Distribution of Mycobacterium tuberculosis Lineages and Drug Resistance in Upper Myanmar
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites
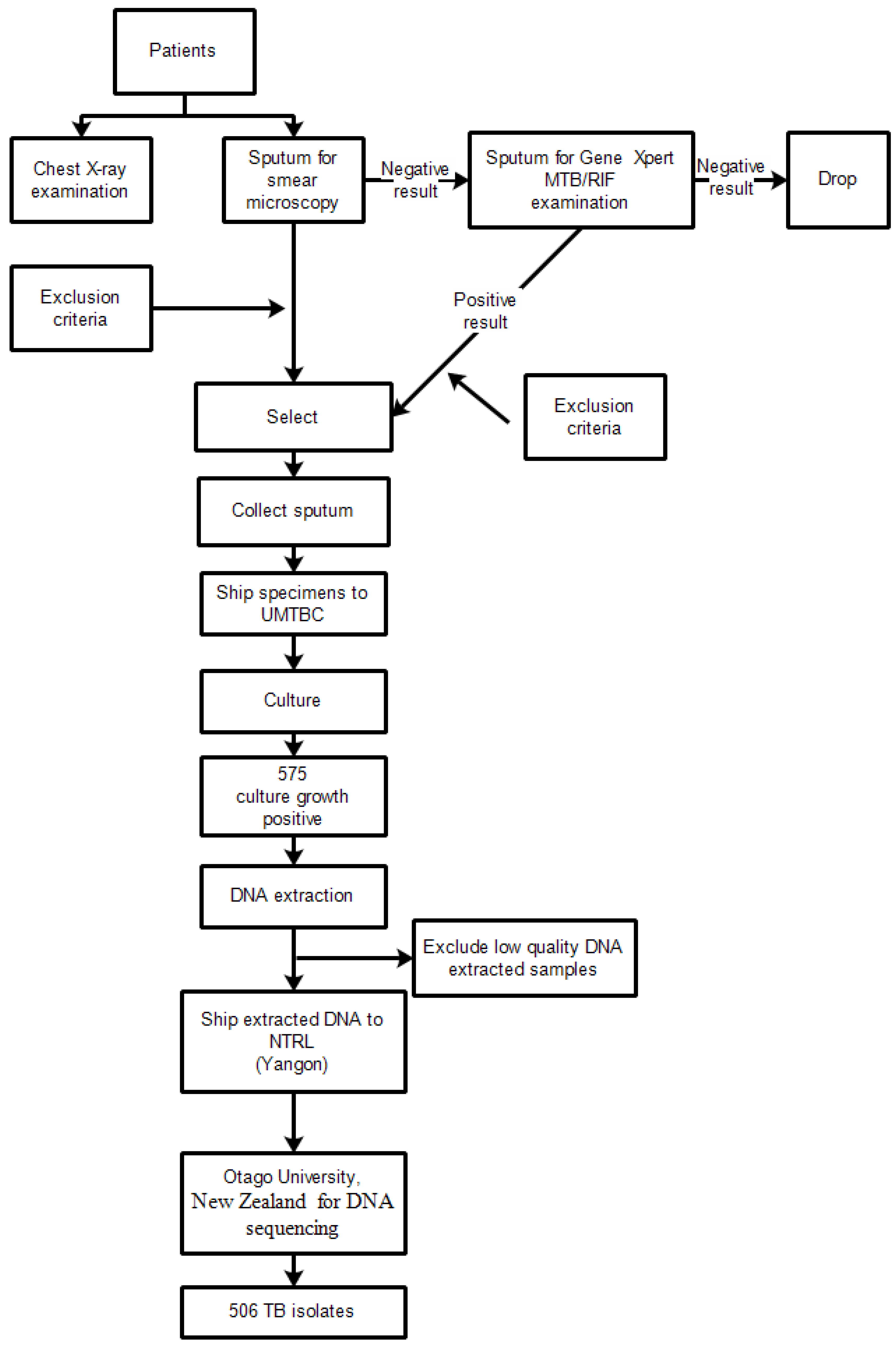
2.2. Study Design
2.3. Participants
2.4. Laboratory Processing of Specimens
2.5. Extraction of Genomic DNA, Whole Genome Sequencing, Variant Calling, Classification of Lineages and Prediction of Drug Resistance
2.6. Statistical Analysis
3. Results
3.1. Background Characteristics of Participants
3.2. Distribution of Lineages
3.3. Type of Anti-TB Drug Resistance by WGS
3.4. Association between Lineages and Drug Resistance Mutations
3.5. Association between Lineages and Drug Resistance Patterns
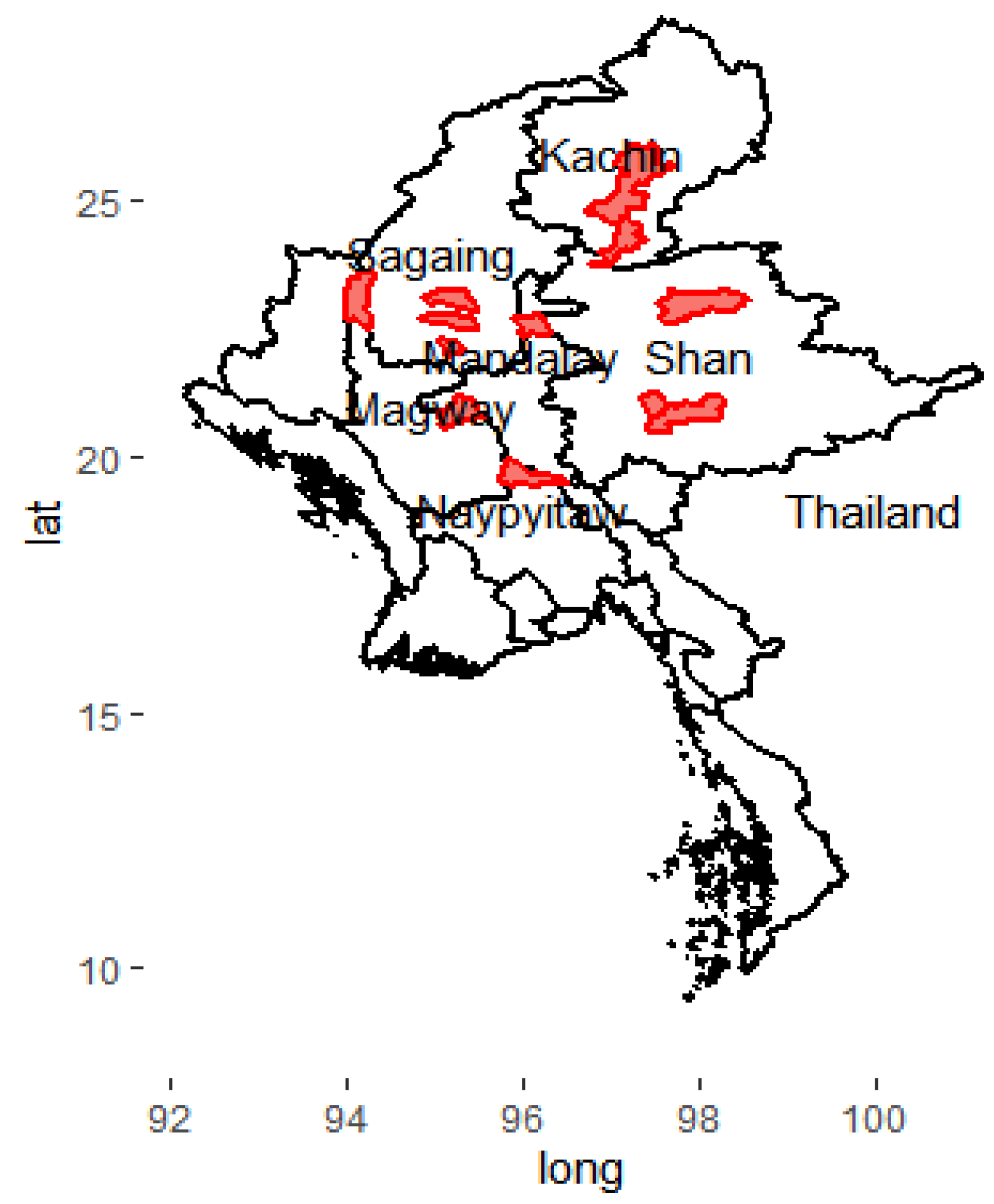
3.6. Geographical Distribution of Lineages and Drug Resistance Patterns
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Netikul, T.; Thawornwattana, Y.; Mahasirimongkol, S.; Yanai, H.; Maung, H.M.W.; Chongsuvivatwong, V.; Palittapongarnpim, P. Whole-genome single nucleotide variant phylogenetic analysis of Mycobacterium tuberculosis lineage 1 in endemic regions of Asia and Africa. Sci. Rep. 2022, 12, 1565. [Google Scholar] [CrossRef] [PubMed]
- Brites, D.; Gagneux, S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol. Rev. 2015, 264, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Netikul, T.; Palittapongarnpim, P.; Thawornwattana, Y.; Plitphonganphim, S. Estimation of the global burden of Mycobacterium tuberculosis lineage 1. Infect. Genet. Evol. 2021, 91, 104802. [Google Scholar] [CrossRef] [PubMed]
- Niemann, S.; Diel, R.; Khechinashvili, G.; Gegia, M.; Mdivani, N.; Tang, Y.-W. Mycobacterium tuberculosis Beijing lineage favors the spread of Multidrug-Resistant Tuberculosis in the Republic of Georgia. J. Clin. Microbiol. 2010, 48, 3544–3550. [Google Scholar] [CrossRef]
- Shuaib, Y.A.; Utpatel, C.; Kohl, T.A.; Barilar, I.; Diricks, M.; Ashraf, N.; Wieler, L.H.; Kerubo, G.; Mesfin, E.A.; Diallo, A.B.; et al. Origin and global expansion of Mycobacterium tuberculosis Complex lineage 3. Genes 2022, 13, 990. [Google Scholar] [CrossRef] [PubMed]
