Marine Microbiome as a Source of Antimalarials
Abstract
1. Introduction
2. Materials and Methods
2.1. Fermentation
2.2. Extraction
2.3. Plasmodium in Vitro Culture
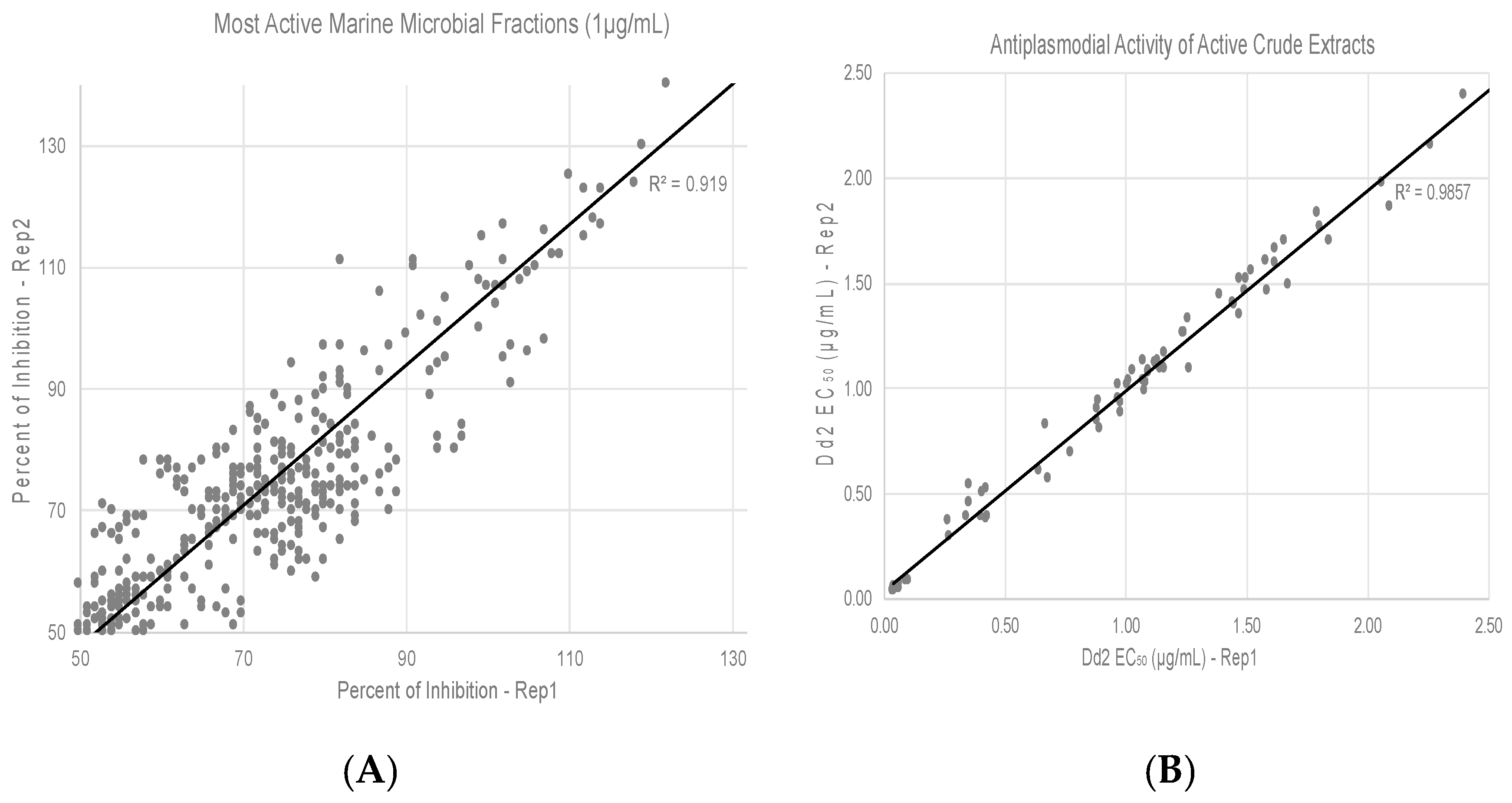
2.4. Antiplasmodium Activity Assay
2.5. Cytotoxicity Assay
2.6. Dereplication of Bioactive Fractions
3. Results
3.1. Extracts of Marine Microbes Used in Screening
3.2. Screening of HBMCC Extracts for Antiplasmodial Activities
3.3. Identification of Bioactive Compounds
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- WHO. World Malaria Report; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Nsanzabana, C. Resistance to Artemisinin Combination Therapies (ACTs): Do Not Forget the Partner Drug! Trop. Med. Infect. Dis. 2019, 4, 26. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Dis. 2009, 8, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, S.J.; Stingl, U. Molecular diversity and ecology of microbial plankton. Nature 2005, 437, 343–348. [Google Scholar] [CrossRef]
- Gloeckner, V.; Wehrl, M.; Moithinho-Silva, L.; Gernert, C.; Schupp, P.; Pawlick, J.R.; Lindquist, N.L.; Erpenbeck, D.; Worheide, G.; Hentschel, U. The HMA-LMA Dichotomy Revisited: An Electron Microscopical Survey of 56 Sponge Species. Biol. Bull. 2014, 227, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Konig, G.M.; Kehraus, S.; Seibert, S.F.; Abdel-Lateff, A.; Muller, D. Natural products from marine organisms and their associated microbes. Chembiochem 2006, 7, 229–238. [Google Scholar] [CrossRef]
- Lackner, G.; Peters, E.E.; Helfrich, E.J.; Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 2017, 114, E347–E356. [Google Scholar] [CrossRef]
- Rao, K.V.; Kasanah, N.; Wahyuono, S.; Tekwani, B.L.; Schinazi, R.F.; Hamann, M.T. Three new manzamine alkaloids from a common Indonesian sponge and their activity against infectious and tropical parasitic diseases. J. Nat. Prod. 2004, 67, 1314–1318. [Google Scholar] [CrossRef]
- Waters, A.L.; Peraud, O.; Kasanah, N.; Sims, J.W.; Kothalawala, N.; Anderson, M.A.; Abbas, S.H.; Rao, K.V.; Jupally, V.R.; Kelly, M.; et al. An analysis of the sponge Acanthostrongylophora igens’ microbiome yields an actinomycete that produces the natural product manzamine A. Front. Mar. Sci. 2014, 1. [Google Scholar] [CrossRef] [PubMed]
- Bultel-Ponce, V.V.; Berge, J.P.; Debitus, C.; Nicolas, J.L.; Guyot, M. Metabolites from the Sponge-Associated Bacterium Pseudomonas Species. Mar. Biotechnol. (N. Y.) 1999, 1, 384–390. [Google Scholar] [CrossRef]
