Identification of Mutations in Antimalarial Resistance Gene Kelch13 from Plasmodium falciparum Isolates in Kano, Nigeria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Drug Treatment and Follow Up
2.3. DNA Extraction and Confirmation of Plasmodium Falciparum Infection Using PCR
2.4. Amplification of Propeller Domains of the Kelch13 Gene and Column Purification
2.5. Cloning of the Kelch13 Fragment and Sequencing
2.6. Analysis of Genetic Variability of Kelch13
3. Results
3.1. Malaria Slide Positivity Rate
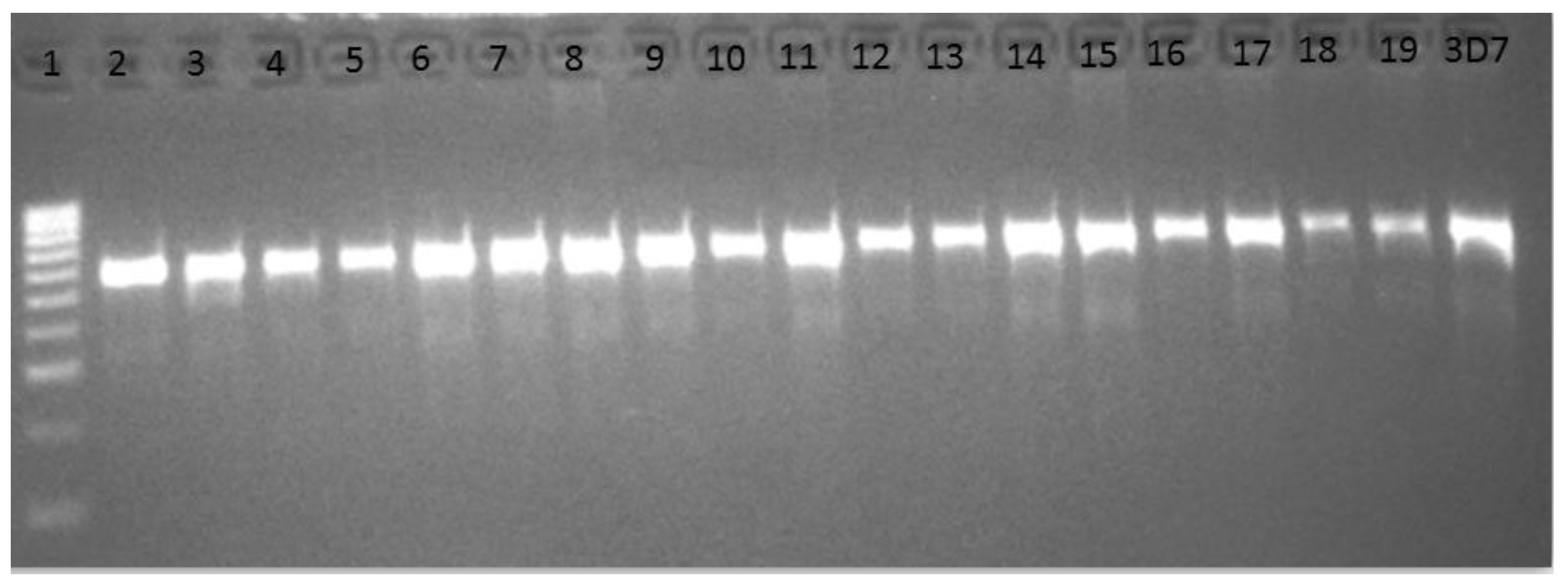
3.2. Cytochrome Oxidase III PCR Confirmation of Plasmodium Infection
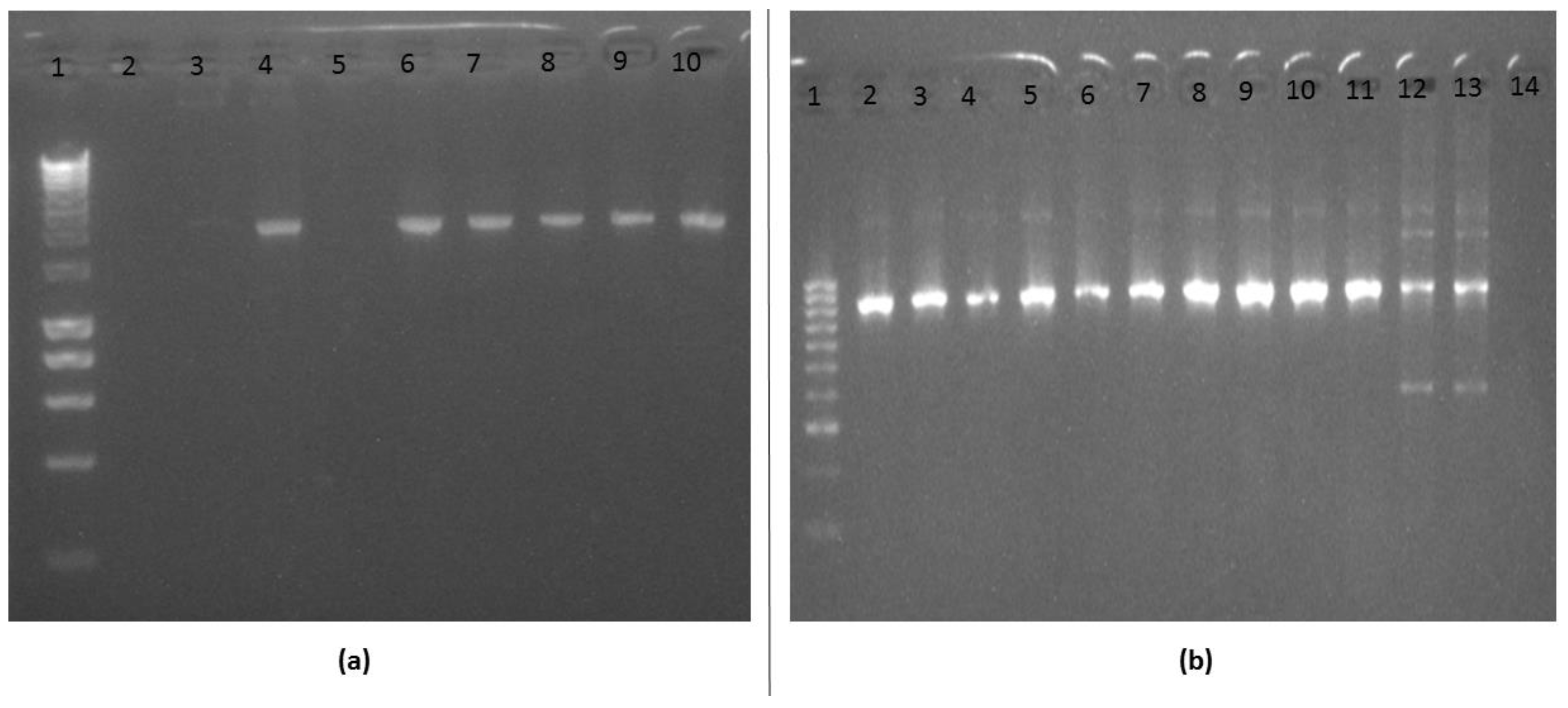
3.3. Amplification and Cloning of the Kelch13 Fragment
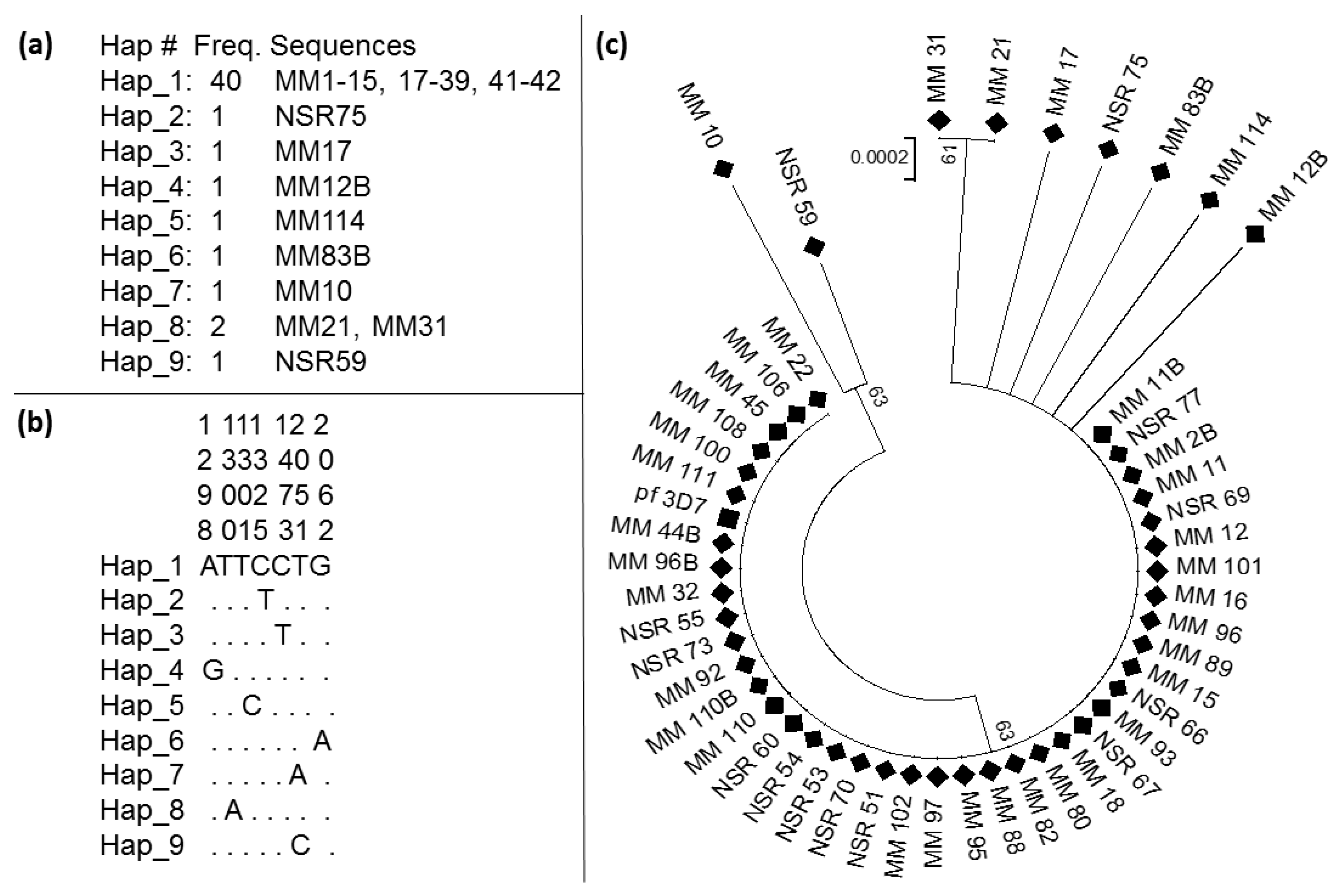
3.4. Pattern of Genetic Variability of the Kelch13 Fragment
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- WHO. World Malaria Report 2019; 9241565721; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- WHO. World Malaria Report; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Bhatt, S.; Weiss, D.J.; Cameron, E.; Bisanzio, D.; Mappin, B.; Dalrymple, U.; Battle, K.E.; Moyes, C.L.; Henry, A.; Eckhoff, P.A.; et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015, 526, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef] [PubMed]
- FMoH. National Antimalarial Treatment Guidelines Policy Federal Ministry of Health, National Malaria and Vector Control Division; Federal Ministry of Health: Abuja-Nigeria, Nigeria, 2005. [Google Scholar]
- WHO. World Malaria Report 2014; World Health Organisation: Geneva, Switzerland, 2015. [Google Scholar]
