Emergence of Multidrug-Resistant Campylobacter jejuni in a Common Variable Immunodeficiency Patient: Evolution of Resistance Under the Selective Antibiotic Pressure
Abstract
1. Introduction
2. Case Presentation
3. Materials and Methods
3.1. Sample Collection and Processing
3.1.1. Sample Collection and Preparation
3.1.2. Bacteriological Cultivation and Identification
3.1.3. Antimicrobial Susceptibility Testing (AST)
3.2. Genomics
3.2.1. Library Preparation and Sequencing
3.2.2. Bioinformatic Tools
4. Results
4.1. Phenotypic Results
4.1.1. Species Identification/Confirmation
4.1.2. Antimicrobial Susceptibility Testing
4.2. WGS Results
4.2.1. Species Identification/Confirmation
4.2.2. Antimicrobial Resistance Genomic Profile
4.2.3. Virulence Genes
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015; ISBN 978-92-4-156516-5. [Google Scholar]
- The European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- Campylobacteriosis Overtakes Salmonellosis as the Most Reported Animal Infection Transmitted to Humans in the EU | EFSA. Available online: https://www.efsa.europa.eu/en/news/campylobacteriosis-overtakes-salmonellosis-most-reported-animal-infection (accessed on 25 March 2025).
- Fischer, G.H.; Hashmi, M.F.; Paterek, E. Campylobacter Infection. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Javid, M.H. Medscape: Campylobacter Infections Treatment & Management. Available online: https://emedicine.medscape.com/article/213720-treatment (accessed on 25 March 2025).
- Barker, C.R.; Painset, A.; Swift, C.; Jenkins, C.; Godbole, G.; Maiden, M.C.J.; Dallman, T.J. Microevolution of Campylobacter Jejuni during Long-Term Infection in an Immunocompromised Host. Sci. Rep. 2020, 10, 10109. [Google Scholar] [CrossRef]
- Nunes, A.; Oleastro, M.; Alves, F.; Liassine, N.; Lowe, D.M.; Benejat, L.; Ducounau, A.; Jehanne, Q.; Borges, V.; Gomes, J.P.; et al. Recurrent Campylobacter Jejuni Infections with In Vivo Selection of Resistance to Macrolides and Carbapenems: Molecular Characterization of Resistance Determinants. Microbiol. Spectr. 2023, 11, e0107023. [Google Scholar] [CrossRef] [PubMed]
- Gharamti, A.A.; Moukalled, N.; Taher, A.; Kanafani, Z.A. Recurrent Campylobacter Bacteremia as the First Manifestation of Hypogammaglobulinemia: A Case Report and Literature Review. Infect. Chemother. 2020, 52, 415–420. [Google Scholar] [CrossRef]
- Shin, E.; Hong, H.; Oh, Y.; Lee, Y. First Report and Molecular Characterization of a Campylobacter Jejuni Isolate with Extensive Drug Resistance from a Travel-Associated Human Case. Antimicrob. Agents Chemother. 2015, 59, 6670–6672. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moore, J.E.; Barton, M.D.; Blair, I.S.; Corcoran, D.; Dooley, J.S.G.; Fanning, S.; Kempf, I.; Lastovica, A.J.; Lowery, C.J.; Matsuda, M.; et al. The Epidemiology of Antibiotic Resistance in Campylobacter. Microbes Infect. 2006, 8, 1955–1966. [Google Scholar] [CrossRef]
- Burch, K.L.; Saeed, K.; Sails, A.D.; Wright, P.A. Successful Treatment by Meropenem of Campylobacter Jejuni Meningitis in a Chronic Alcoholic Following Neurosurgery. J. Infect. 1999, 39, 241–243. [Google Scholar] [CrossRef]
