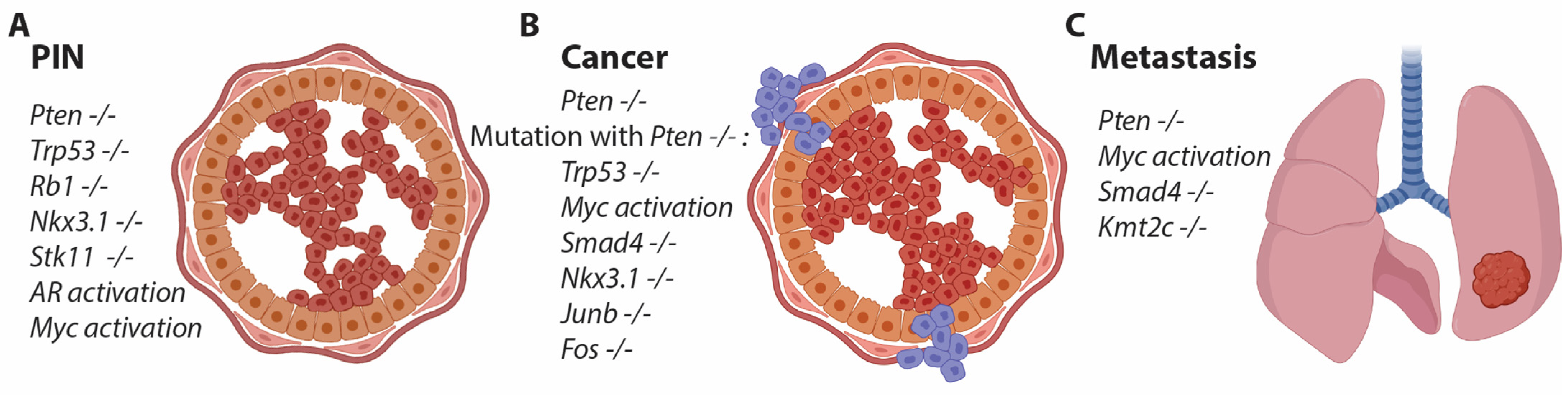
2. Evolution of Mouse Models for Prostate Cancer
2.1. Prostate-Specific Promoters
2.2. Prostate-Specific Gene Alteration
2.3. Targeting Multiple Genes in the Mouse Prostate
2.4. Gain-of-Function Studies for PCa
3. Orthotopic-Manipulation-Based PCa Models
CRISPR-Generated PCa Models
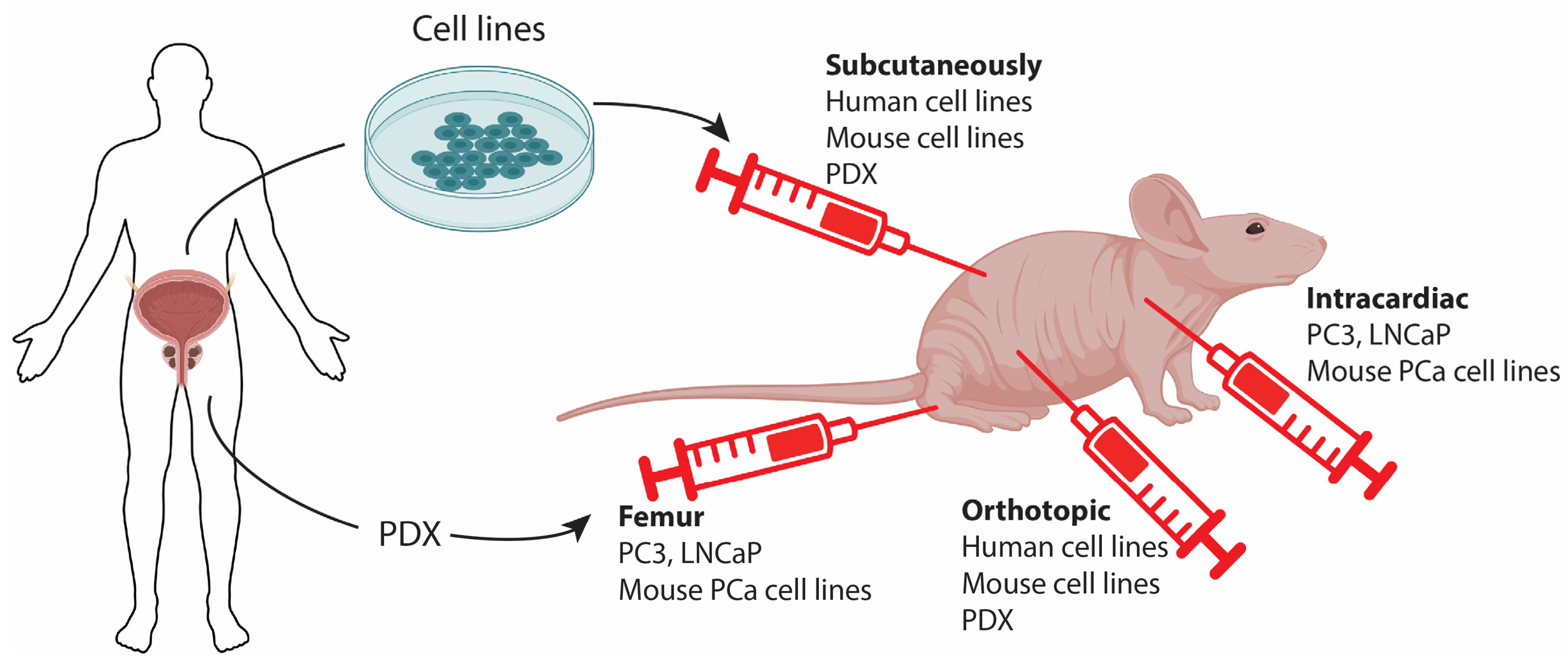
4. Pre-Clinical Models by Cell Implantation
4.1. Classical Cell Line-Derived Xenografts
4.2. Patient-Derived Xenografts
4.3. Allograft and Metastatic Models for PCa
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donin, N.M.; Reiter, R.E. Why Targeting PSMA Is a Game Changer in the Management of Prostate Cancer. J. Nucl. Med. 2018, 59, 177–182. [Google Scholar] [CrossRef]
- McNeal, J.E. The zonal anatomy of the prostate. Prostate 1981, 2, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Price, D. Comparative Aspects of Development and Structure in the Prostate. Natl. Cancer Inst. Monogr. 1963, 12, 1–27. [Google Scholar] [PubMed]
- Russell, P.J.; Voeks, D.J. Animal models of prostate cancer. Prostate Cancer Methods Protoc. 2003, 81, 89–112. [Google Scholar] [CrossRef]
- Bryan, J.N.; Keeler, M.R.; Henry, C.J.; Bryan, M.E.; Hahn, A.W.; Caldwell, C.W. A population study of neutering status as a risk factor for canine prostate cancer. Prostate 2007, 67, 1174–1181. [Google Scholar] [CrossRef]
- McNeal, J.E. The anatomic heterogeneity of the prostate. Prog. Clin. Biol. Res. 1980, 37, 149–160. [Google Scholar]
- Donehower, L.A.; Harvey, M.; Slagle, B.L.; McArthur, M.J.; Montgomery, C.A., Jr.; Butel, J.S.; Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992, 356, 215–221. [Google Scholar] [CrossRef]
- Liaw, D.; Marsh, D.J.; Li, J.; Dahia, P.L.; Wang, S.I.; Zheng, Z.; Bose, S.; Call, K.M.; Tsou, H.C.; Peacocke, M.; et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997, 16, 64–67. [Google Scholar] [CrossRef]
- Greenberg, N.M.; DeMayo, F.J.; Sheppard, P.C.; Barrios, R.; Lebovitz, R.; Finegold, M.; Angelopoulou, R.; Dodd, J.G.; Duckworth, M.L.; Rosen, J.M.; et al. The rat probasin gene promoter directs hormonally and developmentally regulated expression of a heterologous gene specifically to the prostate in transgenic mice. Mol. Endocrinol. 1994, 8, 230–239. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.; Thomas, T.Z.; Kasper, S.; Matusik, R.J. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology 2000, 141, 4698–4710. [Google Scholar] [CrossRef]
- Wu, X.; Wu, J.; Huang, J.; Powell, W.C.; Zhang, J.; Matusik, R.J.; Sangiorgi, F.O.; Maxson, R.E.; Sucov, H.M.; Roy-Burman, P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech. Dev. 2001, 101, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ziel-van der Made, A.C.; Autar, B.; van der Korput, H.A.; Vermeij, M.; van Duijn, P.; Cleutjens, K.B.; de Krijger, R.; Krimpenfort, P.; Berns, A.; et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005, 65, 5730–5739. [Google Scholar] [CrossRef] [PubMed]
