Molecules to Microbes
Abstract
1. Introduction
2. To Set the Scene
3. The Origin of Life?
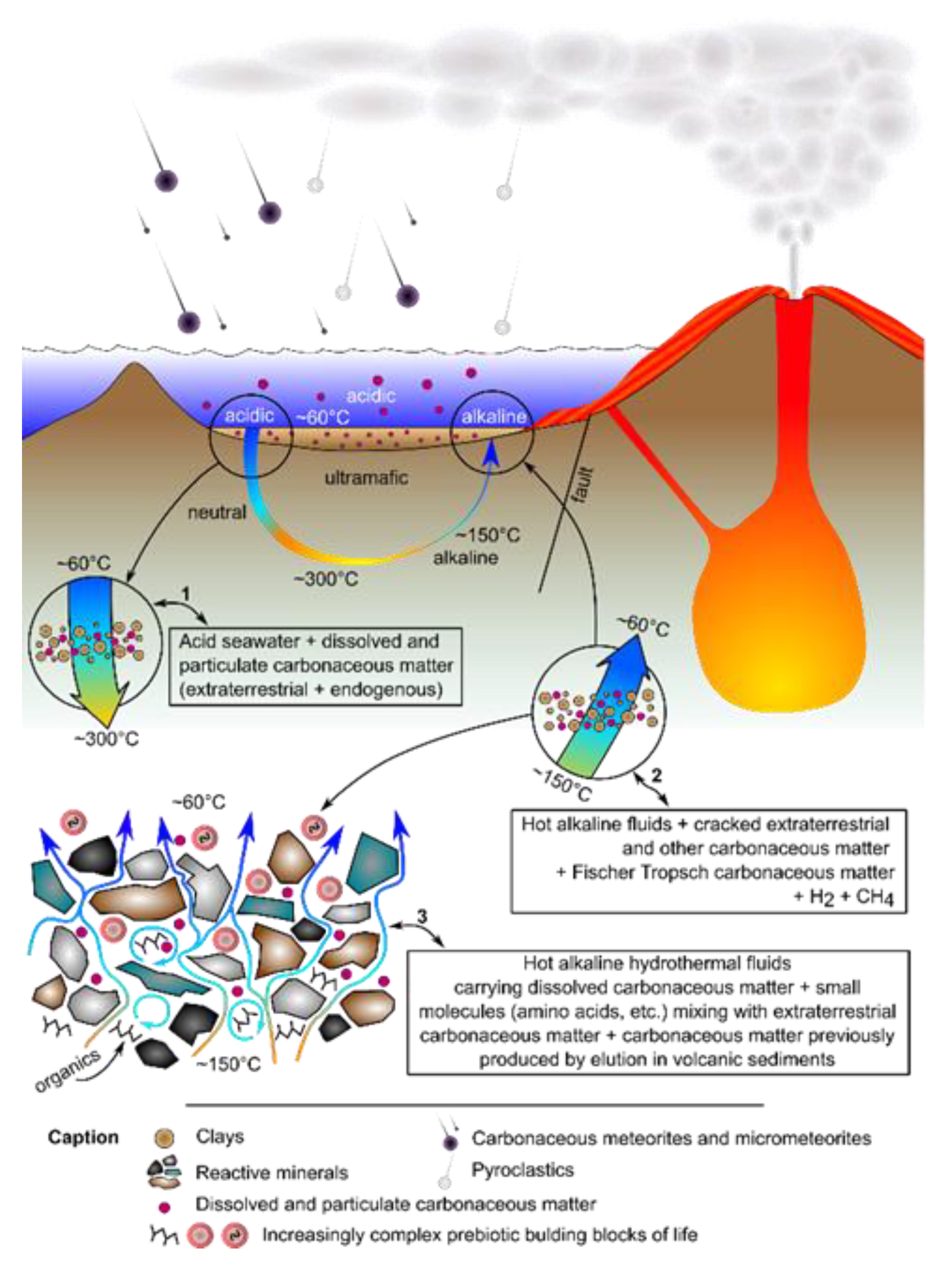
3.1. Metabolism First
3.2. Genetics First
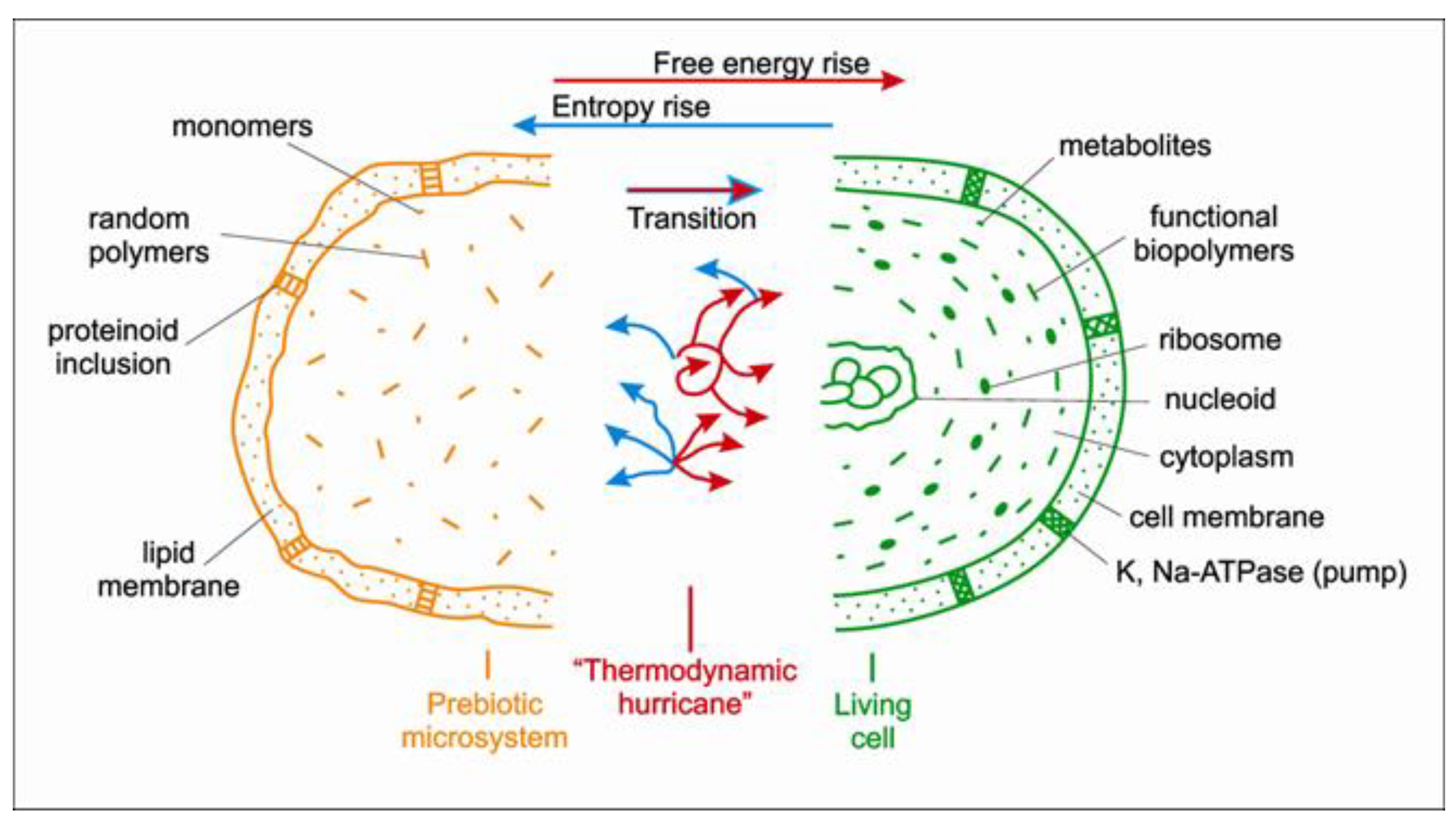
3.3. Vesicles First
4. Gene Transfer
5. Virus System
6. Bio-Signals
7. Space—The Final Frontiers
8. What’s Next?
Funding
Acknowledgments
Conflicts of Interest
References
- Capova, K.A.; Persson, E.; Milligan, T.; Dunér, D. Society, Worldview and Outreach. In Astrobiology and Society in Europe Today; Springer: Cambridge, UK, 2018; pp. 19–24. [Google Scholar]
- Bandyopadhyay, P.S.; Raghavan, R.V.; Dcruz, D.W.; Brittan, G. Truths about Simpson’s Paradox: Saving the paradox from falsity. In ICLA 2015: Logic and Its Applications; Lecture Notes in Computer Science; Banerjee, M., Krishna, S.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 8923. [Google Scholar]
- Bandyopadhyay, P.S.; Beard, T.E.; Greenwood, M.C.; Bertasso, M.P.; Peters, J.W. Why Need a Model? The Debate over the Origin of Life Theories and a Lesson from Simpson’s Paradox. In Proceedings of the Epistemology of Modeling and Simulation Conference, Pittsburgh, PA, USA, 1–3 April 2011; University of Pittsburgh: Pittsburgh, PA, USA. [Google Scholar]