- Portes, A.B.; Panzenhagen, P.; Pereira dos Santos, A.M.; Junior, C.A.C. Antibiotic Resistance in Campylobacter: A Systematic Review of South American Isolates. Antibiotics 2023, 12, 548. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic Resistance in Campylobacter: Emergence, Transmission and Persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef]
- Payot, S.; Bolla, J.-M.; Corcoran, D.; Fanning, S.; Mégraud, F.; Zhang, Q. Mechanisms of Fluoroquinolone and Macrolide Resistance in Campylobacter spp. Microbes Infect. 2006, 8, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Gibreel, A.; Kos, V.N.; Keelan, M.; Trieber, C.A.; Levesque, S.; Michaud, S.; Taylor, D.E. Macrolide Resistance in Campylobacter Jejuni and Campylobacter Coli: Molecular Mechanism and Stability of the Resistance Phenotype. Antimicrob. Agents Chemother. 2005, 49, 2753–2759. [Google Scholar] [CrossRef]
- Hull, D.M.; Harrell, E.; van Vliet, A.H.M.; Correa, M.; Thakur, S. Antimicrobial Resistance and Interspecies Gene Transfer in Campylobacter Coli and Campylobacter Jejuni Isolated from Food Animals, Poultry Processing, and Retail Meat in North Carolina, 2018–2019. PLoS ONE 2021, 16, e0246571. [Google Scholar] [CrossRef]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Ghedira, K.; Ghram, A.; Maaroufi, A. Distribution of Virulence and Antibiotic Resistance Genes in Campylobacter Jejuni and Campylobacter Coli Isolated from Broiler Chickens in Tunisia. J. Microbiol. Immunol. Infect. 2022, 55, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Rokney, A.; Valinsky, L.; Vranckx, K.; Feldman, N.; Agmon, V.; Moran-Gilad, J.; Weinberger, M. WGS-Based Prediction and Analysis of Antimicrobial Resistance in Campylobacter Jejuni Isolates From Israel. Front. Cell. Infect. Microbiol. 2020, 10, 365. [Google Scholar] [CrossRef]
- Redondo, N.; Carroll, A.; McNamara, E. Molecular Characterization of Campylobacter Causing Human Clinical Infection Using Whole-Genome Sequencing: Virulence, Antimicrobial Resistance and Phylogeny in Ireland. PLoS ONE 2019, 14, e0219088. [Google Scholar] [CrossRef]
- Llarena, A.-K.; Taboada, E.; Rossi, M. Whole-Genome Sequencing in Epidemiology of Campylobacter Jejuni Infections. J. Clin. Microbiol. 2017, 55, 1269–1275. [Google Scholar] [CrossRef]
- Painset, A.; Day, M.; Doumith, M.; Rigby, J.; Jenkins, C.; Grant, K.; Dallman, T.J.; Godbole, G.; Swift, C. Comparison of Phenotypic and WGS-Derived Antimicrobial Resistance Profiles of Campylobacter Jejuni and Campylobacter Coli Isolated from Cases of Diarrhoeal Disease in England and Wales, 2015–2016. J. Antimicrob. Chemother. 2020, 75, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Šoprek, S.; Duvnjak, S.; Kompes, G.; Jurinović, L.; Tambić Andrašević, A. Resistome Analysis of Campylobacter Jejuni Strains Isolated from Human Stool and Primary Sterile Samples in Croatia. Microorganisms 2022, 10, 1410. [Google Scholar] [CrossRef]
- Roa-Bautista, A.; Brown, L.-A.K.; Tadros, S.; Burns, S.O.; Godbole, G.; Lowe, D.M. Clinical Features, Immunological Characteristics, and Treatment Outcomes of Campylobacter Spp. Infections in Patients With Common Variable Immunodeficiency. J. Allergy Clin. Immunol. Pract. 2023, 11, 3493–3501.e4. [Google Scholar] [CrossRef]
- Eucast: MIC and Zone Distributions and ECOFFs. Available online: https://www.eucast.org/mic_and_zone_distributions_and_ecoffs (accessed on 7 August 2024).