- Abdulkadir, S.A.; Magee, J.A.; Peters, T.J.; Kaleem, Z.; Naughton, C.K.; Humphrey, P.A.; Milbrandt, J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol. Cell. Biol. 2002, 22, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.K.; Butler, C.M.; Shen, M.M.; Swain, A. Sox9 is required for prostate development. Dev. Biol. 2008, 316, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kruithof-de Julio, M.; Economides, K.D.; Walker, D.; Yu, H.; Halili, M.V.; Hu, Y.P.; Price, S.M.; Abate-Shen, C.; Shen, M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 2009, 461, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, N.M.; DeMayo, F.; Finegold, M.J.; Medina, D.; Tilley, W.D.; Aspinall, J.O.; Cunha, G.R.; Donjacour, A.A.; Matusik, R.J.; Rosen, J.M. Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. USA 1995, 92, 3439–3443. [Google Scholar] [CrossRef]
- Gelman, I.H. How the TRAMP Model Revolutionized the Study of Prostate Cancer Progression. Cancer Res. 2016, 76, 6137–6139. [Google Scholar] [CrossRef] [PubMed]
- Chiaverotti, T.; Couto, S.S.; Donjacour, A.; Mao, J.H.; Nagase, H.; Cardiff, R.D.; Cunha, G.R.; Balmain, A. Dissociation of epithelial and neuroendocrine carcinoma lineages in the transgenic adenocarcinoma of mouse prostate model of prostate cancer. Am. J. Pathol. 2008, 172, 236–246. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Ci, X.; Choi, S.Y.C.; Crea, F.; Lin, D.; Wang, Y. Molecular events in neuroendocrine prostate cancer development. Nat. Rev. Urol. 2021, 18, 581–596. [Google Scholar] [CrossRef]
- Arriaga, J.M.; Abate-Shen, C. Genetically Engineered Mouse Models of Prostate Cancer in the Postgenomic Era. Cold Spring Harb. Perspect. Med. 2019, 9, a030528. [Google Scholar] [CrossRef]
- Bhatia-Gaur, R.; Donjacour, A.A.; Sciavolino, P.J.; Kim, M.; Desai, N.; Young, P.; Norton, C.R.; Gridley, T.; Cardiff, R.D.; Cunha, G.R.; et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999, 13, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Trotman, L.C.; Niki, M.; Dotan, Z.A.; Koutcher, J.A.; Di Cristofano, A.; Xiao, A.; Khoo, A.S.; Roy-Burman, P.; Greenberg, N.M.; Van Dyke, T.; et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003, 1, E59. [Google Scholar] [CrossRef] [PubMed]
- Abate-Shen, C.; Banach-Petrosky, W.A.; Sun, X.; Economides, K.D.; Desai, N.; Gregg, J.P.; Borowsky, A.D.; Cardiff, R.D.; Shen, M.M. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003, 63, 3886–3890. [Google Scholar] [PubMed]