- Napier, G.; Campino, S.; Merid, Y.; Abebe, M.; Woldeamanuel, Y.; Aseffa, A.; Hibberd, M.L.; Phelan, J.; Clark, T.G. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020, 12, 114. [Google Scholar] [CrossRef]
- World Health Organization. Global Tuberculosis Report. 2022. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 30 November 2022).
- National Tuberculosis Programme, Ministry of Health and Sports. Annual National Tuberculosis Report; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- National Tuberculosis Program. Fourth Nationwide Anti-Tuberculosis Drugs Resistance Survey in Myanmar Protocol; NTP: Naypyitaw, Myanmar, 2019. [Google Scholar]
- Aung, H.L.; Devine, T.J.; Mulholland, C.V.; Arcus, V.L.; Cook, G.M. Tackling tuberculosis in the indigenous people of New Zealand. Lancet Public Health 2019, 4, e496. [Google Scholar] [CrossRef]
- Aung, H.L.; Tun, T.; Moradigaravand, D.; Köser, C.U.; Nyunt, W.W.; Aung, S.T.; Lwin, T.; Thinn, K.K.; Crump, J.A.; Parkhill, J.; et al. Whole-genome sequencing of multidrug-resistant Mycobacterium tuberculosis isolates from Myanmar. J. Glob. Antimicrob. Resist. 2016, 6, 113–117. [Google Scholar] [CrossRef]
- Moe Sann, W.; Swe, T.; Swe, K.; Thwin, T.; Sangka, A. The genotypic distribution of drug resistant Mycobacterium tuberculosis strains isolated from Northern region of Myanmar. Arch. AHS 2021, 33, 34–43. [Google Scholar]
- Ajawatanawong, P.; Yanai, H.; Smittipat, N.; Disratthakit, A.; Yamada, N.; Miyahara, R.; Nedsuwan, S.; Imasanguan, W.; Kantipong, P.; Chaiyasirinroje, B.; et al. A novel Ancestral Beijing sublineage of Mycobacterium tuberculosis suggests the transition site to Modern Beijing sublineages. Sci. Rep. 2019, 9, 13718. [Google Scholar] [CrossRef]
- Séraphin, M.N.; Doggett, R.; Johnston, L.; Zabala, J.; Gerace, A.M.; Lauzardo, M. Association between Mycobacterium tuberculosis lineage and site of disease in Florida, 2009–2015. Infect. Genet. Evol. 2017, 55, 366–371. [Google Scholar] [CrossRef]