- European Centre for Disease Prevention and Control. Antimicrobial-Resistance-Salmonella-Campylobacter-Harmonised-Monitoring-Annex-Aug-2021; European Centre for Disease Prevention and Control: Solna, Sweden, 2016. [Google Scholar]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 September 2024).
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinforma. 2020, 70, e102. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinforma. Oxf. Engl. 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome Analysis Using the Kraken Software Suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: A Tool for Fast and Accurate Genomic Comparisons That Enables Improved Genome Recovery from Metagenomes through de-Replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Seeman, T. Mlst, Github. Available online: https://Github.Com/Tseemann/Mlst (accessed on 20 March 2025).
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable Analysis of Bacterial Genome Variation at the Population Level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis--10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Seeman, T. Abricate, Github. Available online: https://Github.Com/Tseemann/Abricate (accessed on 20 March 2025).
- Alfredson, D.A.; Korolik, V. Antibiotic Resistance and Resistance Mechanisms in Campylobacter Jejuni and Campylobacter Coli. FEMS Microbiol. Lett. 2007, 277, 123–132. [Google Scholar] [CrossRef]
- Janssen, R.; Krogfelt, K.A.; Cawthraw, S.A.; van Pelt, W.; Wagenaar, J.A.; Owen, R.J. Host-Pathogen Interactions in Campylobacter Infections: The Host Perspective. Clin. Microbiol. Rev. 2008, 21, 505–518. [Google Scholar] [CrossRef]
- Ferenc, T.; Vujica, M.; Mayer, M. Long Term Management of Complex Patient with Common Variable Immunodeficiency—A Case Report. Reumatizam 2023, 69, 103–109. [Google Scholar] [CrossRef]
- Schürch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole Genome Sequencing Options for Bacterial Strain Typing and Epidemiologic Analysis Based on Single Nucleotide Polymorphism versus Gene-by-Gene–Based Approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Tikhomirova, A.; McNabb, E.R.; Petterlin, L.; Bellamy, G.L.; Lin, K.H.; Santoso, C.A.; Daye, E.S.; Alhaddad, F.M.; Lee, K.P.; Roujeinikova, A. Campylobacter Jejuni Virulence Factors: Update on Emerging Issues and Trends. J. Biomed. Sci. 2024, 31, 45. [Google Scholar] [CrossRef]