- Wang, S.; Gao, J.; Lei, Q.; Rozengurt, N.; Pritchard, C.; Jiao, J.; Thomas, G.V.; Li, G.; Roy-Burman, P.; Nelson, P.S.; et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003, 4, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Svensson, R.U.; Haverkamp, J.M.; Thedens, D.R.; Cohen, M.B.; Ratliff, T.L.; Henry, M.D. Slow disease progression in a C57BL/6 pten-deficient mouse model of prostate cancer. Am. J. Pathol. 2011, 179, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Chen, M.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat. Rev. Mol. Cell Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Riedel, M.; Berthelsen, M.F.; Cai, H.; Haldrup, J.; Borre, M.; Paludan, S.R.; Hager, H.; Vendelbo, M.H.; Wagner, E.F.; Bakiri, L.; et al. In vivo CRISPR inactivation of Fos promotes prostate cancer progression by altering the associated AP-1 subunit Jun. Oncogene 2021, 40, 2437–2447. [Google Scholar] [CrossRef]
- Thomsen, M.K.; Bakiri, L.; Hasenfuss, S.C.; Wu, H.; Morente, M.; Wagner, E.F. Loss of JUNB/AP-1 promotes invasive prostate cancer. Cell Death Differ. 2015, 22, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.K.; Ambroisine, L.; Wynn, S.; Cheah, K.S.; Foster, C.S.; Fisher, G.; Berney, D.M.; Moller, H.; Reuter, V.E.; Scardino, P.; et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010, 70, 979–987. [Google Scholar] [CrossRef]
- Francis, J.C.; McCarthy, A.; Thomsen, M.K.; Ashworth, A.; Swain, A. Brca2 and Trp53 deficiency cooperate in the progression of mouse prostate tumourigenesis. PLoS Genet. 2010, 6, e1000995. [Google Scholar] [CrossRef]
- Chen, Z.; Trotman, L.C.; Shaffer, D.; Lin, H.K.; Dotan, Z.A.; Niki, M.; Koutcher, J.A.; Scher, H.I.; Ludwig, T.; Gerald, W.; et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005, 436, 725–730. [Google Scholar] [CrossRef]
- Ku, S.Y.; Rosario, S.; Wang, Y.; Mu, P.; Seshadri, M.; Goodrich, Z.W.; Goodrich, M.M.; Labbe, D.P.; Gomez, E.C.; Wang, J.; et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Wankowicz, S.A.M.; Liu, D.; Gao, J.; Kundra, R.; Reznik, E.; Chatila, W.K.; Chakravarty, D.; Han, G.C.; Coleman, I.; et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 2018, 50, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Gundem, G.; Van Loo, P.; Kremeyer, B.; Alexandrov, L.B.; Tubio, J.M.C.; Papaemmanuil, E.; Brewer, D.S.; Kallio, H.M.L.; Hognas, G.; Annala, M.; et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015, 520, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wu, C.J.; Chu, G.C.; Xiao, Y.; Ho, D.; Zhang, J.; Perry, S.R.; Labrot, E.S.; Wu, X.; Lis, R.; et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 2011, 470, 269–273. [Google Scholar] [CrossRef]
- Ding, Z.; Wu, C.J.; Jaskelioff, M.; Ivanova, E.; Kost-Alimova, M.; Protopopov, A.; Chu, G.C.; Wang, G.; Lu, X.; Labrot, E.S.; et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 2012, 148, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Agersnap, S.N.; Sjøgren, A.; Simonsen, M.K.; Blaavand, M.S.; Jensen, U.V.; Thomsen, M.K. In Vivo Application of CRISPR/Cas9 Revealed Implication of Foxa1 and Foxp1 in Prostate Cancer Proliferation and Epithelial Plasticity. Cancers 2022, 14, 4381. [Google Scholar] [CrossRef]