- Westall, F.; Hickman-Lewis, K.; Hinman, N.; Gautret, P.; Campbell, K.A.; Bréhéret, J.G.; Foucher, F.; Hubert, A.; Sorieul, S.; Dass, A.V.; et al. A Hydrothermal-Sedimentary Context for the Origin of Life. Astrobiology 2018, 18, 259–293. [Google Scholar] [CrossRef] [PubMed]
- Kompanichenko, V.N. Thermodynamic Inversion: Origin of Living Systems; Springer: Cham, Switzerland, 2017; 275p. [Google Scholar]
- Kompanichenko, V. The Rise of a Habitable Planet: Four Required Conditions for the Origin of Life in the Universe. Geosciences 2019, 9, 92. [Google Scholar] [CrossRef]
- Ball, R.; Brindley, J. The Power Without the Glory: Multiple Roles of Hydrogen Peroxide in Mediating the Origin of Life. Astrobiology 2019, 19, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Ball, R.; Brindley, J. Toy trains, loaded dice and the origin of life: Dimerization on mineral surfaces under periodic drive with Gaussian inputs. R. Soc. Open Sci. 2017, 4, 170141. [Google Scholar] [CrossRef] [PubMed]
- Ball, R.; Brindley, J. The Life Story of Hydrogen Peroxide III: Chirality and Physical Effects at the Dawn of Life. Orig. Life Evol. Biosph. 2016, 46, 81–93. [Google Scholar] [CrossRef]
- Hansma, H.G. Better than Membranes at the Origin of Life? Life 2017, 7, 28. [Google Scholar] [CrossRef]
- Hansma, H.G. The Power of Crowding for the Origins of Life. Orig. Life Evol. Biosph. 2014, 44, 307–311. [Google Scholar] [CrossRef]
