Isolation and Characterization of a Novel Bacteriophage KpCCP1, Targeting Multidrug-Resistant (MDR) Klebsiella Strains
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Strains
2.2. Sample Collection and Processing
2.3. Phage Isolation and Propagation
2.4. Transmission Electron Microscopy
2.5. DNA Extraction
2.6. Sequencing and Assembly
2.7. Phylogenetic and Genomic Analysis
2.8. Host Range
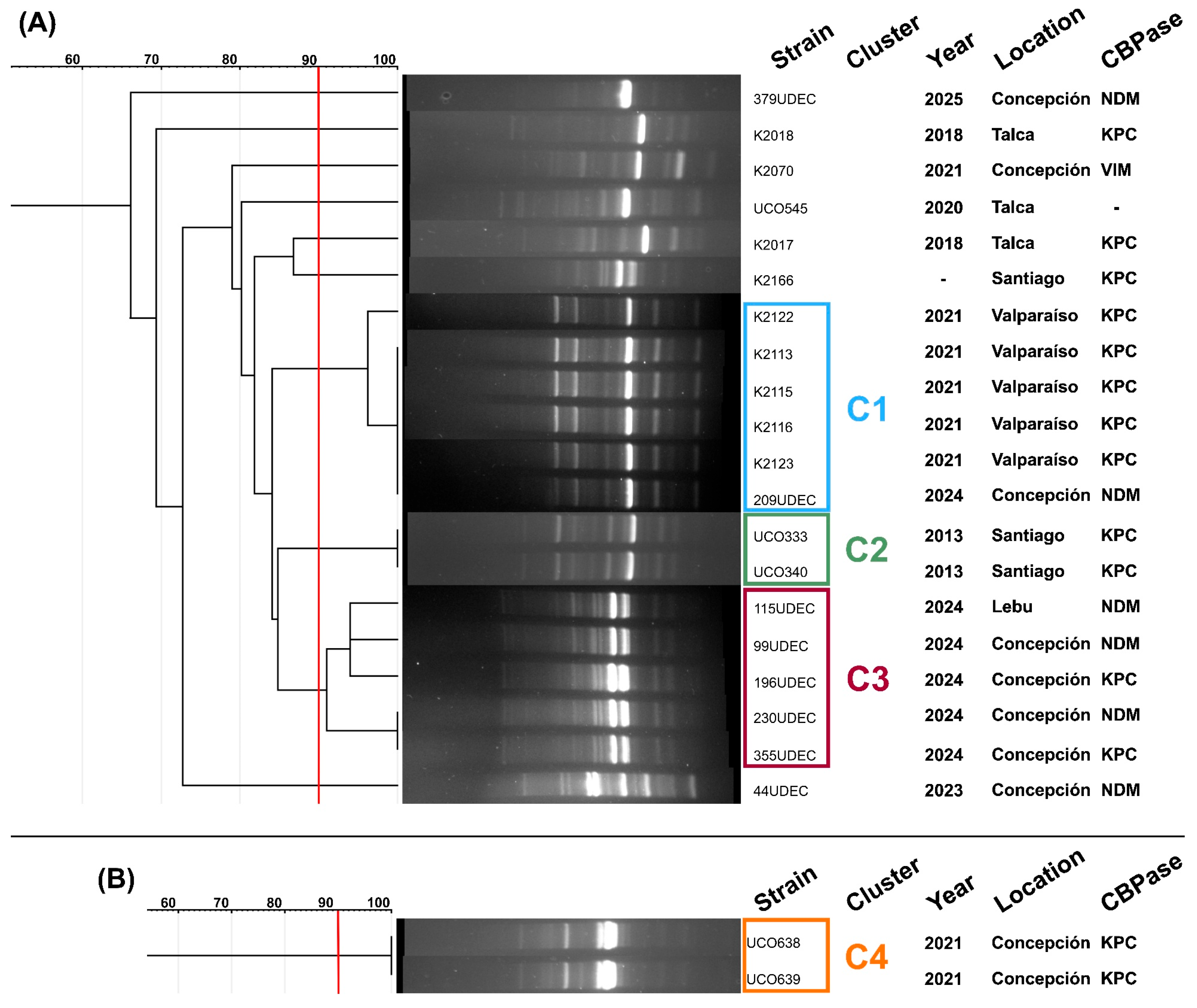
2.9. Carbapenemase-Encoding Genes and Clonality of Phage-Sensitive Strains
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baquero, F.; Martinez, J.L.; FLanza, V.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 2021, 34, e00050-19. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A. Antimicrobial resistance: A growing serious threat for global public health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Allel, K.; Peters, A.; Haghparast-Bidgoli, H.; Spencer-Sandino, M.; Conejeros, J.; Garcia, P.; Pouwels, K.B.; Yakob, L.; Munita, J.M.; Undurraga, E.A. Excess burden of antibiotic-resistant bloodstream infections: Evidence from a multicentre retrospective cohort study in Chile, 2018–2022. Lancet 2024, 40, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Bacterial Priority Pathogens List; World Health Organization: Geneva, Switzerland, 2024; pp. 12–13. [Google Scholar]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef]
- Martin, R.M.; Bachman, M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 2018, 8, 4. [Google Scholar]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Lam, M.M.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Zhou, R.; Fang, X.; Zhang, J.; Zheng, X.; Shangguan, S.; Chen, S.; Shen, Y.; Liu, Z.; Li, J.; Zhang, R.; et al. Impact of carbapenem resistance on mortality in patients infected with Enterobacteriaceae: A systematic review and meta-analysis. BMJ Open 2021, 11, e054971. [Google Scholar] [CrossRef] [PubMed]
- Hovan, M.R.; Narayanan, N.; Cedarbaum, V.; Bhowmick, T.; Kirn, T.J. Comparing mortality in patients with carbapenemase-producing carbapenem resistant Enterobacterales and non-carbapenemase-producing carbapenem resistant Enterobacterales bacteremia. Diagn. Microbiol. Infect. Dis. 2021, 101, 115505. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.S.; Kim, J.H.; Park, J.H.; Kim, T.W.; Jung, H.I.; Kwon, Y.S. Comparison of mortality rates in patients with carbapenem-resistant enterobacterales bacteremia according to carbapenemase production: A multicenter propensity-score matched study. Sci. Rep. 2024, 14, 597. [Google Scholar]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin. Infect. Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef]
- Cifuentes, M.; Garcia, P.; San Martin, P.; Silva, F.; Zuniga, J.; Reyes, S.; Rojas, R.; Ponce, R.; Quintanilla, R.; Delpiano, L.; et al. First isolation of KPC in Chile: From Italy to a public hospital in Santiago. Rev. Chil. Infectol. 2012, 29, 224–228. [Google Scholar] [CrossRef]
- Carrasco-Anabalón, S.; Neto, C.O.C.; Carvalho-Assef, A.P.D.A.; Lima, C.A.; Cifuentes, M.; Silva, F.; Barrera, B.; Domínguez, M.; González-Rocha, G.; Bello-Toledo, H. Introduction of NDM-1 and OXA-370 from Brazil into Chile in strains of Klebsiella pneumoniae isolated from a single patient. Int. J. Infect. Dis. 2019, 81, 28–30. [Google Scholar] [CrossRef]