- WHO. World Malaria Report; World Health Organisation: Geneva, Switzerland, 2017; ISBN 978-92-4-156552-3. [Google Scholar]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.-C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L.; et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2013, 505, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Daily, J.P. K13-Propeller Mutations and Malaria Resistance. N. Engl. J. Med. 2016, 374, 2492–2493. [Google Scholar] [CrossRef] [PubMed]
- Menard, D.; Khim, N.; Beghain, J.; Adegnika, A.A.; Alam, M.S.; Amodu, O.; Rahim-Awab, G.; Barnadas, C.; Berry, A.; Boum, Y.; et al. A Worldwide Map of Plasmodium falciparum K13-Propeller Polymorphisms. N. Engl. J. Med. 2016, 374, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- MalariaGEN Plasmodium falciparum Community Project. Genomic epidemiology of artemisinin resistant malaria. eLife 2016, 5, 17. [Google Scholar] [CrossRef]
- Balikagala, B.; Mita, T.; Ikeda, M.; Sakurai, M.; Yatsushiro, S.; Takahashi, N.; Tachibana, S.-I.; Auma, M.; Ntege, E.H.; Ito, D.; et al. Absence of in vivo selection for K13 mutations after artemether-lumefantrine treatment in Uganda. Malar. J. 2017, 16, 23. [Google Scholar] [CrossRef]
- Laminou, I.M.; Lamine, M.M.; Mahamadou, B.; Ascofare, O.M.; Dieye, A. Polymorphism of pfk13-propeller in Niger: Detection of Novel Mutations. J. Adv. Med. Med. Res. 2017, 22, 1–5. [Google Scholar] [CrossRef]
- Bayih, A.G.; Getnet, G.; Alemu, A.; Getie, S.; Mohon, A.N.; Pillai, D.R. A Unique Plasmodium falciparum Kelch 13 Gene Mutation in Northwest Ethiopia. Am. J. Trop. Med. Hyg. 2016, 94, 132–135. [Google Scholar] [CrossRef]
- Talundzic, E.; Ndiaye, Y.D.; Deme, A.B.; Olsen, C.; Patel, D.S.; Biliya, S.; Daniels, R.; Vannberg, F.O.; Volkman, S.K.; Udhayakumar, V.; et al. Molecular Epidemiology of Plasmodium falciparum kelch13 Mutations in Senegal Determined by Using Targeted Amplicon Deep Sequencing. Antimicrob. Agents Chemother. 2017, 61, e02116-16. [Google Scholar] [CrossRef]