- Matsumoto, K.; Choshi, T.; Hourai, M.; Zamami, Y.; Sasaki, K.; Abe, T.; Ishikura, M.; Hatae, N.; Iwamura, T.; Tohyama, S.; et al. Synthesis and antimalarial activity of calothrixins A and B, and their N-alkyl derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 4762–4764. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.B.; Lord, C.C.; McCarthy, P.J. Improved Recoverability of Microbial Colonies from Marine Sponge Samples. Microb. Ecol. 2000, 40, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.; Harmody, D.; Dang, P.; Ledger, A.; Pomponi, S.; McCarthy, P.; Lopez, J. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Syst. Appl. Microbiol. 2005, 28, 242–264. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, A.S.; Sfanos, K.S.; Harmody, D.K.; Pomponi, S.A.; McCarthy, P.J.; Lopez, J.V. HBMMD: An enhanced database of the microorganisms associated with deeper water marine invertebrates. Appl. Microbiol. Biotechnol. 2005, 66, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, R.; Trincone, A. Bioactive Compounds Produced by Strains of Penicillium and Talaromyces of Marine Origin. Mar. Drugs 2016, 14, 37. [Google Scholar] [CrossRef]
- Trager, W.; Jensen, J.B. Human malaria parasites in continuous culture. Science 1976, 193, 673–675. [Google Scholar] [CrossRef]
- Bennett, T.N.; Paguio, M.; Gligorijevic, B.; Seudieu, C.; Kosar, A.D.; Davidson, E.; Roepe, P.D. Novel, rapid, and inexpensive cell-based quantification of antimalarial drug efficacy. Antimicrob. Agents Chemother. 2004, 48, 1807–1810. [Google Scholar] [CrossRef]
- Johnson, J.D.; Dennull, R.A.; Gerena, L.; Lopez-Sanchez, M.; Roncal, N.E.; Waters, N.C. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 2007, 51, 1926–1933. [Google Scholar] [CrossRef]
- Smilkstein, M.; Sriwilaijaroen, N.; Kelly, J.X.; Wilairat, P.; Riscoe, M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004, 48, 1803–1806. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.B. In vitro culture of Plasmodium parasites. Methods Mol. Med. 2002, 72, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Peyrottes, S.; Caldarelli, S.; Wein, S.; Perigaud, C.; Pellet, A.; Vial, H. Choline analogues in malaria chemotherapy. Curr. Pharm. Des. 2012, 18, 3454–3466. [Google Scholar] [CrossRef][Green Version]
- Houbraken, J.; Samson, R.A.; Yilmaz, N. Taxonomy of Aspergillus, Penicillium and Talaromyces and Its Significance for Biotechnology. In Aspergillus and Penicillium in the Post-Genomic Era; Caister Academic Press: Norfolk, UK, 2016. [Google Scholar]
- Nielsen, J.C.; Grijseels, S.; Prigent, S.; Ji, B.; Dainat, J.; Nielsen, K.F.; Frisvad, J.C.; Workman, M.; Nielsen, J. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2017, 2, 17044. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, R.; Salvatore, M.M.; Andolfi, A. Secondary Metabolites of Mangrove-Associated Strains of Talaromyces. Mar. Drugs 2018, 16, 12. [Google Scholar] [CrossRef]