- Ministerio de Salud. Instituto de Salud Pública de Chile. Boletín de Resistencia Microbiana. Available online: https://www.ispch.cl/wp-content/uploads/2022/09/BoletinRAM_FINAL-1-1.pdf (accessed on 12 May 2024).
- Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lincopan, N.; Esposito, F.; Fuga, B.; Mella-Montecino, S.; Riedel, G.; Lima, C.A.; Bello-Toledo, H.; Cifuentes, M.; et al. Novel megaplasmid driving NDM-1-mediated carbapenem resistance in Klebsiella pneumoniae ST1588 in South America. Antibiotics 2022, 11, 1207. [Google Scholar] [CrossRef]
- Quesille-Villalobos, A.M.; Solar, C.; Martínez, J.R.; Rivas, L.; Quiroz, V.; González, A.M.; Riquelme-Neira, R.; Ugalde, J.A.; Peters, A.; Ortega-Recalde, O.; et al. Multispecies emergence of dual bla KPC/NDM carbapenemase-producing Enterobacterales recovered from invasive infections in Chile. Antimicrob. Agents Chemother. 2025, 69, e01205–e01224. [Google Scholar]
- Gálvez-Silva, M.; Arros, P.; Berríos-Pastén, C.; Villamil, A.; Rodas, P.I.; Araya, I.; Iglesias, R.; Araya, P.; Hormazábal, J.C.; Bohle, C.; et al. Carbapenem-resistant hypervirulent ST23 Klebsiella pneumoniae with a highly transmissible dual-carbapenemase plasmid in Chile. Biol. Res. 2024, 57, 7. [Google Scholar]
- Veloso, M.; Arros, P.; Acosta, J.; Rojas, R.; Berríos-Pastén, C.; Varas, M.; Araya, P.; Hormazábal, J.C.; Allende, M.L.; Chávez, F.P.; et al. Antimicrobial resistance, pathogenic potential, and genomic features of carbapenem-resistant Klebsiella pneumoniae isolated in Chile: High-risk ST25 clones and novel mobile elements. Microbiol. Spectr. 2023, 11, e00399-23. [Google Scholar]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Morales-León, F.; Matus-Köhler, M.; Araya-Vega, P.; Aguilera, F.; Torres, I.; Vera, R.; Ibarra, C.; Venegas, S.; Bello-Toledo, H.; González-Rocha, G.; et al. Molecular characterization of the convergent carbapenem-resistant and hypervirulent Klebsiella pneumoniae strain K1-ST23, collected in Chile during the COVID-19 pandemic. Microbiol. Spectr. 2023, 11, e00540-e23. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, A.R.; Safaee, M.M.; Wuest, W.M.; Furst, A.L. The silent pandemic: Emergent antibiotic resistances following the global response to SARS-CoV-2. iScience 2021, 24, 102304. [Google Scholar] [CrossRef]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front. Pharmacol. 2019, 10, 457104. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Cisek, A.A.; Dąbrowska, I.; Gregorczyk, K.P.; Wyżewski, Z. Phage therapy in bacterial infections treatment: One hundred years after the discovery of bacteriophages. Curr. Microbiol. 2017, 74, 277–283. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, 10–1128. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Wang, J.Y.; Doudna, J.A. CRISPR technology: A decade of genome editing is only the beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef]
- Li, H.Y.; Kao, C.Y.; Lin, W.H.; Zheng, P.X.; Yan, J.J.; Wang, M.C.; Teng, C.H.; Tseng, C.C.; Wu, J.J. Characterization of CRISPR-Cas systems in clinical Klebsiella pneumoniae isolates uncovers its potential association with antibiotic susceptibility. Front. Microbiol. 2018, 9, 1595. [Google Scholar] [CrossRef] [PubMed]
