Development and Application of DNA-Based Tools to Authenticate Marketed Salvia officinalis Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample DNA Preparation and Analysis
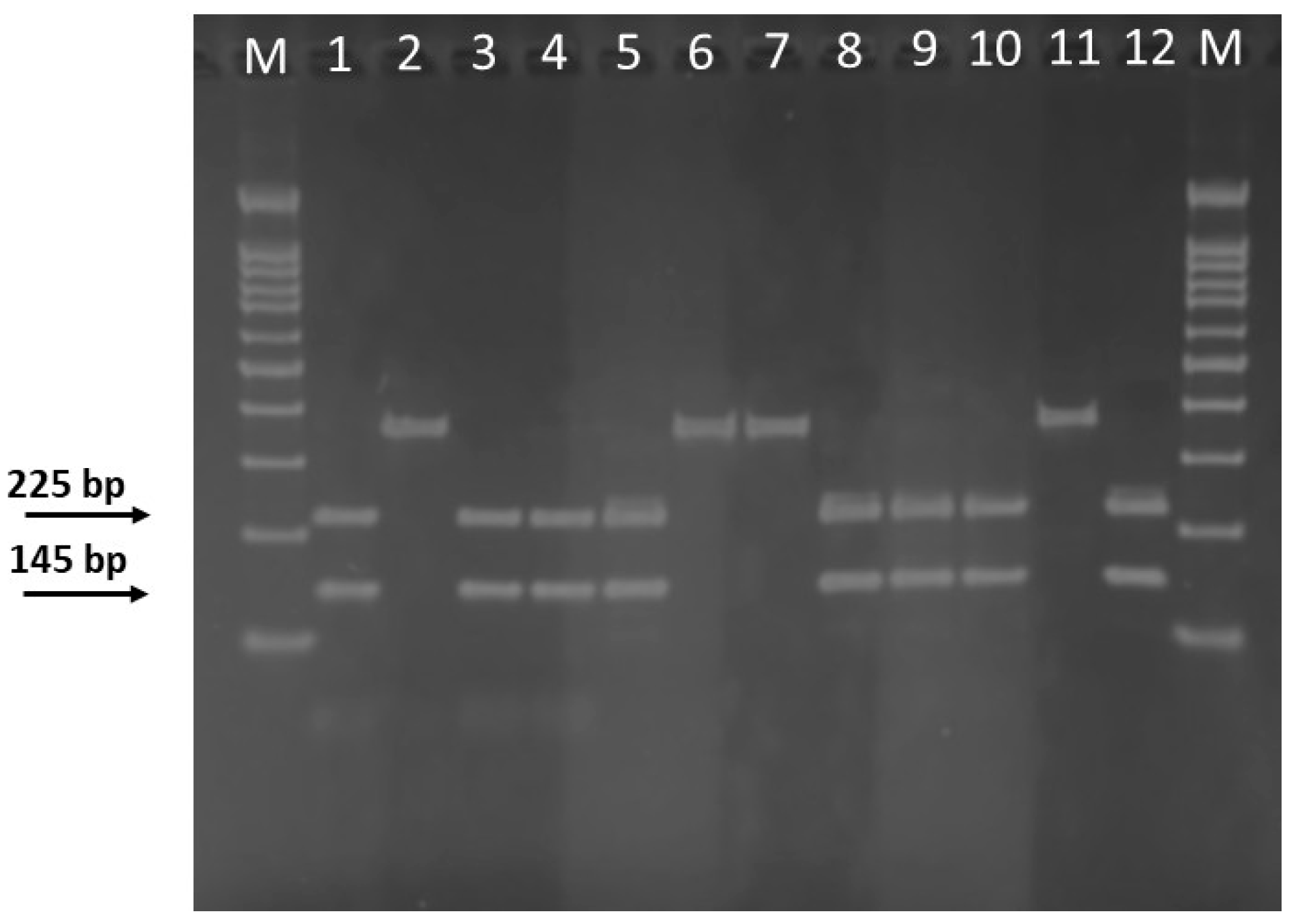
2.3. PCR-RFLP Assay
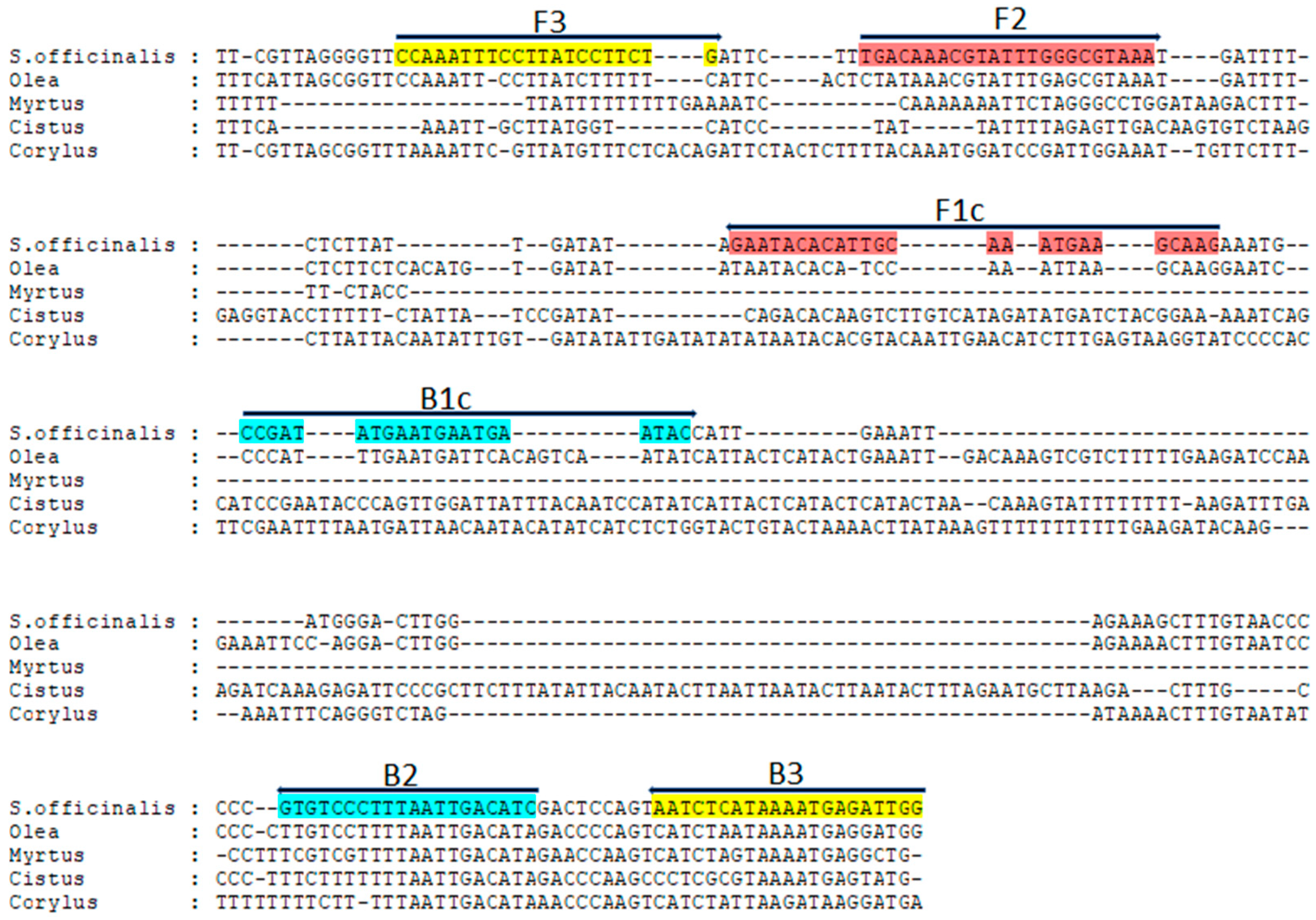
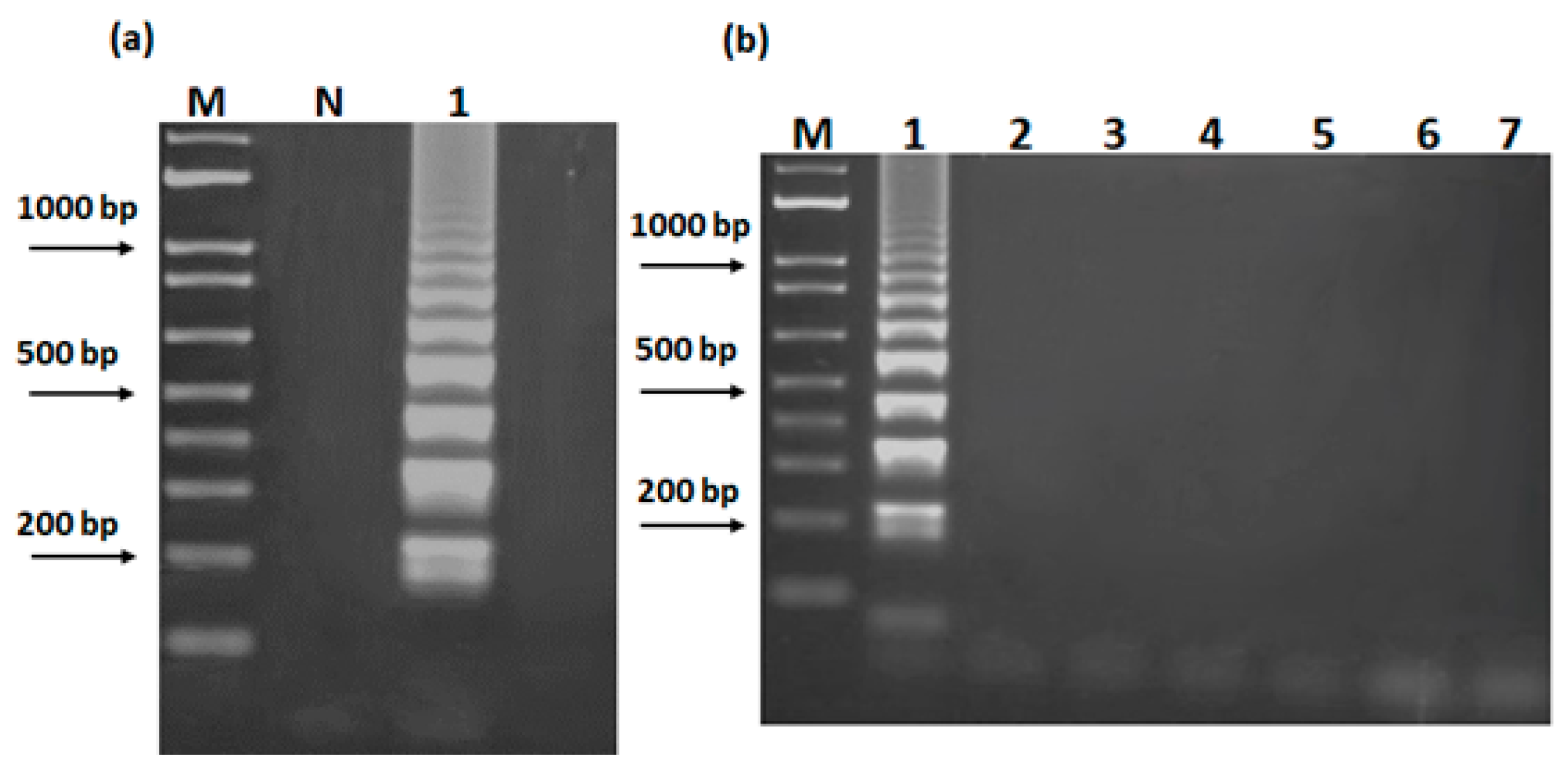
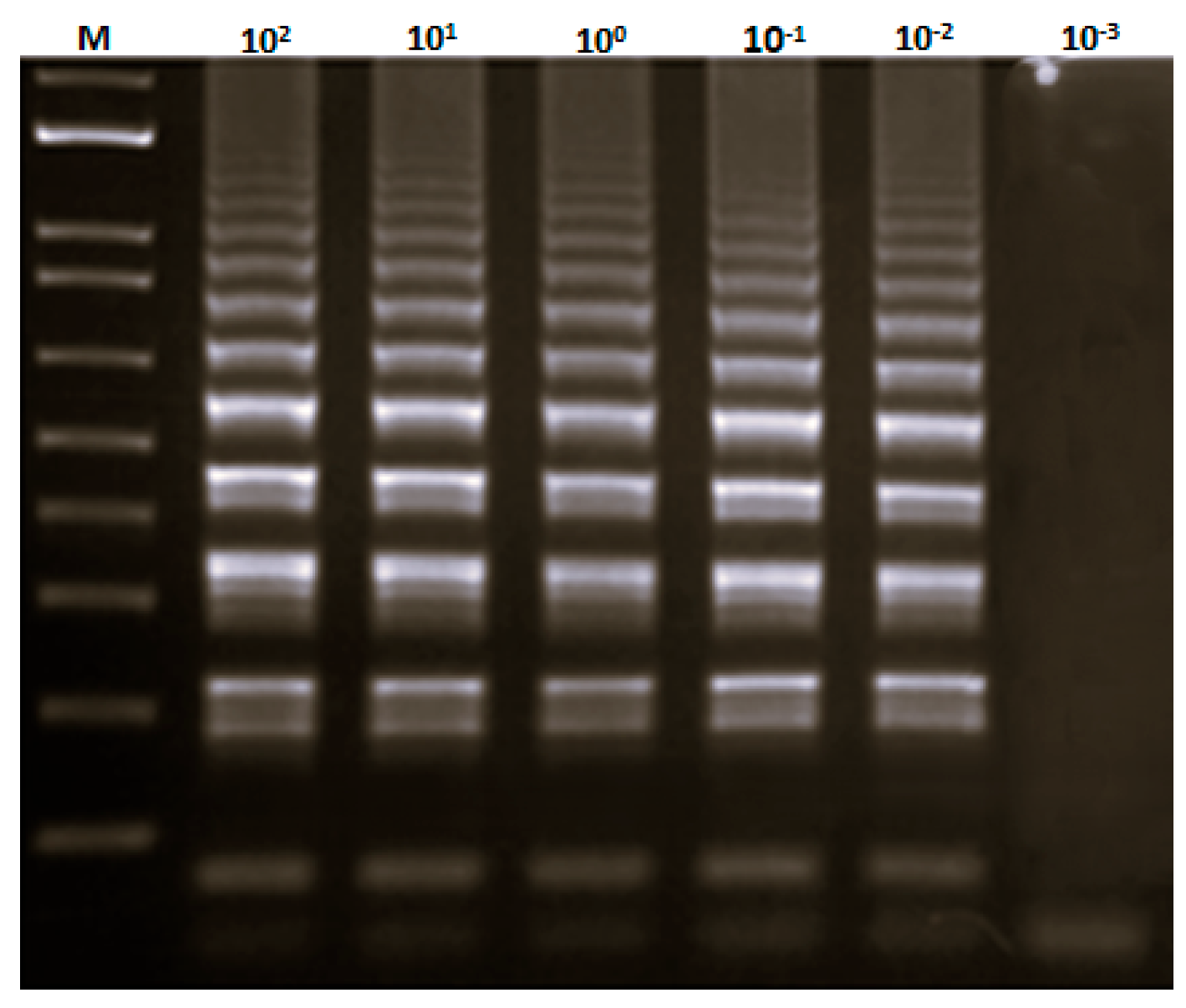
2.4. LAMP Primers and Reaction
3. Results and Discussion
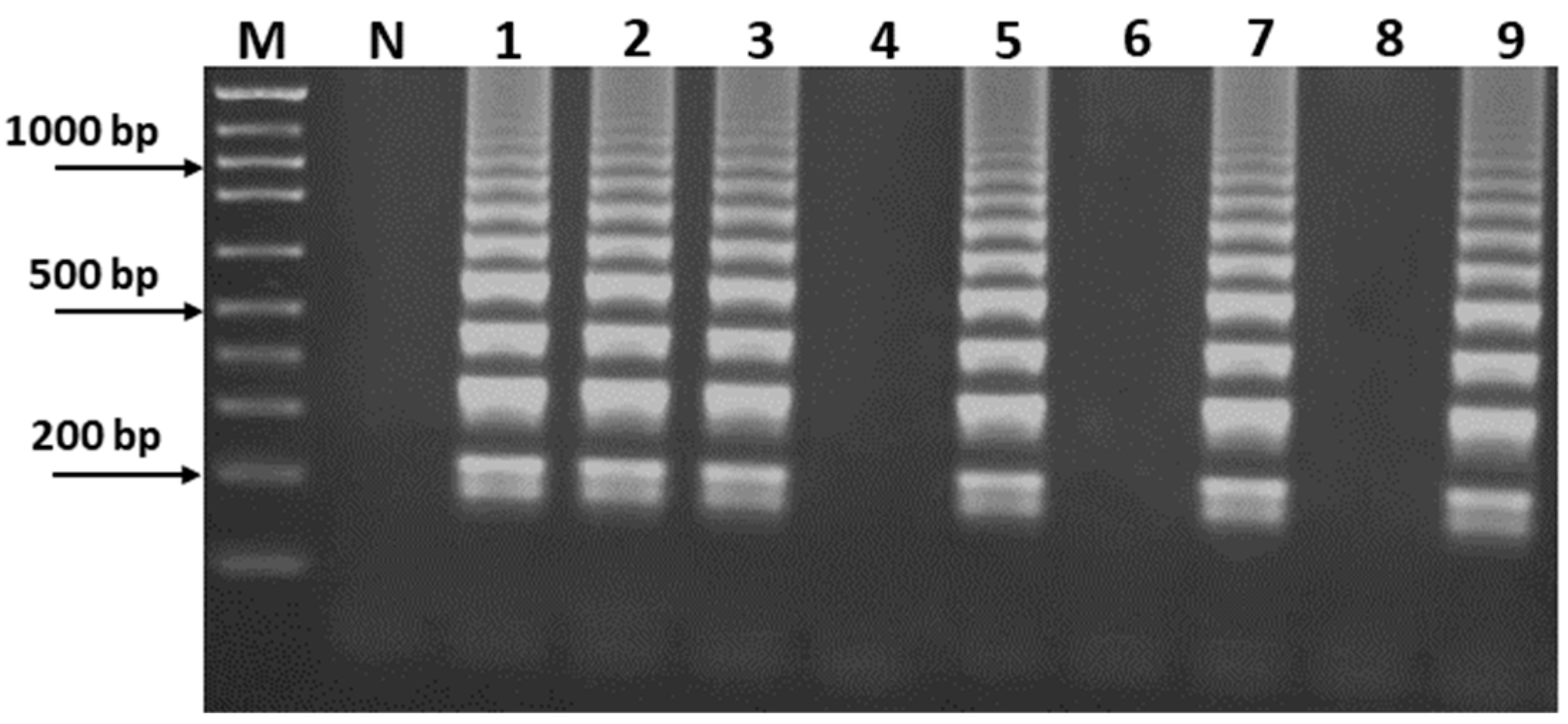
3.1. PCR-RFLP Analysis for S. officinalis Authentication
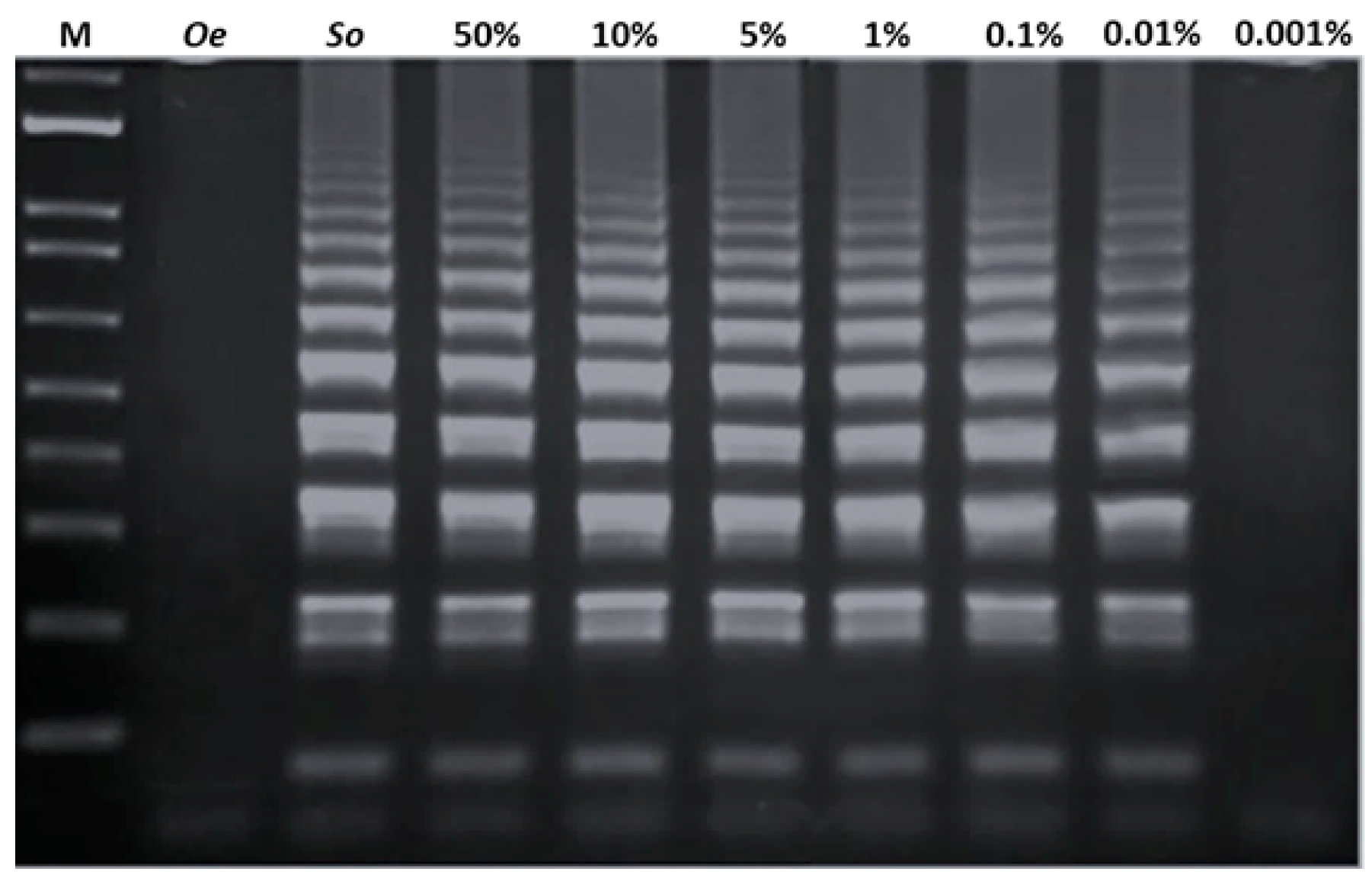
3.2. Development of LAMP Assay for the Identification of S. officinalis DNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jakovljević, M.; Jokić, S.; Molnar, M.; Jašić, M.; Babić, J.; Jukić, H.; Banjari, I. Bioactive profile of various Salvia officinalis L. preparations. Plants 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Kintzios, S.E. Sage: The Genus Salvia; Harwood Academic Publishers: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Ghorbani, A.; Esmaeilizadeh, M. Pharmacological properties of Salvia officinalis and its components. J. Tradit. Complement. Med. 2017, 7, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Topçu, G. Bioactive triterpenoids from Salvia species. J. Nat. Prod. 2006, 69, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Akacha, B.B.; Kačániová, M.; Mekinić, I.G.; Kukula-Koch, W.; Koch, W.; Orhan, I.E.; Čmiková, N.; Taglieri, I.; Venturi, F.; Samartin, C.; et