Proton-Cluster-Beam Lethality and Mutagenicity in Bacillus subtilis Spores
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Irradiation
2.3. Survival Assay
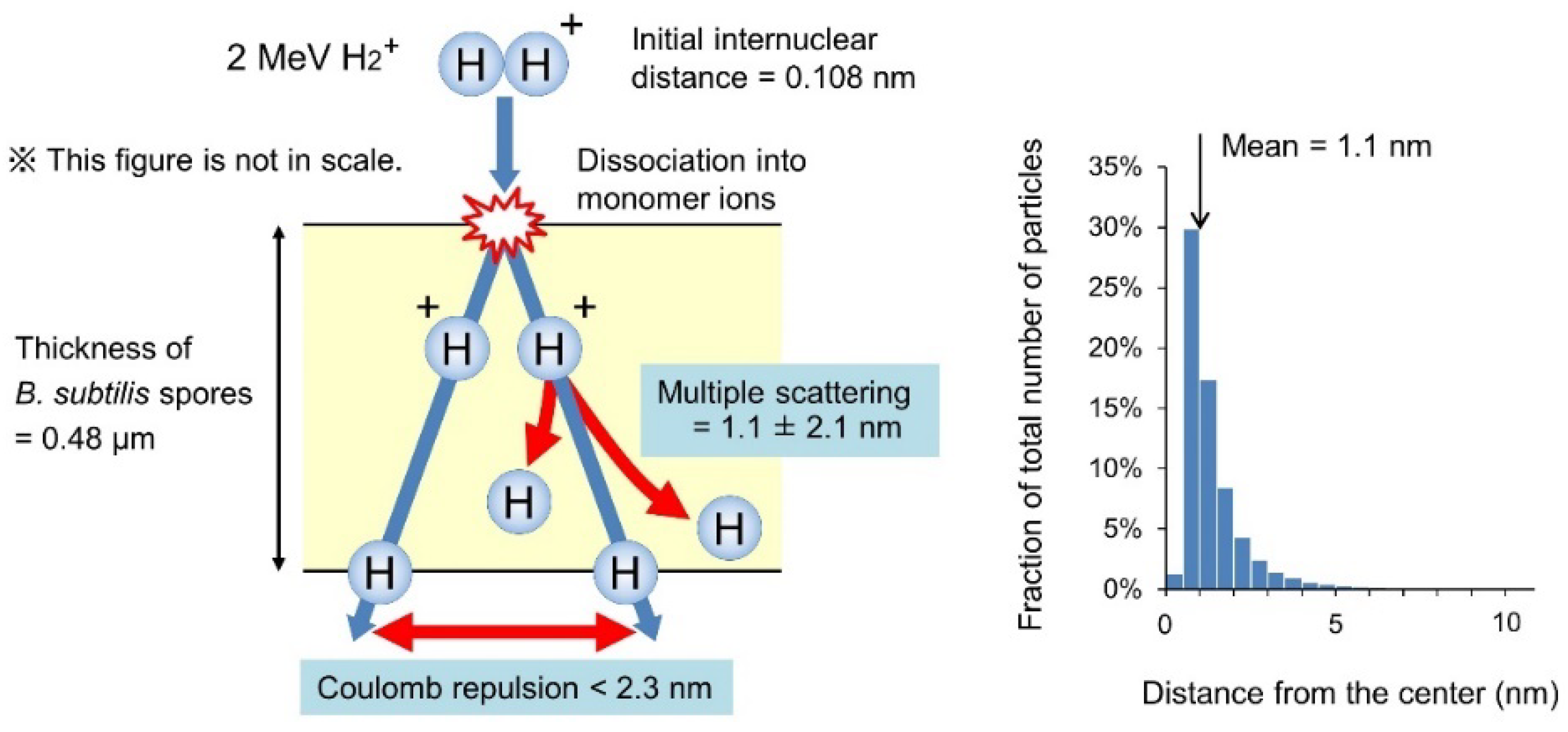
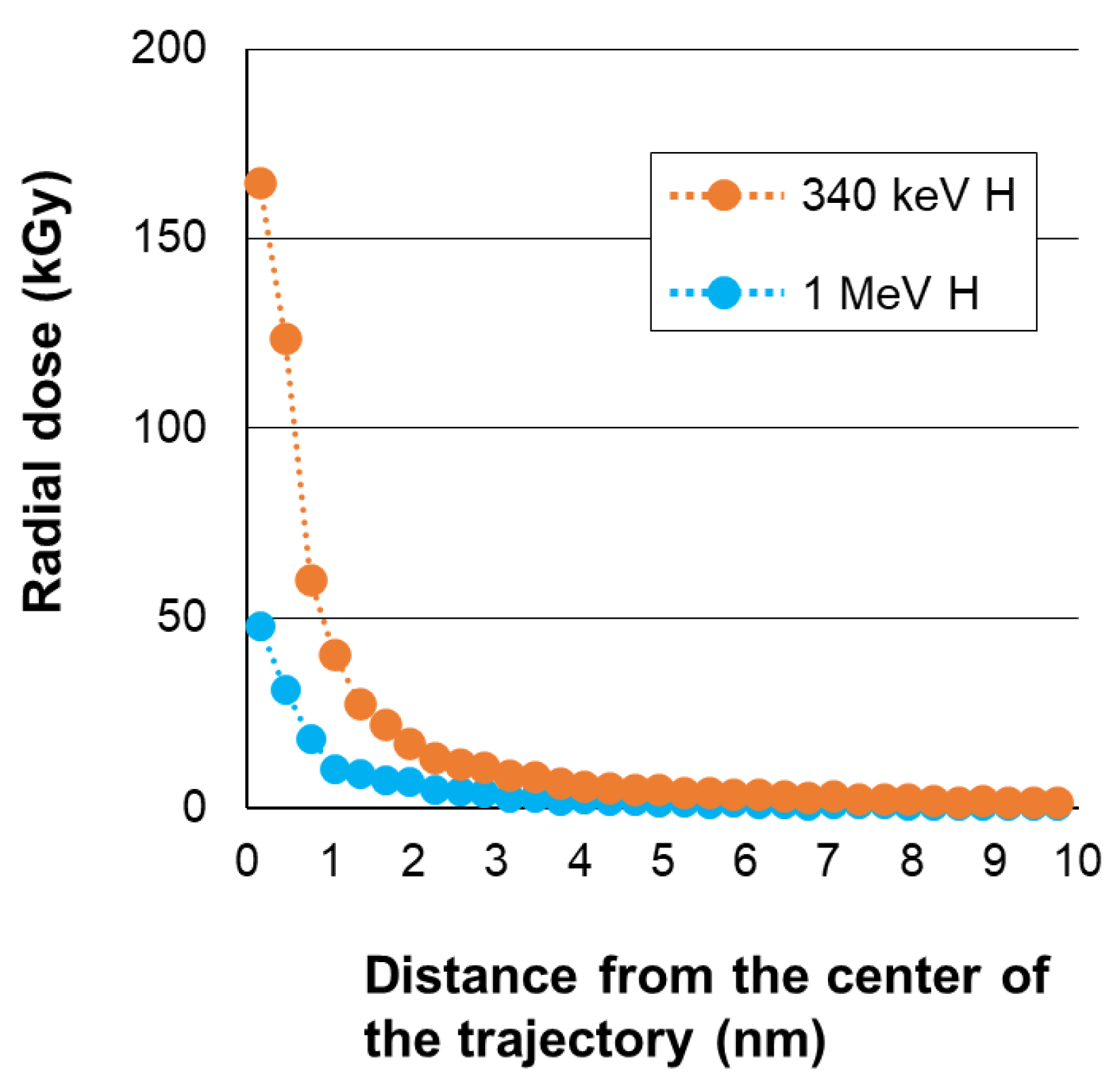
2.4. Calculation of Physical Parameters
3. Results and Discussion
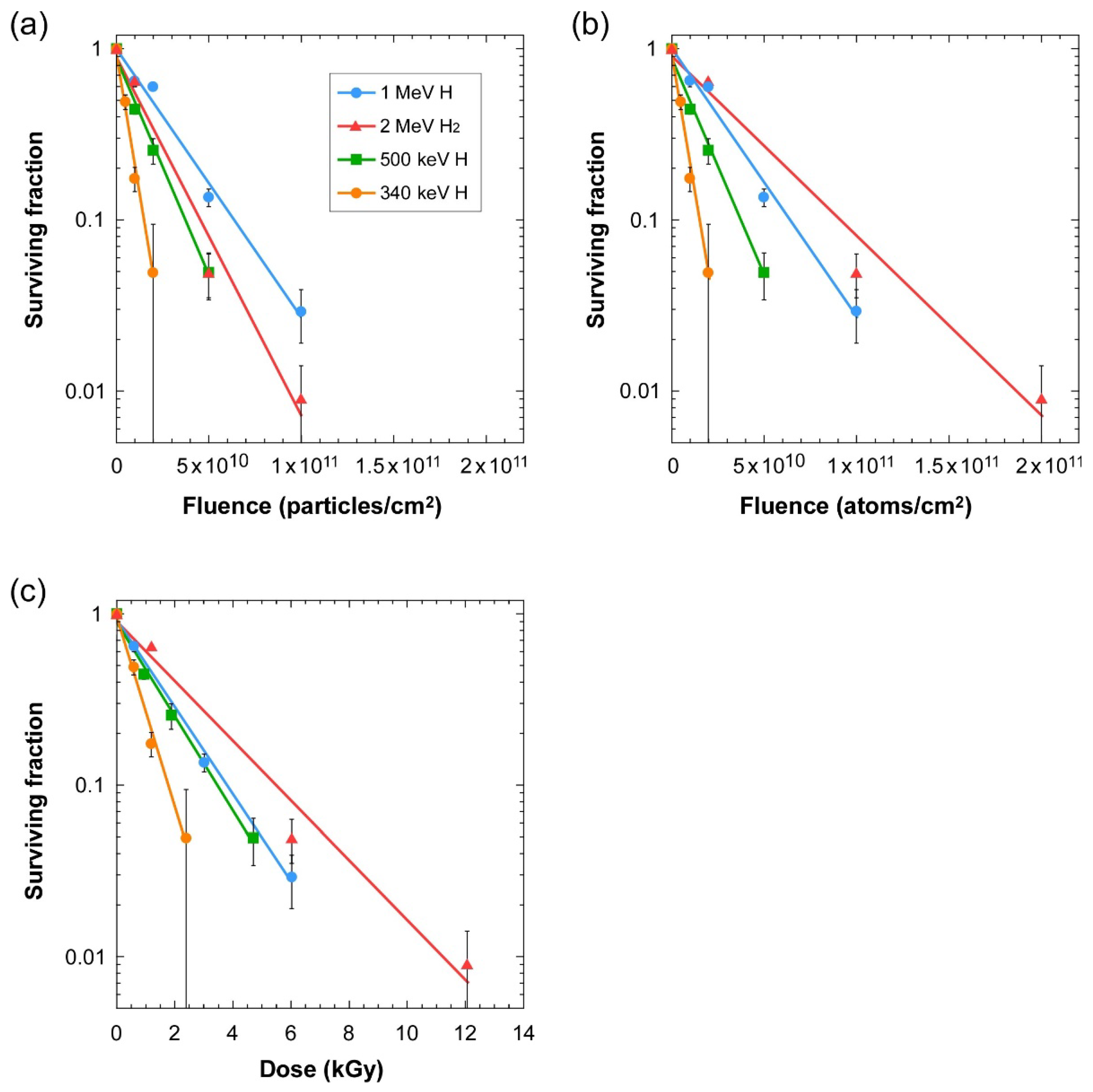
3.1. Lethality of Cluster Ions
3.2. Mutagenicity of Cluster Ions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jo, Y.; Kim, J.B. Frequency and spectrum of radiation-induced mutations revealed by whole-genome sequencing analyses in plants. Quantum Beam Sci. 2019, 3, 7. [Google Scholar] [CrossRef]
- Hu, W.; Li, W.; Chen, J. Recent advances of microbial breeding via heavy-ion mutagenesis at IMP. Lett. Appl. Microbiol. 2017, 65, 274–280. [Google Scholar] [CrossRef]
- Satoh, K.; Oono, Y. Studies on application of ion beam breeding to industrial microorganisms at TIARA. Quantum Beam Sci. 2019, 3, 11. [Google Scholar] [CrossRef]