- Hubner, A.; Mulholland, D.J.; Standen, C.L.; Karasarides, M.; Cavanagh-Kyros, J.; Barrett, T.; Chi, H.; Greiner, D.L.; Tournier, C.; Sawyers, C.L.; et al. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc. Natl. Acad. Sci. USA 2012, 109, 12046–12051. [Google Scholar] [CrossRef]
- Pencik, J.; Schlederer, M.; Gruber, W.; Unger, C.; Walker, S.M.; Chalaris, A.; Marie, I.J.; Hassler, M.R.; Javaheri, T.; Aksoy, O.; et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat. Commun. 2015, 6, 7736. [Google Scholar] [CrossRef]
- Wang, G.; Lunardi, A.; Zhang, J.; Chen, Z.; Ala, U.; Webster, K.A.; Tay, Y.; Gonzalez-Billalabeitia, E.; Egia, A.; Shaffer, D.R.; et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat. Genet. 2013, 45, 739–746. [Google Scholar] [CrossRef]
- Qin, J.; Wu, S.P.; Creighton, C.J.; Dai, F.; Xie, X.; Cheng, C.M.; Frolov, A.; Ayala, G.; Lin, X.; Feng, X.H.; et al. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature 2013, 493, 236–240. [Google Scholar] [CrossRef]
- Nowak, D.G.; Cho, H.; Herzka, T.; Watrud, K.; Demarco, D.V.; Wang, V.M.Y.; Senturk, S.; Fellmann, C.; Ding, D.; Beinortas, T.; et al. MYC Drives Pten/Trp53 Deficient Proliferation and Metastasis due to IL6 Secretion and AKT Suppression via PHLPP2. Cancer Discov. 2015, 5, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Limberger, T.; Schlederer, M.; Trachtova, K.; Garces de Los Fayos Alonso, I.; Yang, J.; Hogler, S.; Sternberg, C.; Bystry, V.; Oppelt, J.; Tichy, B.; et al. KMT2C methyltransferase domain regulated INK4A expression suppresses prostate cancer metastasis. Mol. Cancer 2022, 21, 89. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Buchanan, G.; Ittmann, M.; Harris, J.M.; Yu, X.; Demayo, F.J.; Tilley, W.; Greenberg, N.M. Mutation of the androgen receptor causes oncogenic transformation of the prostate. Proc. Natl. Acad. Sci. USA 2005, 102, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Laxman, B.; Varambally, S.; Cao, X.; Yu, J.; Helgeson, B.E.; Cao, Q.; Prensner, J.R.; Rubin, M.A.; Shah, R.B.; et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008, 10, 177–188. [Google Scholar] [CrossRef]
- Klezovitch, O.; Risk, M.; Coleman, I.; Lucas, J.M.; Null, M.; True, L.D.; Nelson, P.S.; Vasioukhin, V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc. Natl. Acad. Sci. USA 2008, 105, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Graff, R.E.; Pettersson, A.; Lis, R.T.; DuPre, N.; Jordahl, K.M.; Nuttall, E.; Rider, J.R.; Fiorentino, M.; Sesso, H.D.; Kenfield, S.A.; et al. The TMPRSS2:ERG fusion and response to androgen deprivation therapy for prostate cancer. Prostate 2015, 75, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Tretiakova, M.S.; Silvis, M.R.; Lucas, J.; Klezovitch, O.; Coleman, I.; Bolouri, H.; Kutyavin, V.I.; Morrissey, C.; True, L.D.; et al. ERG Activates the YAP1 Transcriptional Program and Induces the Development of Age-Related Prostate Tumors. Cancer Cell 2015, 27, 797–808. [Google Scholar] [CrossRef]