- Welekidan, L.N.; Yimer, S.A.; Skjerve, E.; Dejene, T.A.; Homberset, H.; Tønjum, T.; Brynildsrud, O. Whole Genome Sequencing of drug resistant and drug susceptible Mycobacterium tuberculosis isolates from Tigray region, Ethiopia. Front. Microbiol. 2021, 12, 743198. [Google Scholar] [CrossRef]
- Maung, H.M.W.; Palittapongarnpim, P.; Aung, H.L.; Surachat, K.; Nyunt, W.W.; Chongsuvivatwong, V. Geno-spatial distribution of Mycobacterium Tuberculosis and drug resistance profiles in Myanmar–Thai border area. Trop. Med. Infect. Dis. 2020, 5, 153. [Google Scholar] [CrossRef]
- Phelan, J.E.; Lim, D.R.; Mitarai, S.; de Sessions, P.F.; Tujan, M.; Angelica, A.; Reyes, L.T.; Medado, I.A.P.; Palparan, A.G.; Naim, A.N.M.; et al. Mycobacterium tuberculosis whole genome sequencing provides insights into the Manila strain and drug-resistance mutations in the Philippines. Sci. Rep. 2019, 9, 9305. [Google Scholar] [CrossRef]
- Tun, T. Multidrug-resistant Mycobacterium tuberculosis strains in Myanmar patients. Myanmar Health Sci. Res. J. 2017, 29, 51–57. [Google Scholar]
- Maharjan, B.; Nakajima, C.; Isoda, N.; Thapa, J.; Poudel, A.; Shah, Y.; Yamaguchi, T.; Shrestha, B.; Hoffmann, H.; Avsar, K.; et al. Genetic diversity and distribution dynamics of multidrug-resistant Mycobacterium tuberculosis isolates in Nepal. Sci. Rep. 2018, 8, 16634. [Google Scholar] [CrossRef]
- Glasauer, S.; Altmann, D.; Hauer, B.; Brodhun, B.; Haas, W.; Perumal, N. First-line tuberculosis drug resistance patterns and associated risk factors in Germany, 2008-2017. Escobar-Gutiérrez A, editor. PLoS ONE 2019, 14, e0217597. [Google Scholar] [CrossRef]
- Aung, H.L.; Nyunt, W.W.; Fong, Y.; Biggs, P.J.; Winkworth, R.C.; Lockhart, P.J.; Yeo, T.W.; Hill, P.C.; Cook, G.M.; Aung, S.T. Genomic profiling of Mycobacterium tuberculosis strains, Myanmar. Emerg. Infect. Dis. 2021, 27, 2847–2855. [Google Scholar] [CrossRef]
- Crofton, J.; Mitchison, D.A. Streptomycin resistance in pulmonary tuberculosis. Br. Med. J. 1948, 2, 1009. [Google Scholar] [CrossRef]
- Manson, A.L.; Cohen, K.A.; Abeel, T.; Desjardins, C.A.; Armstrong, D.T.; Barry, C.E.; Brand, J.; Chapman, S.B.; Cho, S.N.; Gabrielian, A.; et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat. Genet. 2017, 49, 395–402. [Google Scholar] [CrossRef]