- Callahan, S.M.; Dolislager, C.G.; Johnson, J.G. The Host Cellular Immune Response to Infection by Campylobacter spp. and Its Role in Disease. Infect. Immun. 2021, 89, e0011621. [Google Scholar] [CrossRef]
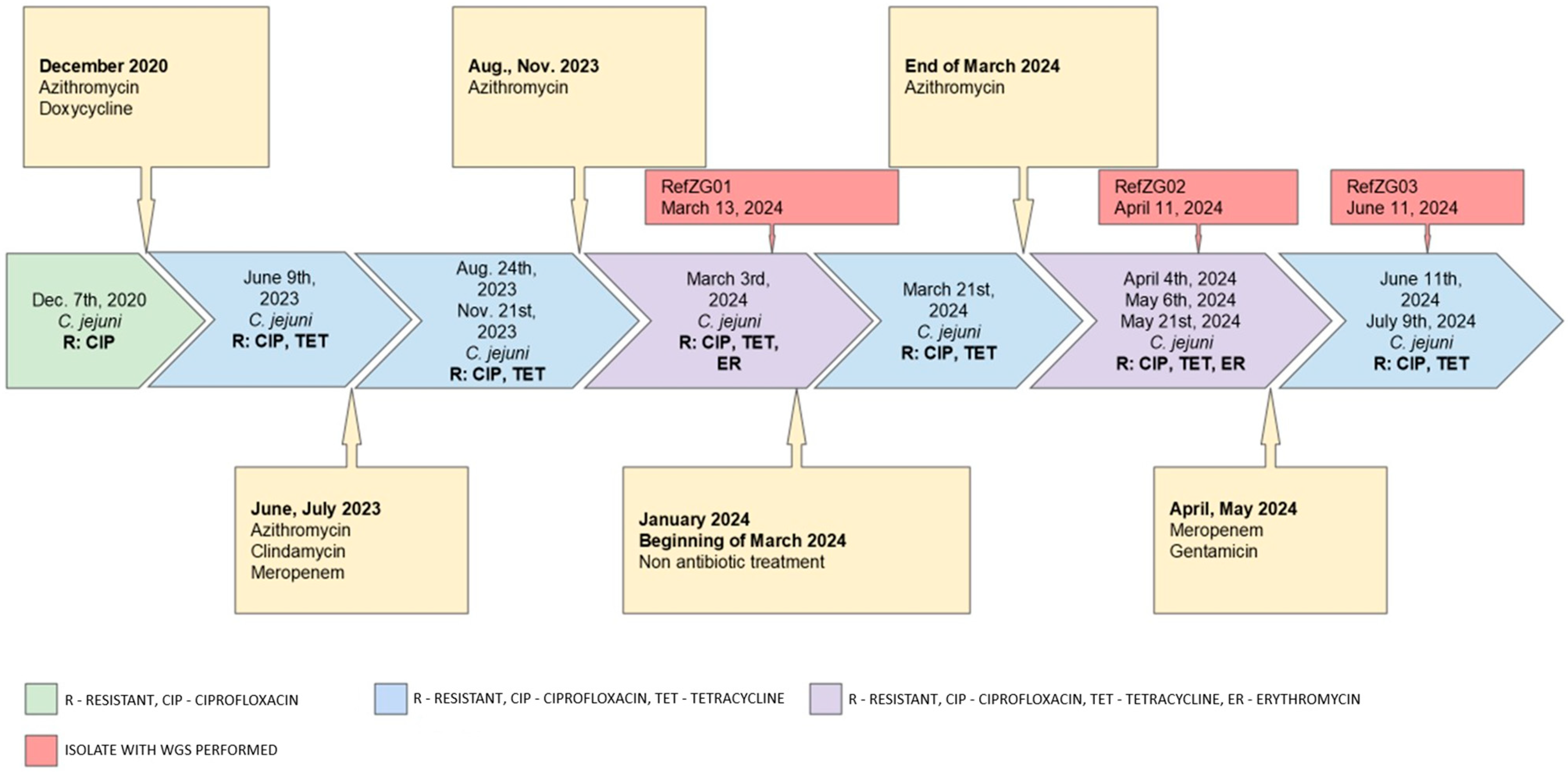
| Date Range | Symptoms | Isolates | Therapy |
|---|---|---|---|
| 2019 | abdominal cramps, diarrhea, bloating and weight loss | C. jejuni negative | oral ciprofloxacin (CIP) (0.5 g twice daily) and subcutaneous IG therapy (80 g/800 mL per month) |
| 01/2020 | worsening GI symptoms (diarrhea, abdominal cramps and fever) | Toxin-producing C. difficile | oral vancomycin (0.125 g four times daily), metronidazole (0.4 g three times daily), corticosteroids (0.024 g/day), and mesalazine (1 g three times daily), along with an increased Ig dose to 100 g/1000 mL per month |
| 12/2020 | worsening GI symptoms | C. jejuni | oral azithromycin (0.5 g once daily) |
| 2021–2022 | intermittent diarrhea | C. jejuni negative | no reported treatment |
| 12/2022 | intermittent diarrhea | Toxin-producing C. difficile | oral vancomycin (0.125 g four times daily) |
| 02/2023 | diarrhea (25 watery stools/day), fever of 38 °C, ICU admission for treatment of malabsorption, electrolyte imbalance, and subsequent pneumonia | C. jejuni negative | oral vancomycin (0.125 g four times daily), levofloxacin for pneumonia |
| 06/2023 | diarrhea, fever | C. jejuni R: CIP 1, TET 2 | oral azithromycin (0.5 g once daily) followed by oral clindamycin (0.3 g four times daily). Hospitalization with fever and treated with intravenous (IV) meropenem (1 g three times daily) |
| 08/2023 | diarrhea | C. jejuni R: CIP, TET | oral azithromycin (0.5 g once daily) |
| 11/2023 | diarrhea | C. jejuni R: CIP, TET | oral azithromycin (0.5 g once daily) |
| 03/2024 | diarrhea with 3–5 s stools per day, abdominal discomfort, and low-grade fever | C. jejuni R: CIP, TET, ER 3 C. jejuni R: CIP, TET | oral azithromycin (0.5 g once daily) |
| 04–05/2024 | persistent diarrhea | C. jejuni R: CIP, TET, ER | supportive therapy only |