- Jheeta, S. The Landscape of the Emergence of Life. Life 2017, 7, 27. [Google Scholar] [CrossRef]
- Hansma, H.G. Possible origin of life between mica sheets. J. Theor. Biol. 2010, 266, 175–188. [Google Scholar] [CrossRef]
- Cech, T.R. Self-splicing of group I introns. Annu. Rev. Biochem. 1990, 59, 543–568. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.J.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H.A. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [PubMed]
- Altman, S. Structural biology. Nature 2000, 7, 827–828. [Google Scholar]
- Cech, T.R.; Zaug, A.J.; Grabowski, P.J. In Vitro splicing of the ribosomal RNA precursor of Tetrahymena: Involvement of a guanosine nucleotides in the excision of the intervening sequence. Cell 1981, 27, 487–496. [Google Scholar] [CrossRef]
- Prosdocimia, F.; Jheeta, S.; Farias, S.T. Conceptual challenges for the emergence of the biological system: Cell theory and self-replication. Med. Hypotheses 2018, 119, 79–83. [Google Scholar] [CrossRef]
- Hordijk, W.; Steel, M. Chasing the tail: The emergence of autocatalytic networks. Biosystems 2017, 152, 1–10. [Google Scholar] [CrossRef]
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef]
- Ameta, S.; Arsene, S.; Lehman, N.; Griffiths, A.D.; Nghe, P. Evolution Using Autocatalytic Sets of RNA, Manuscript under preparation.