- Okamoto, S.; Tamaru, A.; Nakajima, C.; Nishimura, K.; Tanaka, Y.; Tokuyama, S.; Suzuki, Y.; Ochi, K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007, 63, 1096–1106. [Google Scholar] [CrossRef]
- ZZhao, L.L.; Liu, H.C.; Sun, Q.; Xiao, T.Y.; Zhao, X.Q.; Li, G.L.; Zeng, C.Y.; Wan, K.L. Identification of mutations conferring streptomycin resistance in multidrug-resistant tuberculosis of China. Diagn. Microbiol. Infect. Dis. 2015, 85, 150–153. [Google Scholar] [CrossRef]
- Wong, S.Y.; Lee, J.S.; Kwak, H.K.; Via, L.E.; Boshoff, H.I.M.; Barry, C.E. Mutations in gidB Confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2011, 55, 2515–2522. [Google Scholar] [CrossRef]
- Feuerriegel, S.; Oberhauser, B.; George, A.G.; Dafae, F.; Richter, E.; Rüsch-Gerdes, S.; Niemann, S. Sequence analysis for detection of first-line drug resistance in Mycobacterium tuberculosis strains from a high-incidence setting. BMC Microbiol. 2012, 12, 90. [Google Scholar] [CrossRef]
- Spies, F.S.; da Silva, P.E.A.; Ribeiro, M.O.; Rossetti, M.L.; Zaha, A. Identification of mutations related to streptomycin resistance in clinical isolates of Mycobacterium tuberculosis and possible involvement of efflux mechanism. Antimicrob. Agents Chemother. 2008, 52, 2947–2949. [Google Scholar] [CrossRef]
- Smittipat, N.; Juthayothin, T.; Billamas, P.; Jaitrong, S.; Rukseree, K.; Dokladda, K.; Chaiyasirinroje, B.; Disratthakit, A.; Chaiprasert, A.; Mahasirimongkol, S.; et al. Mutations in rrs, rpsL and gidB in streptomycin-resistant Mycobacterium tuberculosis isolates from Thailand. J. Glob. Antimicrob. Resist. 2016, 4, 5–10. [Google Scholar] [CrossRef]
- Nath, H.; Heekyung, J.; Sungweon, Y.; Koh, K.; Kim, C.; Kil, Y. Molecular genetics of Mycobacterium tuberculosis resistant to aminoglycosides and cyclic peptide capreomycin antibiotics in Korea. World J. Microbiol. Biotechnol. 2013, 29, 975–982. [Google Scholar]
- Jagielski, T.; Ignatowska, H.; Bakula, Z.; Dziewit, L.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Zwolska, Z.; Bielecki, J. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS ONE 2014, 9, e100078. [Google Scholar] [CrossRef]