- Taylor, S.M.; Parobek, C.; DeConti, D.K.; Kayentao, K.; Coulibaly, S.O.; Greenwood, B.M.; Tagbor, H.; Williams, J.; Bojang, K.A.; Njie, F.; et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: A molecular epidemiologic study. J. Infect. Dis. 2014, 211, 680–688. [Google Scholar] [CrossRef]
- WWARN K13 Genotype-Phenotype Study Group; Amaratunga, C. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments—A WWARN individual patient data meta-analysis. BMC Med. 2019, 17, 1. [Google Scholar] [CrossRef]
- Lu, F.; Culleton, R.; Zhang, M.; Ramaprasad, A.; Von Seidlein, L.; Zhou, H.; Zhu, G.; Tang, J.; Liu, Y.; Wang, W.; et al. Emergence of Indigenous Artemisinin-Resistant Plasmodium falciparum in Africa. N. Engl. J. Med. 2017, 376, 991–993. [Google Scholar] [CrossRef]
- Hawkes, M.T.; Conroy, A.L.; Opoka, R.O.; Namasopo, S.; Zhong, K.; Liles, W.C.; John, C.C.; Kain, K. Slow Clearance of Plasmodium falciparum in Severe Pediatric Malaria, Uganda, 2011–2013. Emerg. Infect. Dis. 2015, 21, 1237–1239. [Google Scholar] [CrossRef]
- Oboh, M.A.; Ndiaye, D.; Antony, H.A.; Badiane, A.S.; Singh, U.; Ali, N.A.; Bharti, P.K.; Das, A. Status of Artemisinin Resistance in Malaria Parasite Plasmodium falciparum from Molecular Analyses of the Kelch13 Gene in Southwestern Nigeria. BioMed Res. Int. 2018, 2018, 1–5. [Google Scholar] [CrossRef]
- Cheesbrough, M. District Laboratory Practice in Tropical Countries by Monica Cheesbrough. In District Laboratory Practice in Tropical Countries; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Ohrt, C.; Purnomo; Sutamihardja, M.A.; Tang, D.; Kain, K. Impact of Microscopy Error on