- Ameta, S.; Arsene, S.; Lehman, N.; Nghe, P.; Griffiths, A.D. (Conference Abstract) Autocatalytic sets of RNA replicators in origin of life. In Proceedings of the XVIIIth International Conference on the Origin of Life, San Diego, CA, USA, 16–21 July 2017; Volume 1967. [Google Scholar]
- Arsène, S.; Ameta, S.; Lehman, N.; Griffiths, A.D.; Nghe, P. Coupled catabolism and anabolism in autocatalytic RNA sets. Nucleic Acids Res. 2018, 46, 9660–9666. [Google Scholar] [CrossRef]
- Iqubal, M.A.; Sharma, R.; Jheeta, S. Thermal condensation of glycine and alanine on metal ferrite surface: Primitive peptide bond formation scenario. Life 2017, 7, 15. [Google Scholar] [CrossRef]
- Sharma, R.; Iqubal, M.A.; Jheeta, S. Adsorption and Oxidation of Aromatic Amines on Metal(II) Hexacyanocobaltate(III) Complexes: Implication for Oligomerization of Exotic Aromatic Compounds. Inorganics 2017, 5, 18. [Google Scholar] [CrossRef]
- Kotakis, C. Non-coding RNAs’ partitioning in the evolution of photosynthetic organisms via energy transduction and redox signalling. RNA Biol. 2015, 12, 101–104. [Google Scholar] [CrossRef]
- Smith, D.R. RNA-Seq data: A goldmine for organelle research. Brief. Funct. Genom. 2013, 12, 454–456. [Google Scholar] [CrossRef] [PubMed]
- Kahana, A.; Lancet, D. Protobiotic Systems Chemistry Analyzed by Molecular Dynamics. Life 2019, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Lancet, D.; Zidovetzki, R.; Markovitch, O. Systems protobiology: Origin of life in lipid catalytic networks. J. R. Soc. Interface 2018, 15, 20180159. [Google Scholar] [CrossRef] [PubMed]
- Schopf, J.W.; Kitajima, K.; Spicuzza, M.J.; Kudryavtsev, A.B.; Valley, J.W. SIMS analyses of the oldest known assemblage of microfossils document their taxon-correlated carbon isotope compositions. Proc. Natl. Acad. Sci. USA 2018, 115, 53–58. [Google Scholar] [CrossRef]
- Jheeta, S. The Routes of Emergence of Life from LUCA during the RNA and Viral World: A Conspectus. Life 2015, 5, 1445–1453. [Google Scholar] [CrossRef]
- Koumandou, V.L.; Kossida, S. Evolution of the F0F1 ATP Synthase Complex in Light of the Patchy Distribution of Different Bioenergetic Pathways across Prokaryotes. PLoS Comput. Biol. 2014, 10, e1003821. [Google Scholar] [CrossRef][Green Version]
- Agioutantis, P.; Koumandou, V.L. Bioenergetic diversity of the human gut microbiome. Meta Gene 2018, 16, 10–14. [Google Scholar] [CrossRef]
- Koumandou, V.L.; Kossida, S. Evolution of b-type cytochromes in prokaryotes. PeerJ Prepr. 2015. [Google Scholar] [CrossRef]
- Lang, A.S.; Westbye, A.B.; Beatty, J.T. The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Annu. Rev. Virol. 2017, 4, 87–104. [Google Scholar] [CrossRef]
- Westbye, A.B.; Beatty, J.T.; Lang, A.S. Guaranteeing a captive audience: Coordinated regulation of gene transfer agent (GTA) production and recipient capability by cellular regulators. Curr. Opin. Microbiol. 2017, 38, 122–129. [Google Scholar] [CrossRef]
- Ayre, D.C.C.; Elstner, M.; Smith, N.C.; Moores, E.S.; Hogan, A.M.; Christian, S.L. Dynamic regulation of CD24 expression and release of CD24-containing microvesicles in immature B cells in response to CD24 engagement. Immunology 2015, 146, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Ayre, D.C.C.; Chute, I.C.; Joy, A.P.; Barnett, D.A.; Hogan, A.M.; Gruel, M.P.; Christian, S.L. CD24 induces changes to the surface receptors of B cell microvesicles with variable effects on their RNA and protein cargo. Sci. Rep. 2017, 7, 8642. [Google Scholar] [CrossRef] [PubMed]