- Sun, Y.J.; Luo, J.T.; Wong, S.Y.; Lee, A.S.G. Analysis of rpsL and rrs mutations in Beijing and non-Beijing streptomycin-resistant Mycobacterium tuberculosis isolates from Singapore. Clin. Microbiol. Infect. 2010, 16, 287–289. [Google Scholar] [CrossRef]
- Huang, H.N.; Han, J.X.; Wang, J.H.; Song, C.Z.; Liang, H.; Zhang, Z.L. rpsL gene analysis associated with streptomycin resistance in Mycobacterium tuberculosis. Yi Chuan Xue Bao 2003, 30, 376–381. [Google Scholar]
- Ramaswamy, S.V.; Dou, S.-J.; Rendon, A.; Yang, Z.; Cave, M.D.; Graviss, E.A. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J. Med. Microbiol. 2004, 53, 107–113. [Google Scholar] [CrossRef]
- Bauskenieks, M.; Pole, I.; Skenders, G.; Jansone, I.; Broka, L.; Nodieva, A.; Ozere, I.; Kalvisa, A.; Ranka, R.; Baumanis, V. Genotypic and phenotypic characteristics of aminoglycoside-resistant Mycobacterium tuberculosis isolates in Latvia. Diagn. Microbiol. Infect. Dis. 2015, 81, 177–182. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, C.; Xiang, L.; Pi, R.; Guo, Z.; Zheng, C.; Li, S.; Zhao, Y.; Tang, K.; Luo, M.; et al. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis isolates in Sichuan, China and the association between Beijing-lineage and dual-mutation in gidB. Tuberculosis 2016, 96, 102–106. [Google Scholar] [CrossRef]
- Oo, N.A.T.; San, L.L.; Thapa, J.; Aye, K.S.; Aung, W.W.; Nakajima, C.; Suzuki, Y. Characterization of mutations conferring streptomycin resistance to multidrug-resistant Mycobacterium tuberculosis isolates from Myanmar. Tuberculosis 2018, 111, 8–13. [Google Scholar]
- Holt, K.E.; McAdam, P.; Thai, P.V.K.; Thuong, N.T.T.; Ha, D.T.M.; Lan, N.N.; Lan, N.H.; Nhu, N.T.Q.; Hai, H.T.; Ha, V.T.N.; et al. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat. Genet. 2018, 50, 849–856. [Google Scholar] [CrossRef]