| 05/2024 | fever, 15 diarrheal stools daily, and abdominal cramps | C. jejuni R: CIP, TET, ER C. jejuni R: CIP, TET | IV meropenem (1 g three times daily)- discontinued after 10 days due to leukopenia. IV gentamicin (0.24 g once daily) for four days, with prophylactic vancomycin. |
| 06–07/2024 | abdominal pain, spasms, and flatulence | C. jejuni R: CIP, TET | Supportive therapy, nutritional supplementation, probiotics |
| Antimicrobial Agent (Class) | |||||
|---|---|---|---|---|---|
| Isolate/ Ref. Isolate | Erythromycin (Macrolide) | Ciprofloxacin (Fluoroquinolone) | Tetracycline (Tetracycline) | Gentamycin (Aminoglycoside) | Meropenem (Beta-Lactam) |
| KC01 | S | R | S | - | - |
| KC02 | S | R | S | - | - |
| KC03 | S | R | R | - | - |
| KC04 | S | R | R | - | - |
| KC05/RefZG01 | R/R | R/R | R/R | -/WT | - |
| ZG01 | S | R | R | - | - |
| KC06/RefZg02 | R/R | R/R | R/R | -/WT | |
| ZG02 | R | R | R | WT | WT |
| ZG03 | R | R | R | WT | WT |
| ZG04/RefZG03 | S/S | R/R | R/R | - | - |
| ZG05 | S | R | R | - | - |
| ZG06 | S | R | R | - | - |
| ZG07 | S | R | R | - | - |
| RefZG01 vs RefZG02 | RefZG01 vs RefZG03 | RefZG02 vs RefZG03 | |
|---|---|---|---|
| MASH 1 distance | 0.000024 | 0.000024 | 0.000024 |
| ANI 2 (%) | 99.9 | 99.9 | 99.9 |
| DDH 3 (%) | 100 | 99.9 | 99.9 |
| RefZG01 | RefZG02 | RefZG03 | |
|---|---|---|---|
| RefZG01 | 0 | 9 | 9 |
| RefZG02 | 9 | 0 | 3 |
| RefZG03 | 9 | 3 | 0 |
| Isolates | AMR Genes | Point Mutations Associated with AMR |
|---|---|---|
| RefZG01 | blaOXA-461 | blaOXA-61_G-57T |
| tet(O) | gyrA_T86I | |
| cmeA, cmeB, cmeC, cmeR | 23S_A2074C | |
| RefZG02 | blaOXA-461 | blaOXA-61_G-57T |
| tet(O) cmeA, cmeB, cmeC, cmeR | gyrA_T86I | |
| RefZG03 | blaOXA-461 | blaOXA-61_G-57T |
| tet(O) cmeA, cmeB, cmeC, cmeR | gyrA_T86I |
| Isolate | Adherence Related Genes | Invasion Related Genes | Motility Related Genes | Exotoxin Related Genes | Immune Modulation Related Genes |
|---|---|---|---|---|---|
| RefZG01 | cadF, jlpA, pebA, porA, cj1279c | ciaB, ciaC | fla(C, -G, -A), flg(B, -C, -D, -E, -G, -G2, -H, -I, -J, -K, -L, -M, -P, -Q, -R, -S), flh(A, -B, -F, -G), fli(A, -D, -E, -F, -G, -H, -I, -K, -L, -M, -N, -P, -Q, -R, -S, -W, -Y) pse(B, -C, -F) rpoN che(A, -V, -W, -Y) eptC Cj0371, Cj0883c | cdtA, cdtB, cdtC | Cj1135 |