- Mackow, N.A.; Shen, J.; Adnan, M.; Khan, A.S.; Fries, B.C.; Diago-Navarro, E. CRISPR-Cas influences the acquisition of antibiotic resistance in Klebsiella pneumoniae. PLoS ONE 2019, 14, e0225131. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Song, G.; Xu, Y. Association of CRISPR/Cas system with the drug resistance in Klebsiella pneumoniae. Infect. Drug Resist. 2020, 13, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Jwair, N.A.; Al-Ouqaili, M.T.; Al-Marzooq, F. Inverse association between the existence of CRISPR/Cas systems with antibiotic resistance, extended spectrum β-lactamase and carbapenemase production in multidrug, extensive drug and pandrug-resistant Klebsiella pneumoniae. Antibiotics 2023, 12, 980. [Google Scholar] [CrossRef]
- Pinilla-Redondo, R.; Shehreen, S.; Marino, N.D.; Fagerlund, R.D.; Brown, C.M.; Sørensen, S.J.; Fineran, P.C.; Bondy-Denomy, J. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat. Commun. 2020, 11, 5652. [Google Scholar] [CrossRef]
- Wang, C.; Sun, Z.; Hu, Y.; Li, D.; Guo, Q.; Wang, M. A novel anti-CRISPR AcrIE9. 2 is associated with dissemination of bla KPC plasmids in Klebsiella pneumoniae sequence type 15. Antimicrob. Agents Chemother. 2023, 67, e01547-e22. [Google Scholar] [CrossRef]
- Jiang, C.; Yu, C.; Sun, S.; Lin, J.; Cai, M.; Wei, Z.; Feng, L.; Li, J.; Zhang, Y.; Dong, K.; et al. A new anti-CRISPR gene promotes the spread of drug-resistance plasmids in Klebsiella pneumoniae. Nucleic Acids Res. 2024, 52, 8370–8384. [Google Scholar] [CrossRef]
- Jiang, J.; Cienfuegos-Galletd, A.V.; Long, T.; Peirano, G.; Chu, T.; Pitout, J.D.; Kreiswirth, B.N.; Chen, L. Intricate interplay of CRISPR-Cas systems, anti-CRISPR proteins, and antimicrobial resistance genes in a globally successful multi-drug resistant Klebsiella pneumoniae clone. Genome Med. 2025, 17, 9. [Google Scholar] [CrossRef]
- Herridge, W.P.; Shibu, P.; O’Shea, J.; Brook, T.C.; Hoyles, L. Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J. Med. Microbiol. 2020, 69, 176–194. [Google Scholar]
- Li, N.; Zeng, Y.; Bao, R.; Zhu, T.; Tan, D.; Hu, B. Isolation and characterization of novel phages targeting pathogenic Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2021, 11, 792305. [Google Scholar] [CrossRef] [PubMed]
- Zurabov, F.; Zhilenkov, E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol. J. 2021, 18, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ballesté, E.; Blanch, A.R.; Muniesa, M.; García-Aljaro, C.; Rodríguez-Rubio, L.; Martín-Díaz, J.; Pascual-Benito, M.; Jofre, J. Bacteriophages in sewage: Abundance, roles, and applications. FEMS Microbes 2022, 3, xtac009. [Google Scholar] [CrossRef] [PubMed]
- Parra, B.; Sandoval, M.; Arriagada, V.; Amsteins, L.; Aguayo, C.; Opazo-Capurro, A.; Dechesne, A.; González-Rocha, G. Isolation and Characterization of Lytic Bacteriophages Capable of Infecting Diverse Multidrug-Resistant Strains of Pseudomonas aeruginosa: PaCCP1 and PaCCP2. Pharmaceuticals 2024, 17, 1616. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. In Bacteriophages: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2009; Volume 1, pp. 69–76. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 December 2024).