al. Sage (Salvia officinalis L.): A botanical marvel with versatile pharmacological properties and sustainable applications in functional foods. S. Afr. J. Botany 2024, 169, 361–382. [Google Scholar] [CrossRef]
- Black, C.; Haughey, S.A.; Chevallier, O.P.; Galvin-King, P.; Elliott, C.T. A comprehensive strategy to detect the fraudulent adulteration of herbs: The oregano approach. Food Chem. 2016, 210, 551–557. [Google Scholar] [CrossRef]
- Bononi, M.; Tateo, F. LC-ESI-MS/MS identification of oleuropein as marker of Olea europaea L. leaves used as a bulking agent in ground oregano and sage. Ital. J. Food Sci. 2011, 23, 245–251. [Google Scholar]
- Tomčić, N.; Jankov, M.; Ristivojevic, P.; Trifkovic, J.; Andrić, F. Assessment of adulteration of sage (Salvia sp.) with olive leaves using high-performance thin-layer chromatography, image analysis, and multivariate linear modeling. J. Chemom. 2024, 38, e3533. [Google Scholar] [CrossRef]
- Velázquez, R.; Rodríguez, A.; Hernández, A.; Casquete, R.; Benito, M.J.; Martín, A. Spice and herb frauds: Types, incidence, and detection: The state of the art. Foods 2023, 12, 3373. [Google Scholar] [CrossRef]
- Avula, B.; Bae, J.Y.; Chittiboyina, A.G.; Wang, Y.H.; Wang, M.; Srivedavyasasri, R.; Ali, Z.; Li, J.; Wu, C.; Khan, I.A. Comparative analysis of five Salvia species using LC-DAD-QToF. J. Pharm. Biomed. Anal. 2022, 209, 114520. [Google Scholar] [CrossRef]