| RefZG02 | cadF, jlpA, pebA, porA, cj1279c | ciaB, ciaC | fla(C, -G, -A), flg(B, -C, -D, -E, -G, -G2, -H, -I, -J, -K, -L, -M, -P, -Q, -R, -S), flh(A, -B, -F, -G), fli(A, -D, -E, -F, -G, -H, -I, -K, -L, -M, -N, -P, -Q, -R, -S, -W, -Y) pse(B, -C, -F) rpoN che(A, -V, -W, -Y) eptC Cj0371, Cj0883c | cdtA, cdtB, cdtC | Cj1135 |
| RefZG03 | cadF, jlpA, pebA, porA, cj1279c | ciaB, ciaC | fla(C, -G, -A), flg(B, -C, -D, -E, -G, -G2, -H, -I, -J, -K, -L, -M, -P, -Q, -R, -S), flh(A, -B, -F, -G), fli(A, -D, -E, -F, -G, -H, -I, -K, -L, -M, -N, -P, -Q, -R, -S, -W, -Y) pse(B, -C, -F) rpoN che(A, -V, -W, -Y) eptC Cj0371, Cj0883c | cdtA, cdtB, cdtC | Cj1135 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juzbašić, T.; Andrijašević, N.; Ferenčak, I.; Jurić, D.; Šoprek, S.; Poje Janeš, V.; Žmak, L.; Tambić Andrašević, A.; Gverić Grginić, A. Emergence of Multidrug-Resistant Campylobacter jejuni in a Common Variable Immunodeficiency Patient: Evolution of Resistance Under the Selective Antibiotic Pressure. Trop. Med. Infect. Dis. 2025, 10, 165. https://doi.org/10.3390/tropicalmed10060165
Juzbašić T, Andrijašević N, Ferenčak I, Jurić D, Šoprek S, Poje Janeš V, Žmak L, Tambić Andrašević A, Gverić Grginić A. Emergence of Multidrug-Resistant Campylobacter jejuni in a Common Variable Immunodeficiency Patient: Evolution of Resistance Under the Selective Antibiotic Pressure. Tropical Medicine and Infectious Disease. 2025; 10(6):165. https://doi.org/10.3390/tropicalmed10060165
Chicago/Turabian StyleJuzbašić, Tajana, Nataša Andrijašević, Ivana Ferenčak, Dragan Jurić, Silvija Šoprek, Vlatka Poje Janeš, Ljiljana Žmak, Arjana Tambić Andrašević, and Ana Gverić Grginić. 2025. "Emergence of Multidrug-Resistant Campylobacter jejuni in a Common Variable Immunodeficiency Patient: Evolution of Resistance Under the Selective Antibiotic Pressure" Tropical Medicine and Infectious Disease 10, no. 6: 165. https://doi.org/10.3390/tropicalmed10060165
APA StyleJuzbašić, T., Andrijašević, N., Ferenčak, I., Jurić, D., Šoprek, S., Poje Janeš, V., Žmak, L., Tambić Andrašević, A., & Gverić Grginić, A. (2025). Emergence of Multidrug-Resistant Campylobacter jejuni in a Common Variable Immunodeficiency Patient: Evolution of Resistance Under the Selective Antibiotic Pressure. Tropical Medicine and Infectious Disease, 10(6), 165. https://doi.org/10.3390/tropicalmed10060165