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef]
- Ellwood-Yen, K.; Graeber, T.G.; Wongvipat, J.; Iruela-Arispe, M.L.; Zhang, J.; Matusik, R.; Thomas, G.V.; Sawyers, C.L. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 2003, 4, 223–238. [Google Scholar] [CrossRef]
- Nandana, S.; Ellwood-Yen, K.; Sawyers, C.; Wills, M.; Weidow, B.; Case, T.; Vasioukhin, V.; Matusik, R. Hepsin cooperates with MYC in the progression of adenocarcinoma in a prostate cancer mouse model. Prostate 2010, 70, 591–600. [Google Scholar] [CrossRef]
- Cho, H.; Herzka, T.; Zheng, W.; Qi, J.; Wilkinson, J.E.; Bradner, J.E.; Robinson, B.D.; Castillo-Martin, M.; Cordon-Cardo, C.; Trotman, L.C. RapidCaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of Pten-mutant metastasis. Cancer Discov. 2014, 4, 318–333. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, G.K.; Mutton, L.N.; Khalili, M.; McMullin, R.P.; Hicks, J.L.; Bianchi-Frias, D.; Horn, L.A.; Kulac, I.; Moubarek, M.S.; Nelson, P.S.; et al. Combined MYC Activation and Pten Loss Are Sufficient to Create Genomic Instability and Lethal Metastatic Prostate Cancer. Cancer Res. 2016, 76, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Arriaga, J.M.; Panja, S.; Alshalalfa, M.; Zhao, J.; Zou, M.; Giacobbe, A.; Madubata, C.J.; Kim, J.Y.; Rodriguez, A.; Coleman, I.; et al. A MYC and RAS co-activation signature in localized prostate cancer drives bone metastasis and castration resistance. Nat. Cancer 2020, 1, 1082–1096. [Google Scholar] [CrossRef] [PubMed]
- Blattner, M.; Liu, D.; Robinson, B.D.; Huang, D.; Poliakov, A.; Gao, D.; Nataraj, S.; Deonarine, L.D.; Augello, M.A.; Sailer, V.; et al. SPOP Mutation Drives Prostate Tumorigenesis In Vivo through Coordinate Regulation of PI3K/mTOR and AR Signaling. Cancer Cell 2017, 31, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Shoag, J.; Liu, D.; Blattner, M.; Sboner, A.; Park, K.; Deonarine, L.; Robinson, B.D.; Mosquera, J.M.; Chen, Y.; Rubin, M.A.; et al. SPOP mutation drives prostate neoplasia without stabilizing oncogenic transcription factor ERG. J. Clin. Investig. 2018, 128, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Riedel, M.; Berthelsen, M.F.; Bakiri, L.; Wagner, E.F.; Thomsen, M.K. Virus Delivery of CRISPR Guides to the Murine Prostate for Gene Alteration. J. Vis. Exp. 2018, 134, e57525. [Google Scholar] [CrossRef]
- Leow, C.C.; Wang, X.D.; Gao, W.Q. Novel method of generating prostate-specific Cre-LoxP gene switching via intraductal delivery of adenovirus. Prostate 2005, 65, 1–9. [Google Scholar] [CrossRef]
- Munteanu, R.; Feder, R.I.; Onaciu, A.; Munteanu, V.C.; Iuga, C.A.; Gulei, D. Insights into the Human Microbiome and Its Connections with Prostate Cancer. Cancers 2023, 15, 2539. [Google Scholar] [CrossRef]
- Wong, L.; Hutson, P.R.; Bushman, W. Prostatic inflammation induces fibrosis in a mouse model of chronic bacterial infection. PLoS ONE 2014, 9, e100770. [Google Scholar] [CrossRef]