- Shen, A.; Millard, A. Phage genome annotation: Where to begin and end. Phage 2021, 2, 183–193. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Tolstoy, I.; Kropinski, A.M. Phage annotation guide: Guidelines for assembly and high-quality annotation. Phage 2021, 2, 170–182. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Millard, A.; Denise, R.; Lestido, M.; Thomas, M.T.; Webster, D.; Turner, D.; Sicheritz-Pontén, T. taxmyPHAGE: Automated taxonomy of dsDNA phage genomes at the genus and species level. Phage 2025, 6, 5–11. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Brister, J.R. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- McNair, K.; Zhou, C.; Dinsdale, E.A.; Souza, B.; Edwards, R.A. PHANOTATE: A novel approach to gene identification in phage genomes. Bioinformatics 2019, 35, 4537–4542. [Google Scholar] [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Pharokka: A fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.; Rasche, H.; Maughmer, C.; Criscione, A.; Mijalis, E.; Liu, M.; Hu, J.C.; Young, R.; Gill, J.J. Galaxy and Apollo as a biologist-friendly interface for high-quality cooperative phage genome annotation. PLoS Comput. Biol. 2020, 16, e1008214. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Shang, J.; Tang, X.; Sun, Y. PhaTYP: Predicting the lifestyle for bacteriophages using BERT. Brief. Bioinform. 2023, 24, bbac487. [Google Scholar] [CrossRef]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI-bacteriophage life cycle recognition with machine learning and natural language processing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Rajandas, H.; Parimannan, S.; Manickam, R.; Marimuthu, K.; Petersen, B.; Clokie, M.R.; Millard, A.; Sicheritz-Pontén, T. PhageLeads: Rapid assessment of phage therapeutic suitability using an ensemble machine learning approach. Viruses 2022, 14, 342. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dai, W.; Li, J.; Xie, R.; Dunstan, R.A.; Stubenrauch, C.; Zhang, Y.; Lithgow, T. PaCRISPR: A server for predicting and visualizing anti-CRISPR proteins. Nucleic Acids Res. 2020, 48, W348–W357. [Google Scholar] [CrossRef] [PubMed]
- Eitzinger, S.; Asif, A.; Watters, K.E.; Iavarone, A.T.; Knott, G.J.; Doudna, J.A.; Minhas, F.U.A.A. Machine learning predicts new anti-CRISPR proteins. Nucleic Acids Res. 2020, 48, 4698–4708. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 34th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2024. [Google Scholar]