Estimates of Protective Efficacy in Malaria? Prevention Trials. J. Infect. Dis. 2002, 186, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Echeverry, D.F.; Deason, N.A.; Davidson, J.; Makuru, V.; Xiao, H.; Niedbalski, J.; Kern, M.; Russell, T.L.; Burkot, T.R.; Collins, F.H.; et al. Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene. Malar. J. 2016, 15, 128. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1998; Volume 41, pp. 95–98. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Boil. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Laminou, I.M.; Lamine, M.M.; Arzika, I.; Mahamadou, B.; Gora, D.; Dieye, A. Detection of Plasmodium falciparum K13 Propeller A569G Mutation after Artesunate-amodiaquine Treatment Failure in Niger. J. Adv. Boil. Biotechnol. 2018, 18, 1–8. [Google Scholar] [CrossRef]
- Dama, S.; Niangaly, H.; Ouattara, A.; Sagara, I.; Sissoko, S.; Traore, O.B.; Bamadio, A.; Dara, N.; Djimde, A.; Alhousseini, M.L.; et al. Reduced ex vivo susceptibility of Plasmodium falciparum after oral artemether-lumefantrine treatment in Mali. Malar. J. 2017, 16, 59. [Google Scholar] [CrossRef] [PubMed]
- Kamau, E.; Campino, S.; Amenga-Etego, L.; Drury, E.; Ishengoma, D.S.; Johnson, K.; Mumba, D.; Kekre, M.; Yavo, W.; Mead, D.; et al. K13-Propeller Polymorphisms in Plasmodium falciparum Parasites from Sub-Saharan Africa. J. Infect. Dis. 2014, 211, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Boussaroque, A.; Fall, B.; Madamet, M.; Wade, K.A.; Fall, M.; Nakoulima, A.; Fall, K.B.; Dionne, P.; Benoit, N.; Diatta, B.; et al. Prevalence of anti-malarial resistance genes in Dakar, Senegal from 2013 to 2014. Malar. J. 2016, 15, 347. [Google Scholar] [CrossRef]