- Sabi, R.; Tuller, T. Modelling and measuring intracellular competition for finite resources during gene expression. J. R. Soc. Interface 2019, 16, 20180887. [Google Scholar] [CrossRef] [PubMed]
- Zur, H.; Tuller, T. Predictive biophysical modeling and understanding of the dynamics of mRNA translation and its evolution. Nucleic Acids Res. 2016, 44, 9031–9049. [Google Scholar] [CrossRef] [PubMed]
- Goz, E.; Zafrir, Z.; Tuller, T. Universal evolutionary selection for high dimensional silent patterns of information hidden in the redundancy of viral genetic code. Bioinformatics 2018, 34, 3241–3248. [Google Scholar] [CrossRef] [PubMed]
- Jheeta, S.; Ptasinska, S.; Sivaraman, B.; Mason, N.J. The irradiation of 1:1 mixture of ammonia:carbon dioxide ice at 30 K using 1 kev Electrons. Chem. Phys. Lett. 2012, 543, 208–212. [Google Scholar] [CrossRef]
- Jheeta, S.; Domaracka, A.; Ptasinska, S.; Mason, N.J. The irradiation of pure CH3OH and 1:1 mixture of NH3:CH3OH ices at 30 K using low energy electrons. Chem. Phys. Lett. 2013, 556, 359–364. [Google Scholar] [CrossRef]
- Jheeta, S. Final frontiers: The hunt for life elsewhere in the Universe. Astrophys. Space Sci. 2013, 348, 1–10. [Google Scholar] [CrossRef]
- Le Roy, L.; Briani, G.; Briois, C.; Cottin, H.; Fray, N.; Thirkell, L.; Poulet, G.; Hilchenbach, M. On the prospective detection of polyoxymethylene in comet 67P/Churyumov–Gerasimenko with the COSIMA instrument onboard Rosetta. Planet. Space Sci. 2012, 65, 83–92. [Google Scholar] [CrossRef]
- Kissel, J.; Altwegg, K.; Clark, B.C.; Colangeli, L.; Cottin, H.; Czempiel, S.; Eibl, J.; Engrand, C.; Fehringer, H.M.; Feuerbacher, B.; et al. COSIMA—High Resolution Time-of-Flight Secondary Ion Mass Spectrometer for the Analysis of Cometary Dust Particles onboard Rosetta. Space Sci. Rev. 2007, 128, 823–867. [Google Scholar] [CrossRef]
- Pleyer, H.L.; Strasdeit, H.; Fox, S. A Possible Prebiotic Ancestry of Porphyrin-Type Protein Cofactors. Orig. Life Evol. Biosph. 2018, 48, 347–371. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.; Strasdeit, H. A Possible Prebiotic Origin on Volcanic Islands of Oligopyrrole-Type Photopigments and Electron Transfer Cofactors. Astrobiology 2013, 13, 578–595. [Google Scholar] [CrossRef]
- Fox, S.; Strasdeit, H. Inhabited or Uninhabited? Pitfalls in the Interpretation of Possible Chemical Signatures of Extraterrestrial Life. Front. Microbiol. 2017, 8, 1622. [Google Scholar] [CrossRef] [PubMed]
- Schwieterman, E.W.; Kiang, N.Y.; Parenteau, M.N.; Harman, C.E.; DasSarma, S.; Fisher, T.M.; Arney, G.N.; Hilairy, E.; Hartnett, H.E.; Reinhard, C.T.; et al. Exoplanet Biosignatures: A Review of Remotely Detectable Signs of Life. Astrobiology 2018, 18, 663–708. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [Google Scholar] [CrossRef]
- Georgiou, C.D. Functional properties of amino acid side chains as biomarkers of extraterrestrial life. Astrobiology 2018, 18, 1479–1496. [Google Scholar] [CrossRef]