- Lee, J.; Wang, M.; Zhao, J.; Avula, B.; Chittiboyina, A.G.; Li, J.; Wu, C.; Khan, I.A. Chemical authentication and speciation of Salvia botanicals: An investigation utilizing GC/Q-TOF and chemometrics. Foods 2022, 11, 2132. [Google Scholar] [CrossRef]
- de Aguiar, T.R.; Dorocz, E.L.; do Santos, L.D.; Tanamati, A.A.C.; Gozzo, A.M.; Bona, E. Mid-infrared spectroscopy and chemometrics in the detection of adulteration in chia oil (Salvia hispanica L) and α-linolenic acid content prediction. Food Control 2024, 165, 110687. [Google Scholar] [CrossRef]
- Braglia, L.; Casabianca, V.; De Benedetti, L.; Pecchioni, N.; Mercuri, A.; Cervelli, C.; Ruffoni, B. Amplified fragment length polymorphism markers for DNA fingerprinting in the genus Salvia. Plant Biosyst. 2011, 145, 274–277. [Google Scholar] [CrossRef]
- Sunar, S.; Korkmaz, M.; SiĞmaz, B.; AĞar, G. Determination of the genetic relationships among Salvia species by RAPD and ISSR analyses. Turk. J. Pharm. Sci. 2020, 17, 480–485. [Google Scholar] [CrossRef]
- Bahadirli, N.P.; Ayanoglu, F. Genetic diversity of Salvia species from Turkey assessed by microsatellite markers. J. Appl. Res. Med. Aromat. Plants 2021, 20, 100281. [Google Scholar] [CrossRef]
- Ibrahim, R.I.H.; Sakamoto, M.; Azuma, J.I. PCR-RFLP and genetic diversity analysis of cpDNA in some species of the genus Salvia L. Chromosome Bot. 2012, 7, 1–8. [Google Scholar] [CrossRef][Green Version]