- Ruesen, C.; Chaidir, L.; Van Laarhoven, A.; Dian, S.; Ganiem, A.R.; Nebenzahl-Guimaraes, H.; Huynen, M.A.; Alisjahbana, B.; Dutilh, B.E.; Van Crevel, R. Large-scale genomic analysis shows association between homoplastic genetic variation in Mycobacterium tuberculosis genes and meningeal or pulmonary tuberculosis. BMC Genom. 2018, 19, 122. [Google Scholar] [CrossRef]
- Abascal, E.; Pérez-Lago, L.; Martínez-Lirola, M.; Chiner-Oms, Á.; Herranz, M.; Chaoui, I.; Comas, I.; El Messaoudi, M.D.; Cárdenas, J.A.G.; Santantón, S.; et al. Whole genome sequencing–based analysis of tuberculosis (TB) in migrants: Rapid tools for cross-border surveillance and to distinguish between recent transmission in the host country and new importations. Eurosurveillance 2019, 24, 1800005. [Google Scholar] [CrossRef]
| Variables | N = 506 (%) |
|---|---|
| Age in year | |
| <20 | 40 (7.9) |
| 20–30 | 93 (18.4) |
| 31–40 | 113 (22.3) |
| 41–50 | 106 (20.9) |
| 51–60 | 90 (17.8) |
| >60 | 64 (12.6) |
| Gender | |
| Male | 346 (68.4) |
| Female | 160 (31.6) |
| Diabetes mellitus status | |
| Positive | 59 (11.7) |
| Negative | 348 (68.8) |
| Unknown | 99 (19.6) |
| Smoking status | |
| Yes | 115 (22.7) |
| Never | 268 (53.0) |
| Ex-smoker | 123 (24.3) |
| Previously treated for TB | |
| Yes | 40 (7.9) |
| No | 465 (91.9) |
| Unknown | 1 (0.2) |
| Lineages | N = 506 (%) |
|---|---|
| Lineage 1 | 201 (39.7) |
| Lineage 2 | 223 (44.1) |
| Lineage 3 | 20 (4.0) |
| Lineage 4 | 62 (12.3) |
| Type of Anti-TB Drug Resistance | N (%) |
|---|---|
| Sensitive to all drugs | 446 (88.1) |
| Any drug resistance | 60 (11.9) |
| Mono drug resistance to streptomycin | 26 (5.1) |
| Mono drug resistance to isoniazid | 8 (1.6) |
| Mono drug resistance to levofloxacin | 2 (0.4) |
| Poly drug resistance | 24 (4.7) |
| Multi-drug resistance | 9 (1.8) |
| Pre-XDR-TB | 5 (1.0) |
| Drug Resistance Mutation | Total | Lineages | Fisher’s Exact Test, p Value | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| N | (N = 201) | (N = 223) | (N = 20) | (N = 62) | ||
| Drug resistant mutation | 0.017 | |||||
| Isoniazid | ||||||
| katG_S315T | 22 | 3 | 14 | 1 | 4 | |
| InhA_C15T | 9 | 2 | 4 | 1 | 2 | |
| oxyR-ahpC_G-48A ap | 1 | 0 | 1 | 0 | 0 | |
| Rifampicin | ||||||
| rpoB_S450L | 7 | 1 | 5 | 0 | 1 | |