- Ueda, J.Y.; Takagi, M.; Shin-ya, K. New xanthoquinodin-like compounds, JBIR-97, -98 and -99, obtained from marine sponge-derived fungus Tritirachium sp. SpB081112MEf2. J. Antibiot. (Tokyo) 2010, 63, 615–618. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Masschelein, J.; Jenner, M.; Challis, G.L. Antibiotics from Gram-negative bacteria: A comprehensive overview and selected biosyntheitic highlights. Nat. Prod. Rep. 2017, 34, 712–783. [Google Scholar] [CrossRef]
- Homann, V.V.; Edwards, K.J.; Webb, E.A.; Butler, A. Siderophores of Marinobacter aquaeolei: Petrobactin and its sulfonated derivatives. Biometals 2009, 22, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Golyshin, P.N.; Lang, S.; Moore, E.R.B.; Abraham, W.-R.; Lünsdorf, H.; Timmis, K.N. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 1998, 48, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, K.; Adachi, K.; Katsuta, A.; Shizuri, Y. Structural determination and proposed biosynthesis of alcanivorone, a novel a-pyrone produced by Alcanivorax jadensis. J. Antibiotics 2008, 61, 70–74. [Google Scholar] [CrossRef]
- Bayer, T.; Neave, M.J.; Alsheikh-Hussain, A.; Aranda, M.; Yum, L.K.; Mincer, T.; Hughen, K.; Apprill, A.; Voolstra, C.R. The Microbiome of the Red Sea Coral Stylophora pistillata Is Dominated by Tissue-Associated Endozoicomonas Bacteria. Appl. Environ. Microbiol. 2013, 79, 4759–4762. [Google Scholar] [CrossRef] [PubMed]
- Rua, C.P.J.; Trindade-Silva, A.E.; Appolinario, L.R.; Venas, T.M.; Garcia, G.D.; Carvalho, L.S.; Lima, A.; Kruger, R.; Pereira, R.C.; Berlinck, R.G.S.; et al. Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine spongeArenosclera brasiliensis. PeerJ 2014, 2, e419. [Google Scholar] [CrossRef] [PubMed]
- Pike, R.E.; Haltli, B.; Kerr, R.G. Description of Endozoicomonas euniceicola sp. nov. and Endozoicomonas gorgoniicola sp. nov., bacteria isolated from the octocorals Eunicea fusca and Plexaura sp., and an emended description of the genus Endozoicomonas. Int. J. Syst. Evol. Microbiol. 2013, 63, 4294–4302. [Google Scholar] [CrossRef]
| Taxonomic Affiliation | Number of Isolates | Number Isolated from Sponges |
|---|---|---|
| Actinomycetes | 1037 | 416 |
| Other bacteria | 16,129 | 11,281 |
| Fungi | 2074 | 1427 |
| Isolate # | Taxonomy | Category | Medium | Incubation Period | Growth Conditions | Extraction System | Dd2 IC50 µg/mL | Cytotoxicity IC50 µg/mL |
|---|---|---|---|---|---|---|---|---|