- Bielecka, M.; Pencakowski, B.; Stafiniak, M.; Jakubowski, K.; Rahimmalek, M.; Gharibi, S.; Matkowski, A.; Ślusarczyk, S. Metabolomics and DNA-based authentication of two traditional Asian medicinal and aromatic species of Salvia subg. Perovskia. Cells 2021, 10, 112. [Google Scholar] [CrossRef]
- Lo, Y.-T.; Shaw, P.-C. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018, 240, 767–774. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Sakaridis, I.; Argiriou, A.; Tsaftaris, A. Advances of DNA-based methods for tracing the botanical origin of food products. Food Res. Int. 2014, 60, 163–172. [Google Scholar] [CrossRef]
- Zeng, L.; Wen, J.; Fan, S.; Chen, Z.; Xu, Y.; Sun, Y.; Chen, D.; Zhao, J.; Xu, L.; Li, Y. Identification of sea cucumber species in processed food products by PCR-RFLP method. Food Control 2018, 90, 166–171. [Google Scholar] [CrossRef]
- Griffiths, A.M.; Sotelo, C.G.; Mendes, R.; Pérez-Martín, R.I.; Schröder, U.; Shorten, M.; Silva, H.A.; Verrez-Bagnis, V.; Mariani, S. Current methods for seafood authenticity testing in Europe: Is there a need for harmonisation? Food Control 2014, 45, 95–100. [Google Scholar] [CrossRef]
- Wu, K.; Liu, Y.; Yang, B.; Kung, Y.; Chang, K.; Lee, M. Rapid discrimination of the native medicinal plant Adenostemma lavenia from its adulterants using PCR-RFLP. Peer J. 2022, 10, e13924. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.C.; Lee, M.S.; Sun, F.C.; Chao, H.H.; Chang, W.T.; Lin, M.K.; Chen, H.J.; Lee, M.S. Rapid identification of the indigenous medicinal crop Cinnamomum osmophloeum from various adulterant Cinnamomum species by DNA polymorphism analysis. Pharmacogn. Mag. 2020, 16, 64–68. [Google Scholar]