| rpoB_I491F | 1 | 0 | 1 | 0 | 0 | |
| rpoB_H445D | 1 | 0 | 1 | 0 | 0 | |
| Pyrazinamide (Z) | ||||||
| ncA_408_ins_1_a_at | 1 | 0 | 1 | 0 | 0 | |
| Ethambutol (E) | ||||||
| embB_M306V | 3 | 0 | 2 | 0 | 1 | |
| embB_M306I | 1 | 0 | 1 | 0 | 0 | |
| embC_c-516t | 1 | 0 | 1 | 0 | 0 | |
| Streptomycin (S) | ||||||
| rpsL_K43R | 32 | 2 | 28 | 1 | 1 | |
| rpsL_K88R | 7 | 1 | 6 | 0 | 0 | |
| gidB_A138V | 3 | 2 | 0 | 0 | 1 | |
| rrs_C517T | 1 | 1 | 0 | 0 | 0 | |
| gidB_L91P_gid | 1 | 0 | 1 | 0 | 0 | |
| Levofloxacin (Lfx) | ||||||
| gyrA_D94G | 3 | 0 | 2 | 0 | 1 | |
| gyrA_D94N | 1 | 0 | 1 | 0 | 0 | |
| gyrA_A90V | 3 | 1 | 1 | 0 | 1 | |
| Ethionamide (Eto) | ||||||
| inhA_C15T | 6 | 1 | 3 | 1 | 1 | |
| Drug Resistance Pattern | Lineages | Fisher’s Exact Test, p Value | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| (N = 201) | (N = 223) | (N = 20) | (N = 62) | ||
| Drug resistance patterns | <0.001 | ||||
| H0 R0 Z0 E0 S0 Lfx0 Eto0 | 193 | 181 | 18 | 54 | |
| H0 R0 Z0 E0 SR Lfx0 Eto0 | 3 | 22 | 0 | 1 | |
| HR R0 Z0 E0 S0 Lfx0 Eto0 | 2 | 2 | 0 | 4 | |
| HR R0 Z0 E0 SR Lfx0 Eto0 | 1 | 6 | 1 | 0 | |
| HR R0 Z0 E0 S0 Lfx0 EtoR | 0 | 2 | 1 | 1 | |
| HR RR Z0 E0 SR Lfx0 Eto0 | 0 | 3 | 0 | 0 | |
| Others including HR RR | 1 | 4 | 0 | 1 | |
| Others not including HR RR | 1 | 3 | 0 | 1 | |
| Lineages and Drug Resistance Patterns | Regions and States of Upper Myanmar | Fisher’s Exact Test, p Value | |||||
|---|---|---|---|---|---|---|---|
| Regions | States | ||||||
| Sagaing (N = 105) | Magway (N = 72) | Naypyitaw (N = 34) | Mandalay (N = 174) | Shan (N = 56) | Kachin (N = 65) | ||
| (A) Lineages | <0.001 | ||||||
| Lineage 1 | 51 | 33 | 15 | 79 | 6 | 17 | |
| Lineage 2 | 39 | 28 | 13 | 63 | 40 | 40 | |
| Lineage 3 | 4 | 3 | 0 | 10 | 1 | 2 | |
| Lineage 4 | 11 | 8 | 6 | 22 | 9 | 6 | |
| (B) Drug Resistance Patterns | 0.123 | ||||||
| H0 R0 Z0 E0 S0Lfx0 Eto0 | 91 | 66 | 31 | 158 | 43 | 57 | |
| H0 R0 Z0 E0 SR Lfx0 Eto0 | 4 | 3 | 2 | 5 | 8 | 4 | |
| HR R0 Z0 E0 S0 Lfx0 Eto0 | 3 | 2 | 1 | 2 | 0 | 0 | |
| HR R0 Z0 E0 SR Lfx0 Eto0 | 2 | 0 | 0 | 3 | 1 | 2 | |