- Shinohara, D.B.; Vaghasia, A.M.; Yu, S.H.; Mak, T.N.; Bruggemann, H.; Nelson, W.G.; De Marzo, A.M.; Yegnasubramanian, S.; Sfanos, K.S. A mouse model of chronic prostatic inflammation using a human prostate cancer-derived isolate of Propionibacterium acnes. Prostate 2013, 73, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.K. Application of CRISPR for In Vivo Mouse Cancer Studies. Cancers 2022, 14, 5014. [Google Scholar] [CrossRef] [PubMed]
- Daya, S.; Berns, K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008, 21, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Chiou, S.H.; Winters, I.P.; Wang, J.; Naranjo, S.; Dudgeon, C.; Tamburini, F.B.; Brady, J.J.; Yang, D.; Grüner, B.M.; Chuang, C.H.; et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015, 29, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef]
- Berthelsen, M.F.; Leknes, S.L.; Riedel, M.; Pedersen, M.A.; Joseph, J.V.; Hager, H.; Vendelbo, M.H.; Thomsen, M.K. Comparative Analysis of Stk11/Lkb1 versus Pten Deficiency in Lung Adenocarcinoma Induced by CRISPR/Cas9. Cancers 2021, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Busk, M.; Horsman, M.R.; Overgaard, J. Resolution in PET hypoxia imaging: Voxel size matters. Acta Oncol. 2008, 47, 1201–1210. [Google Scholar] [CrossRef]
- Zhang, W.; Fan, W.; Rachagani, S.; Zhou, Z.; Lele, S.M.; Batra, S.K.; Garrison, J.C. Comparative Study of Subcutaneous and Orthotopic Mouse Models of Prostate Cancer: Vascular Perfusion, Vasculature Density, Hypoxic Burden and BB2r-Targeting Efficacy. Sci. Rep. 2019, 9, 11117. [Google Scholar] [CrossRef]
- Graves, E.E.; Vilalta, M.; Cecic, I.K.; Erler, J.T.; Tran, P.T.; Felsher, D.; Sayles, L.; Sweet-Cordero, A.; Le, Q.T.; Giaccia, A.J. Hypoxia in models of lung cancer: Implications for targeted therapeutics. Clin. Cancer Res. 2010, 16, 4843–4852. [Google Scholar] [CrossRef]
- Overgaard, J.; Overgaard, M.; Nielsen, O.S.; Pedersen, A.K.; Timothy, A.R. A comparative investigation of nimorazole and misonidazole as hypoxic radiosensitizers in a C3H mammary carcinoma in vivo. Br. J. Cancer 1982, 46, 904–911. [Google Scholar] [CrossRef]
- Mortensen, L.S.; Busk, M.; Nordsmark, M.; Jakobsen, S.; Theil, J.; Overgaard, J.; Horsman, M.R. Accessing radiation response using hypoxia PET imaging and oxygen sensitive electrodes: A preclinical study. Radiother. Oncol. 2011, 99, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Lilja-Fischer, J.K.; Ulhoi, B.P.; Alsner, J.; Stougaard, M.; Thomsen, M.S.; Busk, M.; Lassen, P.; Steiniche, T.; Nielsen, V.E.; Overgaard, J. Characterization and radiosensitivity of HPV-related oropharyngeal squamous cell carcinoma patient-derived xenografts. Acta Oncol. 2019, 58, 1489–1494. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Chen, X.; Tan, D. Development of patient-derived xenograft models of prostate cancer for maintaining tumor heterogeneity. Transl. Androl. Urol. 2019, 8, 519–528. [Google Scholar] [CrossRef]