- Spaniolas, S.; May, S.T.; Bennett, M.J.; Tucker, G.A. Authentication of coffee by means of PCR-RFLP analysis and lab-on-a-chip capillary electrophoresis. J. Agric. Food Chem. 2006, 54, 7466–7470. [Google Scholar] [CrossRef] [PubMed]
- Souframanien, J.; Joshi, A.; Gopalakrishna, T. Intraspecific variation in the internal transcribed region of rDNA in black gram (Vigna mungo (L.) Hepper). Curr. Sci. 2003, 85, 798–802. [Google Scholar]
- Kim, O.T.; Bang, K.H.; In, D.S.; Lee, J.W.; Kim, Y.C.; Shin, Y.S.; Hyun, D.Y.; Lee, S.S.; Cha, S.W.; Seong, N.S. Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5.8S rDNA sequences. Plant Biotechnol. Rep. 2007, 1, 163–167. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.W.; Sung, J.S.; Bang, K.H.; Moon, S.G. Molecular authentication of 21 Korean Artemisia species (Compositae) by polymerase chain reaction-restriction fragment length polymorphism based on trnL-F region of chloroplast DNA. Biol. Pharm. Bull. 2009, 32, 1912–1916. [Google Scholar] [CrossRef]
- Murphy, T.M.; Bola, G. DNA identification of Salvia divinorum samples. Forensic Sci. Int. Genet. 2013, 7, 189–193. [Google Scholar] [CrossRef]
- Lee, S.C.; Wang, C.H.; Yen, C.E.; Chang, C. DNA barcode and identification of the varieties and provenances of Taiwan’s domestic and imported made teas using ribosomal internal transcribed spacer 2 sequences. J. Food Drug Anal. 2016, 25, 260–274. [Google Scholar] [CrossRef]