- Ouattara, A.; Bjorkman, A.; Kone, A.; Adams, M.; Diallo, N.; Dara, A.; Maiga, A.W.; Takala-Harrison, S.; Fofana, B.; Plowe, C.V.; et al. Polymorphisms in the K13-Propeller Gene in Artemisinin-Susceptible Plasmodium falciparum Parasites from Bougoula-Hameau and Bandiagara, Mali. Am. J. Trop. Med. Hyg. 2015, 92, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Somé, A.; Sorgho, H.; Zongo, I.; Bazié, T.; Nikiéma, F.; Sawadogo, A.; Zongo, M.; Compaore, Y.D.; Ouedraogo, J.B. Polymorphisms inK13, pfcrt, pfmdr1, pfdhfr, andpfdhpsin parasites isolated from symptomatic malaria patients in Burkina Faso. Parasite 2016, 23, 60. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Olasehinde, G.; Ojurongbe, D.; Akinjogunl, O.; Egwari, L.; Adeyeba, A. Prevalence of Malaria and Predisposing Factors to Antimalarial Drug Resistance in Southwestern Nigeria. Res. J. Parasitol. 2015, 10, 92–101. [Google Scholar] [CrossRef]
- Kalu, K.M.; Obasi, N.A.; Nduka, F.O.; Oko, M.O. Prevalence of Malaria Parasitaemia in Umuchieze and Uturu Communities of Abia State, Nigeria. Asian J. Epidemiol. 2012, 5, 95–102. [Google Scholar] [CrossRef]

| Day of Treatment | No. of Positive | No. of Negative | Total No. of Samples |
|---|---|---|---|
| Day 0 | 114 (74.0%) | 40 (26.0%) | 154 (100%) |
| Day 14 (Follow Up) | 11 (22.0%) | 39 (78.0%) | 50 (100%) |
| Total | 125 | 79 | 204 |
| Domain/ Propeller | Wild Type Codon | Polymorphic Site | Position (nt) | Observed Mutation | Sequence (s) with Mutation | Substitution Type | Day of Collection |
|---|---|---|---|---|---|---|---|
| BTB/POZ BTB/POZ | GAA TTT | GGA ATT | 1298 1300 | Glu433Gly Phe434Ile | 1 (MM_12B) 2 (MM_31, MM_21) | NS NS | 14th 0 |
| BTB/POZ | TTT | TCT | 1301 | Phe434Ser | 1 (MM_114) | NS | 0 |
| Blade 1 | TTT | TTC | 1325 | Phe442Phe | 1 | S | 0 |