- Russell, P.J.; Russell, P.; Rudduck, C.; Tse, B.W.; Williams, E.D.; Raghavan, D. Establishing prostate cancer patient derived xenografts: Lessons learned from older studies. Prostate 2015, 75, 628–636. [Google Scholar] [CrossRef]
- Abdolahi, S.; Ghazvinian, Z.; Muhammadnejad, S.; Saleh, M.; Asadzadeh Aghdaei, H.; Baghaei, K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022, 20, 206. [Google Scholar] [CrossRef]
- Elbadawy, M.; Abugomaa, A.; Yamawaki, H.; Usui, T.; Sasaki, K. Development of Prostate Cancer Organoid Culture Models in Basic Medicine and Translational Research. Cancers 2020, 12, 777. [Google Scholar] [CrossRef]
- Slovin, S.F. Immunotherapy combinations for metastatic castration-resistant prostate cancer—Failed trials and future aspects. Current Opinion in Urology 2023, 33, 390–395. [Google Scholar] [CrossRef]
- Jin, K.T.; Du, W.L.; Lan, H.R.; Liu, Y.Y.; Mao, C.S.; Du, J.L.; Mou, X.Z. Development of humanized mouse with patient-derived xenografts for cancer immunotherapy studies: A comprehensive review. Cancer Sci. 2021, 112, 2592–2606. [Google Scholar] [CrossRef] [PubMed]
- Simons, B.W.; Kothari, V.; Benzon, B.; Ghabili, K.; Hughes, R.; Zarif, J.C.; Ross, A.E.; Hurley, P.J.; Schaeffer, E.M. A mouse model of prostate cancer bone metastasis in a syngeneic immunocompetent host. Oncotarget 2019, 10, 6845–6854. [Google Scholar] [CrossRef]
- Park, S.H.; Eber, M.R.; Shiozawa, Y. Models of Prostate Cancer Bone Metastasis. Methods Mol. Biol. 2019, 1914, 295–308. [Google Scholar] [CrossRef]
- Wu, T.T.; Sikes, R.A.; Cui, Q.; Thalmann, G.N.; Kao, C.; Murphy, C.F.; Yang, H.; Zhau, H.E.; Balian, G.; Chung, L.W. Establishing human prostate cancer cell xenografts in bone: Induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int. J. Cancer 1998, 77, 887–894. [Google Scholar] [CrossRef]
| Biological Inoculation Material/Induction Technology | Key Advantages | Key Limitations | |
|---|---|---|---|
| Classical cell line-based xenograft models | Cell lines: LNCaP DU145 PC3 |
|
|
| PDX models | Patient biopsy |
|
|
| Mouse derived cell lines | Cell lines: B6CaP NPK |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomsen, M.K.; Busk, M. Pre-Clinical Models to Study Human Prostate Cancer. Cancers 2023, 15, 4212. https://doi.org/10.3390/cancers15174212
Thomsen MK, Busk M. Pre-Clinical Models to Study Human Prostate Cancer. Cancers. 2023; 15(17):4212. https://doi.org/10.3390/cancers15174212
Chicago/Turabian StyleThomsen, Martin K., and Morten Busk. 2023. "Pre-Clinical Models to Study Human Prostate Cancer" Cancers 15, no. 17: 4212. https://doi.org/10.3390/cancers15174212
APA StyleThomsen, M. K., & Busk, M. (2023). Pre-Clinical Models to Study Human Prostate Cancer. Cancers, 15(17), 4212. https://doi.org/10.3390/cancers15174212