- Ng, A.E.; Sandoval, E.; Murphy, T.M. Identification and individualization of Lophophora using DNA analysis of the trnL/trnF region and rbcL gene. J. Forensic Sci. 2016, 61, S226–S229. [Google Scholar] [CrossRef]
- Sen, F.; Uncu, A.O.; Uncu, A.T.; Erdeger, S.N. The trnL (UAA)-trnF (GAA) intergenic spacer is a robust marker of green pea (Pisum sativum L.) adulteration in economically valuable pistachio nuts (Pistacia vera L.). J. Sci. Food Agric. 2020, 100, 3056–3061. [Google Scholar] [CrossRef]
- Kitamura, M.; Kazato, A.; Yamamuro, T.; Ando, H.; Sasaki, Y.; Suzuki, R.; Shirataki, Y. Rapid identification of Aconitum plants based on loop-mediated isothermal amplification assay. BMC Res. Notes 2019, 12, 408. [Google Scholar] [CrossRef] [PubMed]
- Focke, F.; Haase, I.; Fischer, M. Loop-mediated isothermal amplification (LAMP): Methods for plant species identification in food. J. Agric. Food Chem. 2013, 61, 2943–2949. [Google Scholar] [CrossRef] [PubMed]
- Holz, N.; Illarionov, B.; Wax, N.; Schmidt, C.; Fischer, M. Point-of-care suitable identification of the adulterants Carthamus tinctorius and Curcuma longa in Crocus sativus based on loop-mediated isothermal amplification (LAMP) and lateral-flow-assay (LFA). Food Control 2023, 148, 109637. [Google Scholar] [CrossRef]