- Pires, J.; Novais, A.; Peixe, L. Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J. Clin. Microbiol. 2013, 51, 4281–4283. [Google Scholar] [CrossRef]
- Vila, J.; Marcos, M.A.; Jimenez de Anta, M.T. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J. Med. Microbiol. 1996, 44, 482–489. [Google Scholar] [CrossRef]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 1–8. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Z.; Tian, C.; Chen, X.; Hu, L.; Wei, X.; Li, H.; Lin, W.; Jiang, A.; Feng, R.; et al. Characterizing the biology of lytic bacteriophage vB_EaeM_φEap-3 infecting multidrug-resistant Enterobacter aerogenes. Front. Microbiol. 2019, 10, 420. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; Van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Amann, R.I.; Bardy, P.; Bartlau, N.; Barylski, J.; Błażejak, S.; Bouzari, M.; Briegel, A.; Briers, Y.; et al. Summary of taxonomy changes ratified by the International Committee on Taxonomy of Viruses (ICTV) from the Bacterial Viruses Subcommittee, 2025. J. Gen. Virol. 2025, 106, 002111. [Google Scholar] [CrossRef]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Łusiak-Szelachowska, M.; Żaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 1–12. [Google Scholar] [CrossRef]
- Provasek, V.E.; Lessor, L.E.; Cahill, J.L.; Rasche, E.S.; Kuty Everett, G.F. Complete genome sequence of carbapenemase-producing Klebsiella pneumoniae myophage Matisse. Genome Announc. 2015, 3, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Koberg, S.; Brinks, E.; Fiedler, G.; Hüsing, C.; Cho, G.S.; Hoeppner, M.P.; Heller, K.J.; Neve, H.; Franz, C.M. Genome sequence of Klebsiella pneumoniae bacteriophage PMBT1 isolated from raw sewage. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Moreirinha, C.; Lewicka, M.; Almeida, P.; Clemente, C.; Romalde, J.L.; Nunes, M.L.; Almeida, A. Characterization and in vitro evaluation of new bacteriophages for the biocontrol of Escherichia coli. Virus Res. 2017, 227, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Wesevich, A.; Sutton, G.; Ruffin, F.; Park, L.P.; Fouts, D.E.; Fowler, V.G., Jr.; Thaden, J.T. Newly named Klebsiella aerogenes (formerly Enterobacter aerogenes) is associated with poor clinical outcomes relative to other Enterobacter species in patients with bloodstream infection. J. Clin. Microbiol. 2020, 58, 1–9. [Google Scholar] [CrossRef]