| HR R0 Z0 E0 S0 Lfx0 EtoR | 1 | 0 | 0 | 1 | 2 | 0 | |
| HR RR Z0 E0 SR Lfx0 Eto0 | 2 | 0 | 0 | 0 | 1 | 0 | |
| Others | 2 | 1 | 0 | 5 | 1 | 2 | |
| Our Study | Yangon, Myanmar | Kayin, Myanmar | Thailand | Philippines | Nepal | Tigray, Ethiopia | |
|---|---|---|---|---|---|---|---|
| Lineage 1 | 201 (39.7) | 9 (13) | 73 (67) | 480 (41) | 143 (80) | 32 (6) | 1 (1.5) |
| Lineage 2 | 223 (44.1) | 55 (76) | 26 (23) | 521 (45) | 2 (1) | 241 (48) | 1 (1.5) |
| Lineage 3 | 20 (4.0) | 4 (5) | 4 (4) | 11 (1) | 0 (0) | 153 (32) | 28 (41.2) |
| Lineage 4 | 62 (12.3) | 4 (5) | 6 (6) | 158 (13) | 33 (19) | 72 (14) | 38 (55.8) |
| Total | 506 (100.0) | 72 (100) | 109 (100) | 1170 (100) | 178 (100) | 498 (100) | 68 (100.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phyu, A.N.; Aung, S.T.; Palittapongarnpim, P.; Htet, K.K.K.; Mahasirimongkol, S.; Aung, H.L.; Chaiprasert, A.; Chongsuvivatwong, V. Distribution of Mycobacterium tuberculosis Lineages and Drug Resistance in Upper Myanmar. Trop. Med. Infect. Dis. 2022, 7, 448. https://doi.org/10.3390/tropicalmed7120448
Phyu AN, Aung ST, Palittapongarnpim P, Htet KKK, Mahasirimongkol S, Aung HL, Chaiprasert A, Chongsuvivatwong V. Distribution of Mycobacterium tuberculosis Lineages and Drug Resistance in Upper Myanmar. Tropical Medicine and Infectious Disease. 2022; 7(12):448. https://doi.org/10.3390/tropicalmed7120448
Chicago/Turabian StylePhyu, Aye Nyein, Si Thu Aung, Prasit Palittapongarnpim, Kyaw Ko Ko Htet, Surakameth Mahasirimongkol, Htin Lin Aung, Angkana Chaiprasert, and Virasakdi Chongsuvivatwong. 2022. "Distribution of Mycobacterium tuberculosis Lineages and Drug Resistance in Upper Myanmar" Tropical Medicine and Infectious Disease 7, no. 12: 448. https://doi.org/10.3390/tropicalmed7120448
APA StylePhyu, A. N., Aung, S. T., Palittapongarnpim, P., Htet, K. K. K., Mahasirimongkol, S., Aung, H. L., Chaiprasert, A., & Chongsuvivatwong, V. (2022). Distribution of Mycobacterium tuberculosis Lineages and Drug Resistance in Upper Myanmar. Tropical Medicine and Infectious Disease, 7(12), 448. https://doi.org/10.3390/tropicalmed7120448