- Holz, N.; Wax, N.; Illarionov, B.A.; Iskhakova, M.; Fischer, M. Food Authentication: The detection of Arbutus unedo and Olea europaea leaves as an admixture of oregano using LAMP- and Duplex LAMP-based test systems with Lateral-Flow Assays. Agriculture 2024, 14, 597. [Google Scholar] [CrossRef]
- Niessen, L.; Bechtner, J.; Fodil, S.; Taniwaki, M.H.; Vogel, R.F. LAMP-based group specific detection of aflatoxin producers within Aspergillus section Flavi in food raw materials, spices, and dried fruit using neutral red for visible-light signal detection. Int. J. Food Microbiol. 2018, 266, 241–250. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Mori, Y.; Kitao, M.; Tomita, N.; Notomi, T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 2004, 59, 145–157. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B., Jr.; Deerfield, D.W. GeneDoc: Analysis and visualization of genetic variation. Embnew News 1997, 4, 14. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, K.; Mage, P.L.; Csordas, A.T.; Eisenstein, M.; Soh, H.T. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem. Commun. 2014, 50, 3747–3749. [Google Scholar] [CrossRef]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 2015, 58, 59–68. [Google Scholar] [CrossRef]
| S. officinalis Samples | Province Manufacturing | ||
|---|---|---|---|
| Dried Leaves | Teas | Infusions | |
| So1 | So11 | So21 | Torino, Piemonte, Italy |
| So2 | So12 | So22 | Torino, Piemonte, Italy |
| So3 | So13 | So23 | Verona, Veneto, Italy |