- Maciejewska, B.; Roszniowski, B.; Espaillat, A.; Kęsik-Szeloch, A.; Majkowska-Skrobek, G.; Kropinski, A.M.; Briers, Y.; Cava, F.; Lavigne, R.; Drulis-Kawa, Z. Klebsiella phages representing a novel clade of viruses with an unknown DNA modification and biotechnologically interesting enzymes. Appl. Microbiol. Biotechnol. 2017, 101, 673–684. [Google Scholar] [CrossRef]
- Yoo, S.; Lee, K.M.; Kim, N.; Vu, T.N.; Abadie, R.; Yong, D. Designing phage cocktails to combat the emergence of bacteriophage-resistant mutants in multidrug-resistant Klebsiella pneumoniae. Microbiol. Spectr. 2024, 12, e01258-23. [Google Scholar] [CrossRef]
- Duarte, J.; Máximo, C.; Costa, P.; Oliveira, V.; Gomes, N.C.; Romalde, J.L.; Pereira, C.; Almeida, A. Potential of an isolated bacteriophage to inactivate Klebsiella pneumoniae: Preliminary studies to control urinary tract infections. Antibiotics 2024, 13, 195. [Google Scholar] [CrossRef]
- Ichikawa, M.; Nakamoto, N.; Kredo-Russo, S.; Weinstock, E.; Weiner, I.N.; Khabra, E.; Ben-Ishai, N.; Inbar, D.; Kowalsman, N.; Mordoch, R.; et al. Bacteriophage therapy against pathological Klebsiella pneumoniae ameliorates the course of primary sclerosing cholangitis. Nat. Commun. 2023, 14, 3261. [Google Scholar] [CrossRef]
- Yang, J.; Long, H.; Hu, Y.; Feng, Y.; McNally, A.; Zong, Z. Klebsiella oxytoca complex: Update on taxonomy, antimicrobial resistance, and virulence. Clin. Microbiol. Rev. 2022, 35, e00006-21. [Google Scholar] [CrossRef]
- Townsend, E.M.; Kelly, L.; Gannon, L.; Muscatt, G.; Dunstan, R.; Michniewski, S.; Sapkota, H.; Kiljunen, S.J.; Kolsi, A.; Skurnik, M.; et al. Isolation and characterization of Klebsiella phages for phage therapy. Ther. Appl. Res. 2021, 2, 26–42. [Google Scholar] [CrossRef]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef]