- Georgiou, C.D.; Deamer, D.W. Lipids as universal biomarkers of extra-terrestrial life. Astrobiology 2014, 14, 541–549. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Chemical roots of biological evolution: The origins of life as a process of development of autonomous functional systems. Open Biol. 2017, 7, 170050. [Google Scholar] [CrossRef]
- Saitta, M. From Computational Physics to the Origins of Life. In Life Sciences, Information Sciences; Thierry, G., Dominique, L., Marie-Christine, M., Jean-Charles, P., Eds.; Wiley: Hoboken, NJ, USA, 25 March 2018; Chapter 20. [Google Scholar] [CrossRef]
- Uyama, T.; Hashimoto, J.; Kuzuhara, M.; Mayama, S.; Akiyama, E.; Currie, J.L.; Kudo, T.; Kusakabe, N.; Abe, L. The SEEDS High-Contrast Imaging Survey of Exoplanets around Young Stellar Objects. Astron. J. 2017, 153, 106. [Google Scholar] [CrossRef]
- Uyama, T.; Tanigawa, T.; Hashimoto, J.; Tamura, M.; Aoyama, Y.; Brandt, T.D.; Ishizuka, M. Constraining Accretion Signatures of Exoplanets in the TW Hya Transitional Disk. Astron. J. 2017, 154, 90. [Google Scholar] [CrossRef]
- Uyama, T.; Hashimoto, J.; Muto, T.; Akiyama, E.; Dong, R.; de Leon, J.; Sakon, I.; Kudo, T.; Kusakabe, N.; Kuzuhara, M.; et al. Subaru/HiCIAO HKs Imaging of LKHa 330: Multi-band Detection of the Gap and Spiral-like Structures. Astron. J. 2018, 156, 63. [Google Scholar] [CrossRef]
- Wisłocka, A.M.; Kovačević, A.B.; Balbi, A. Comparative analysis of the influence of Sgr A* and nearby active galactic nuclei on the mass loss of known exoplanets. Astron. Astrophys. 2019, 624, A71. [Google Scholar] [CrossRef]
- Luger, R.; Barnes, R.; Lopez, E.; Fortney, J.; Jackson, B.; Meadows, V. Habitable Evaporated Cores: Transforming Mini-Neptunes into Super-Earths in the Habitable Zones of M Dwarfs. Astrobiology 2015, 15, 57–88. [Google Scholar] [CrossRef] [PubMed]
- Valencia, D.; Ikoma, M.; Guillot, T.; Nettelmann, N. Composition and fate of short-period super-Earths:The case of CoRoTx7b. Astron. Astrophys. 2010, 516, A20. [Google Scholar] [CrossRef]
- Maccone, C. Energy of Extra-Terrestrial Civilizations according to Evo-SETI Theory. Acta Astronaut. 2018, 144, 202–213. [Google Scholar] [CrossRef]
- Maccone, C. Life Expectancy and Life Energy according to Evo-SETI Theory. Int. J. Astrobiol. 2019, 189, 36–46. [Google Scholar] [CrossRef]
- Lazcano, A.; Miller, S.L. How long did it take for life to begin and evolve to cyanobacteria? J. Mol. Evol. 1994, 39, 546–554. [Google Scholar] [CrossRef]
- Forterre, P. Three RNA cells for ribosomal lineages and three DNAviruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. USA 2006, 10, 3669–3674. [Google Scholar] [CrossRef]
- Forterre, P. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 2005, 87, 793–803. [Google Scholar] [CrossRef]
- Jheeta, S.; Joshi, P.C. Prebiotic RNA Synthesis by Montmorillonite Catalysis. Life 2014, 4, 318–330. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jheeta, S. Molecules to Microbes. Sci 2020, 2, 86. https://doi.org/10.3390/sci2040086
Jheeta S. Molecules to Microbes. Sci. 2020; 2(4):86. https://doi.org/10.3390/sci2040086
Chicago/Turabian StyleJheeta, Sohan. 2020. "Molecules to Microbes" Sci 2, no. 4: 86. https://doi.org/10.3390/sci2040086
APA StyleJheeta, S. (2020). Molecules to Microbes. Sci, 2(4), 86. https://doi.org/10.3390/sci2040086