| V324 | Streptomyces tendae | A | SYZ | 22 days | Static | Resin, MeOH/CH2Cl2 | 0.35 | >50 |
| V663 | Unidentified actinomycete | A | Rice | 21 days | Static | ASE, Heptane | 0.89 | 10.2 |
| V671 | Nocardiopsis sp. | A | Rice | 21 days | Static | ASE, MeOH | 0.88 | 9.1 |
| W305 | Micromonospora sp. | A | SYZ | 14 days | Shake | Resin, MeOH | 0.42 | 9.3 |
| V881 | Streptomyces sp. | A | SYZ | 14days | Shake | Resin, CH2Cl2 | 0.062 | 29.1 |
| Z691 | Penicillium sp. | F | SYZ | 14 days | Shake | Resin, CH2Cl2 | 0.049 | 27.2 |
| E677 | Streptomyces sp. | A | SYZ | 7 days | Shake | Resin, MeOH/CH2Cl2 | 0.037 | 28.6 |
| H402 | Endozoicomonas numazuensis | B (G-) | KP | 7 days | Shake | Resin, MeOH/CH2Cl2 | 0.978 | >50 |
| N161 | Penicillium sp. | F | SYZ | 22 days | Static | Resin, MeOH/CH2Cl2 | 0.266 | >50 |
| S920 | Talaromyces rotundus | F | KP | 21 days | Static | Resin, MeOH/CH2Cl2 | 0.677 | >50 |
| V170 | Penicillium citrinum | F | KP | 21 days | Static | Resin, MeOH/CH2Cl2 | 1.069 | >50 |
| V174 | Alcanivorax sp. | B (G-) | KP | 7 days | Shake | Resin, MeOH/CH2Cl2 | 0.969 | >50 |
| V184 | Marinobacter sp. | B (G-) | KP | 7 days | Shake | Resin, MeOH/CH2Cl2 | 1.008 | >50 |
| V193 | Alcanivorax sp. | B (G-) | KP | 7 days | Shake | Resin, MeOH/CH2Cl2 | 1.079 | >50 |
| V199 | Tritirachium sp. | F | KP | 7 days | Shake | Resin, MeOH/CH2Cl2 | 0.339 | >50 |
| V201 | Marinobacter sp. | B (G-) | SYZ | 24 days | Static | Resin, MeOH/CH2Cl2 | 1.091 | >50 |
| V208 | Marinobacter sp. | B (G-) | SYZ | 7 days | Shake | Resin, MeOH/CH2Cl2 | 1.091 | >50 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCarthy, P.J.; Roberts, B.F.; Carbonell, A.; Roberts, J.; Wright, A.E.; Chakrabarti, D. Marine Microbiome as a Source of Antimalarials. Trop. Med. Infect. Dis. 2019, 4, 103. https://doi.org/10.3390/tropicalmed4030103
McCarthy PJ, Roberts BF, Carbonell A, Roberts J, Wright AE, Chakrabarti D. Marine Microbiome as a Source of Antimalarials. Tropical Medicine and Infectious Disease. 2019; 4(3):103. https://doi.org/10.3390/tropicalmed4030103
Chicago/Turabian StyleMcCarthy, Peter J., Bracken F. Roberts, Abigail Carbonell, Jill Roberts, Amy E. Wright, and Debopam Chakrabarti. 2019. "Marine Microbiome as a Source of Antimalarials" Tropical Medicine and Infectious Disease 4, no. 3: 103. https://doi.org/10.3390/tropicalmed4030103
APA StyleMcCarthy, P. J., Roberts, B. F., Carbonell, A., Roberts, J., Wright, A. E., & Chakrabarti, D. (2019). Marine Microbiome as a Source of Antimalarials. Tropical Medicine and Infectious Disease, 4(3), 103. https://doi.org/10.3390/tropicalmed4030103