- Lourenço, M.; Osbelt, L.; Passet, V.; Gravey, F.; Megrian, D.; Strowig, T.; Rodrigues, C.; Brisse, S. Phages against noncapsulated Klebsiella pneumoniae: Broader host range, slower resistance. Microbiol. Spectr. 2023, 11, e04812-22. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Bondy-Denomy, J.; Cheung, V.H.; Maxwell, K.L.; Davidson, A.R. A new group of phage anti-CRISPR genes inhibits the type IE CRISPR-Cas system of Pseudomonas aeruginosa. mBio 2014, 5, e00896. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.J.C.M.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Fang, M.; Liu, X.; Zhang, C.; Liu, Y.; Yuan, Y. Isolation and characterization of a novel phage for controlling multidrug-resistant Klebsiella pneumoniae. Microorganisms 2020, 8, 542. [Google Scholar] [CrossRef]
- El-Demerdash, A.S.; Alfaraj, R.; Farid, F.A.; Yassin, M.H.; Saleh, A.M.; Dawwam, G.E. Essential oils as capsule disruptors: Enhancing antibiotic efficacy against multidrug-resistant Klebsiella pneumoniae. Front. Microbiol. 2024, 15, 1467460. [Google Scholar] [CrossRef]
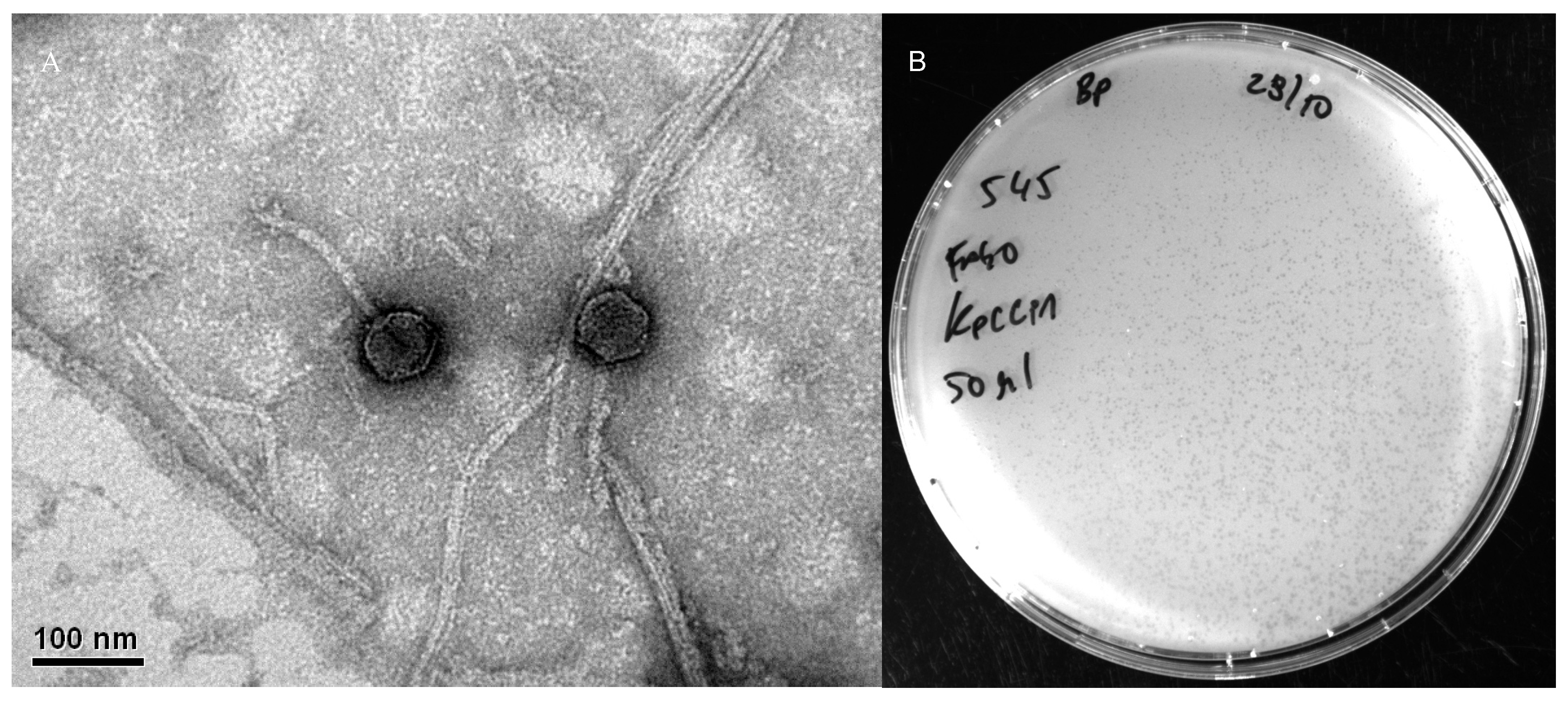
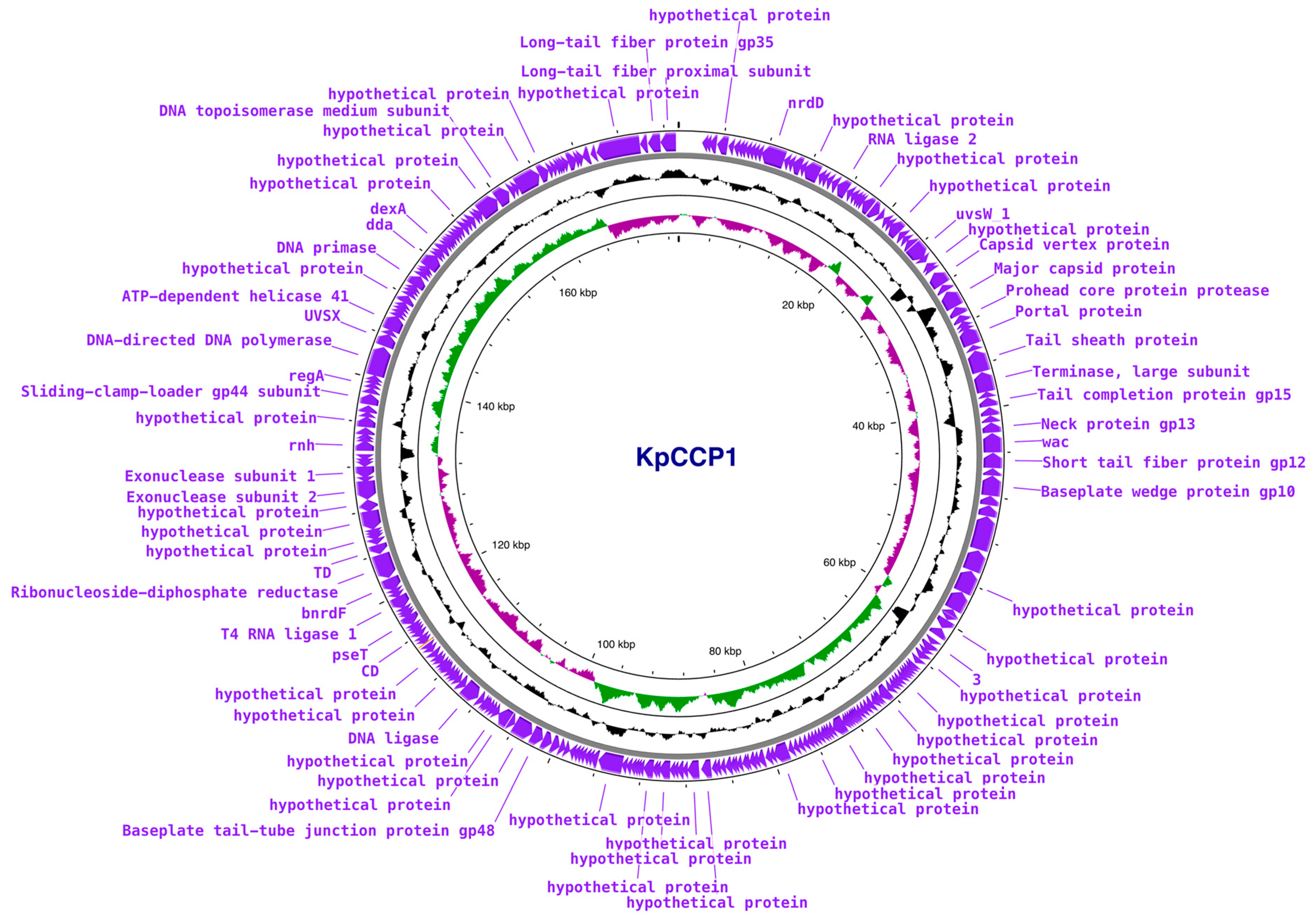
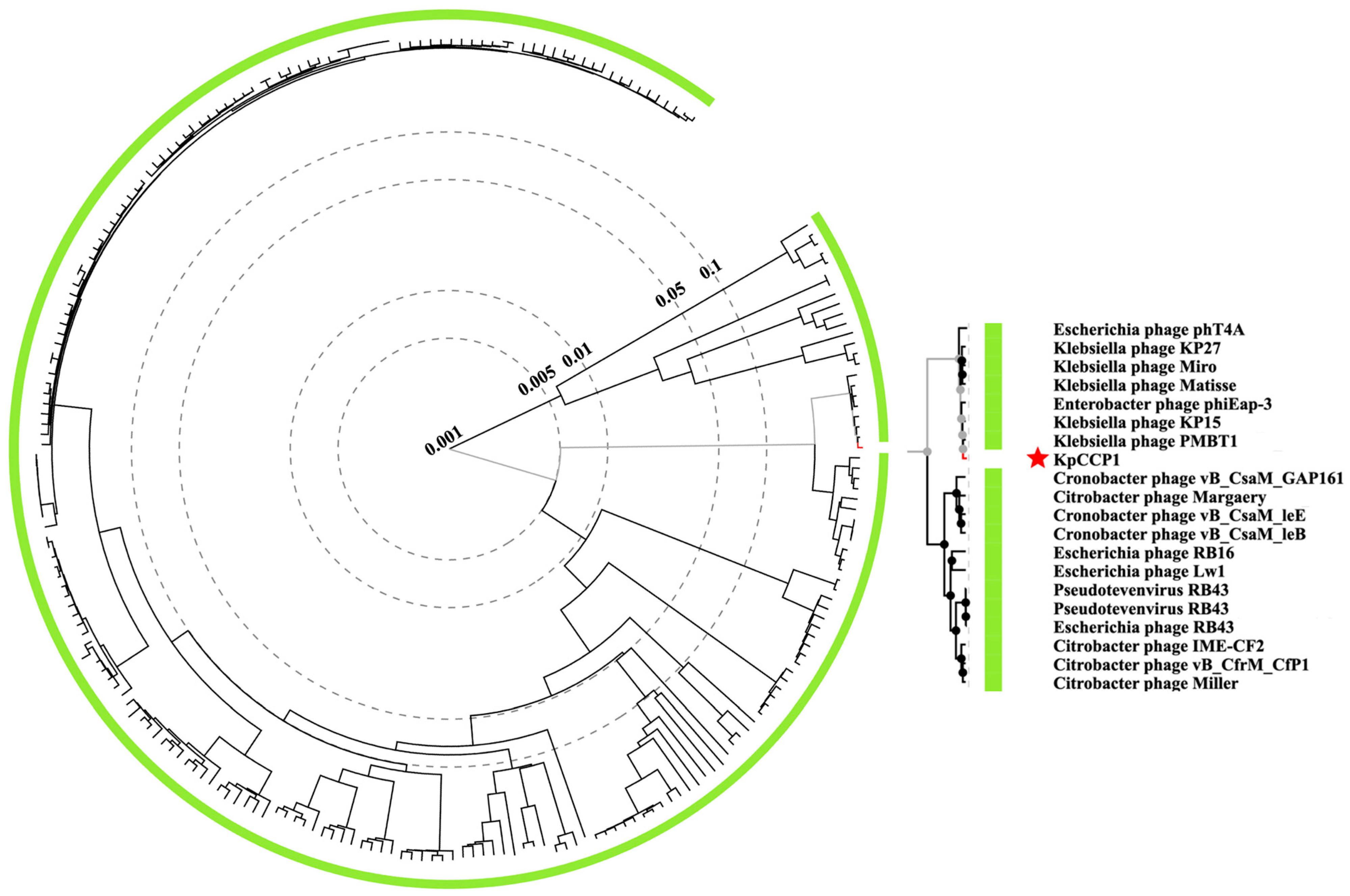
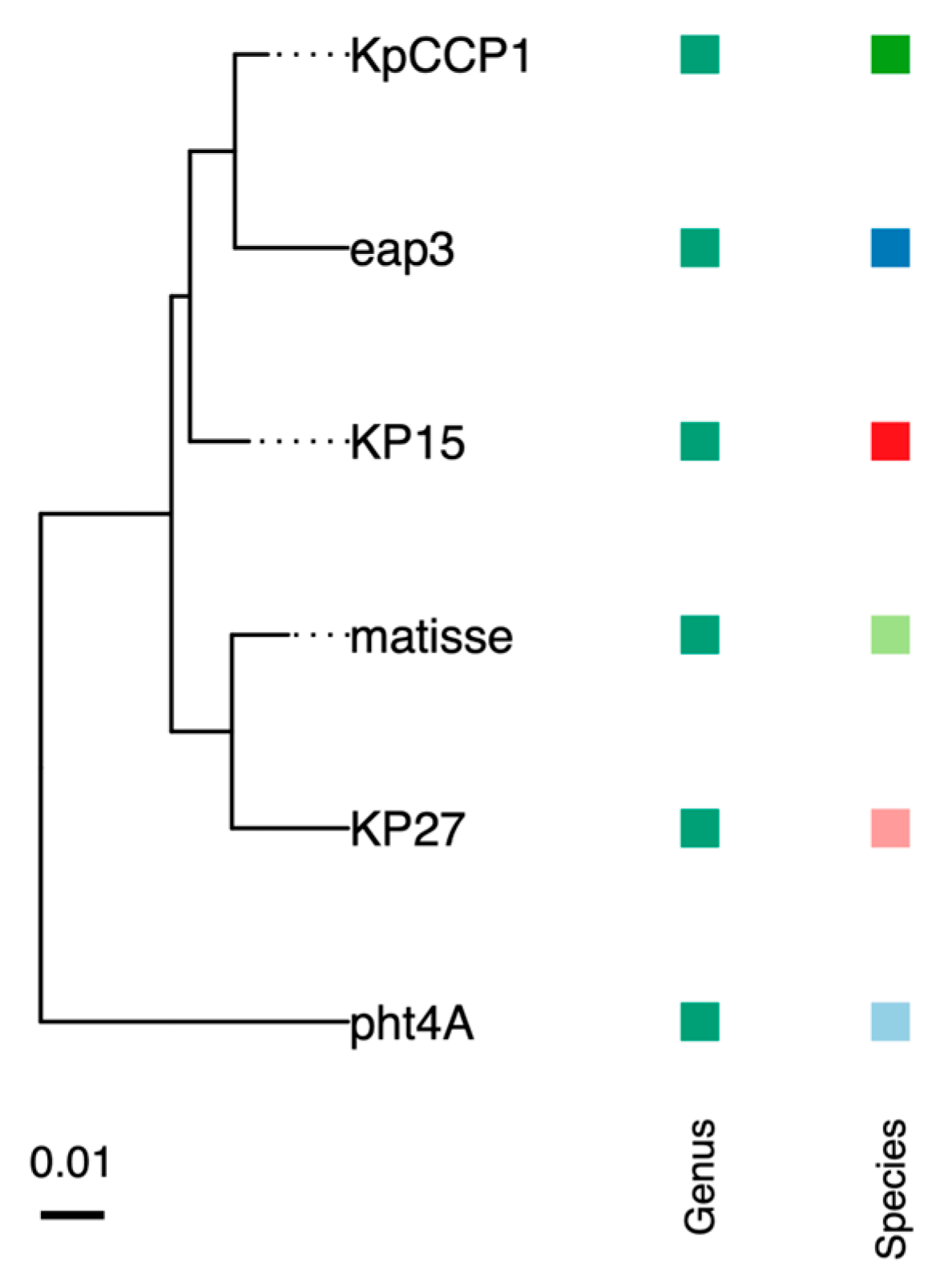

| Phage | IS with KpCCP1 | Species Cluster | Genome Size (bp) | GC Content | No of CDSs | Host | Accession no. |
|---|---|---|---|---|---|---|---|
| KpCCP1 | -- | 1 | 177,276 | 41.8 | 292 | K. pneumoniae | PV402122 |
| Eap3 | 96.0 | 1 | 175,814 | 42 | 278 | K. aerogenes | NC_041980 |
| KP15 | 95.9 | 2 | 174,436 | 42 | 258 | K. pneumoniae | NC_014036 |
| Matisse | 94.8 | 3 | 176,081 | 42 | 280 | K. pneumoniae | NC_028750 |
| KP27 | 93.0 | 3 | 174,413 | 42 | 276 | K. pneumoniae | NC_020080 |
| pht4A | 91 | 4 | 171,598 | 41.5 | 257 | Escherichia coli | NC_055712 |
| CDS | Length (AA) | Sequence |
|---|---|---|
| 160 | 95 | MITIVYWEEVESEMIDGELAEVDGESEMLTATETIENAFDRIKAHQKEMEGRDIQYMMSFWVGGEALCHAVIAESTSVDTCREKIEQYVKSFLH |
| 104 | 66 | MNMYEMLKSENSETQGEKDYNKWKRSVQHMLGSSDYDEDLMYALYSDGCTPEDAVCEYYAQGDEE |
| 205 | 58 | MQIKITKTFFRDITEGKVYTAKKDKEGLWIENDWNRDIWLNLTNEYNKELIEYEIVK |
| Klebsiella spp. Strain | Antibiotic Resistance Profile * |
|---|---|
| K2017 | IMP, MEM, ETP, CRO, CTX, ATM, AMK, SXT, CIP |
| K2018 | IMP, MEM, CTX, ATM, SXT, CIP |
| K2070 | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, ATM, GEN, AMK, CIP |
| K2113 | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, ATM, GEN, SXT, CIP |
| K2115 | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, GEN, SXT, CIP |
| K2116 | MEM, CRO, CAZ, CTX, CZA, SXT, CIP |
| K2122 | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, GEN, SXT, CIP |