| Blade 2 | TTT | TTC | 1473 | Phe492Phe | 1 | S | 0 |
| Blade 6 | ATT | AAT | 2051 | Ile684Asn | 1 (MM_10) | NS | 0 |
| Blade 6 | ATT | ACT | 2051 | Ile684Thr | 1 (NSR_59) | NS | 0 |
| Blade 6 | GAA | AAA | 2062 | Glu688Lys | 1 (MM_83B) | NS | 14th |
| N | S | H | Hd | Syn | Nonsyn | π (k) | D (Tajima) | D* (Fu and Li) |
|---|---|---|---|---|---|---|---|---|
| 49 | 8 | 9 | 0.336 | 2 | 6 | 0.00043(0.364) | −2.18234sig | −3.67210 sig |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abubakar, U.F.; Adam, R.; Mukhtar, M.M.; Muhammad, A.; Yahuza, A.A.; Ibrahim, S.S. Identification of Mutations in Antimalarial Resistance Gene Kelch13 from Plasmodium falciparum Isolates in Kano, Nigeria. Trop. Med. Infect. Dis. 2020, 5, 85. https://doi.org/10.3390/tropicalmed5020085
Abubakar UF, Adam R, Mukhtar MM, Muhammad A, Yahuza AA, Ibrahim SS. Identification of Mutations in Antimalarial Resistance Gene Kelch13 from Plasmodium falciparum Isolates in Kano, Nigeria. Tropical Medicine and Infectious Disease. 2020; 5(2):85. https://doi.org/10.3390/tropicalmed5020085
Chicago/Turabian StyleAbubakar, Umar F., Ruqayya Adam, Muhammad M. Mukhtar, Abdullahi Muhammad, Adamu A. Yahuza, and Sulaiman S. Ibrahim. 2020. "Identification of Mutations in Antimalarial Resistance Gene Kelch13 from Plasmodium falciparum Isolates in Kano, Nigeria" Tropical Medicine and Infectious Disease 5, no. 2: 85. https://doi.org/10.3390/tropicalmed5020085
APA StyleAbubakar, U. F., Adam, R., Mukhtar, M. M., Muhammad, A., Yahuza, A. A., & Ibrahim, S. S. (2020). Identification of Mutations in Antimalarial Resistance Gene Kelch13 from Plasmodium falciparum Isolates in Kano, Nigeria. Tropical Medicine and Infectious Disease, 5(2), 85. https://doi.org/10.3390/tropicalmed5020085