| So4 | So14 | So24 | Verona, Veneto, Italy |
| So5 | So15 | So25 | Verona, Veneto, Italy |
| So6 | So16 | So26 | Roma, Lazio, Italy |
| So7 | So17 | So27 | Roma, Lazio, Italy |
| So8 | So18 | So28 | Roma, Lazio, Italy |
| So9 | So19 | So29 | Cosenza, Calabria, Italy |
| So10 | So20 | So30 | Cosenza, Calabria, Italy |
| Primer | Length (bp) | Sequence (5′–3′) |
|---|---|---|
| F3 | 21 | CCAAATTTCCTTATCCTTCTG |
| B3 | 21 | CCAATCTCATTTTATGAGATT |
| FIP (F2 + F1c) | 48 | TGACAAACGTATTTGGGCGTAAA-CTTGCTTCATTTGCAATGTGTATTC |
| BIP (B2 + B1c) | 41 | GATGTCAATTAAAGGGACAC- CCGATATGAATGAATGAATAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regina, T.M.R.; Calabrese, E. Development and Application of DNA-Based Tools to Authenticate Marketed Salvia officinalis Products. Sci 2025, 7, 70. https://doi.org/10.3390/sci7020070
Regina TMR, Calabrese E. Development and Application of DNA-Based Tools to Authenticate Marketed Salvia officinalis Products. Sci. 2025; 7(2):70. https://doi.org/10.3390/sci7020070
Chicago/Turabian StyleRegina, Teresa Maria Rosaria, and Elisa Calabrese. 2025. "Development and Application of DNA-Based Tools to Authenticate Marketed Salvia officinalis Products" Sci 7, no. 2: 70. https://doi.org/10.3390/sci7020070
APA StyleRegina, T. M. R., & Calabrese, E. (2025). Development and Application of DNA-Based Tools to Authenticate Marketed Salvia officinalis Products. Sci, 7(2), 70. https://doi.org/10.3390/sci7020070