| K2123 | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, GEN, SXT, CIP |
| K2166 | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, GEN, AMK, CIP |
| 44UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, GEN, AMK |
| 99UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, ATM, GEN, AMK, SXT, CIP |
| 115UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, ATM, GEN, AMK, SXT, CIP |
| 196UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, AMK, CIP |
| 209UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, ATM, GEN, AMK, SXT, CIP |
| 230UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, ATM, GEN, AMK, CIP |
| 355UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, AMK |
| 379UDEC | IMP, MEM, ETP, CRO, CAZ, CTX, CZA, AMK |
| UCO333 | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, GEN, AMK, CIP |
| UCO340 | IMP, MEM, ETP, CRO, CAZ, CTX, ATM, GEN, AMK, CIP |
| UCO545 | ETP, CRO, CAZ, CTX, ATM, AMK, SXT, CIP |
| UCO638 | CRO, CTX, SXT |
| UCO639 | CRO, CTX, SXT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra, B.; Matus-Köhler, M.; Cerda-Leal, F.; Suárez-Villota, E.Y.; Hepp, M.I.; Opazo-Capurro, A.; González-Rocha, G. Isolation and Characterization of a Novel Bacteriophage KpCCP1, Targeting Multidrug-Resistant (MDR) Klebsiella Strains. Sci 2025, 7, 157. https://doi.org/10.3390/sci7040157
Parra B, Matus-Köhler M, Cerda-Leal F, Suárez-Villota EY, Hepp MI, Opazo-Capurro A, González-Rocha G. Isolation and Characterization of a Novel Bacteriophage KpCCP1, Targeting Multidrug-Resistant (MDR) Klebsiella Strains. Sci. 2025; 7(4):157. https://doi.org/10.3390/sci7040157
Chicago/Turabian StyleParra, Boris, Maximiliano Matus-Köhler, Fabiola Cerda-Leal, Elkin Y. Suárez-Villota, Matias I. Hepp, Andrés Opazo-Capurro, and Gerardo González-Rocha. 2025. "Isolation and Characterization of a Novel Bacteriophage KpCCP1, Targeting Multidrug-Resistant (MDR) Klebsiella Strains" Sci 7, no. 4: 157. https://doi.org/10.3390/sci7040157
APA StyleParra, B., Matus-Köhler, M., Cerda-Leal, F., Suárez-Villota, E. Y., Hepp, M. I., Opazo-Capurro, A., & González-Rocha, G. (2025). Isolation and Characterization of a Novel Bacteriophage KpCCP1, Targeting Multidrug-Resistant (MDR) Klebsiella Strains. Sci, 7(4), 157. https://doi.org/10.3390/sci7040157