- Goodhead, D.T. Initial events in the cellular effects of ionizing radiations: Clustered damage in DNA. Int. J. Radiat. Biol. 1994, 65, 7–17. [Google Scholar] [CrossRef]
- Sage, E.; Shikazono, N. Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 2017, 107, 125–135. [Google Scholar] [CrossRef]
- Mavragani, I.V.; Nikitaki, Z.; Kalospyros, S.A.; Georgakilas, A.G. Ionizing radiation and complex DNA damage: From prediction to detection challenges and biological significance. Cancers 2019, 11, 1789. [Google Scholar] [CrossRef]
- Dunlop, A.; Jaskierowicz, G.; Della-Negra, S. Latent track formation in silicon irradiation by 30 MeV fullerenes. Nucl. Instr. Meth. Phys. Res. B 1998, 146, 302–308. [Google Scholar] [CrossRef]
- El-Said, A.S. Tracks of 30-MeV C60 clusters in yttrium iron garnet studied by scanning force microscopy. Nucl. Instr. Meth. Phys. Res. B 2009, 267, 953–956. [Google Scholar] [CrossRef][Green Version]
- Koide, T.; Saitoh, Y.; Sakamaki, M.; Amemiya, K.; Iwase, A.; Matsui, T. Change in magnetic and structural properties of FeRh thin films by gold cluster ion beam irradiation with the energy of 1.67 MeV/atom. J. Appl. Phys. 2014, 115, 17B722. [Google Scholar] [CrossRef]
- Hase, Y.; Satoh, K.; Chiba, A.; Hirano, Y.; Tomita, S.; Saito, Y.; Narumi, I. Experimental study on biological effect of cluster ion beams in Bacillus subtilis spores. Quantum Beam Sci. 2019, 3, 8. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Carrera, M.; Zandomeni, R.O.; Fitzgibbon, J.; Sagripanti, J.-L. Difference between the spore size of Bacillus anthracis and other Bacillus species. J. App. Microbiol. 2007, 102, 303–312. [Google Scholar] [CrossRef]
- Kurashima, S.; Satoh, T.; Saitoh, Y.; Yokota, W. Irradiation Facilities of the Takasaki Advanced Radiation Research Institute. Quantum Beam Sci. 2017, 1, 2. [Google Scholar] [CrossRef]
- Ziegler, J.F.; Ziegler, M.D.; Biersack, J.P. SRIM—The stopping and range of ions in matter. Nucl. Instr. Meth. Phys. Res. B 2010, 268, 1818–1823. [Google Scholar] [CrossRef]
- Tanaka, S.; Fukuda, K.; Nishimura, K.; Watanabe, H.; Yamano, N. IRAC M: A Code System to Calculate Induced Radioactivity Produced by Ions and Neutrons. JAERI-Data/Code 97-019; Japan Atomic Energy Research Institute: Ibaraki, Japan, 1997. [Google Scholar]
- Carrera, M.; Zandomeni, R.O.; Sagripanti, J.-L. Wet and dry density of Bacillus anthracis and other Bacillus species. J. App. Microbiol. 2008, 105, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Yanagida, T.; Fujimoto, S.; Saga, K.; Minowa, T. Process Simulation of Yeast Cultivation and Ethanol Fermentation in Bio-ethanol Production. Energy Resour. 2010, 31, 335–340. (In Japanese) [Google Scholar]
- Moribayashi, K. Application of atomic and molecular data for plasma production and cancer therapy by heavy particle irradiation. Jpn. J. Appl. Phys. 2020, 59, SH0801. [Google Scholar] [CrossRef]
- Moeller, R.; Reitz, G.; Berger, T.; Okayasu, R.; Nicholson, W.L.; Horneck, G. Astrobiological aspects of the mutagenesis of cosmic radiation on bacterial spores. Astrobiology 2010, 10, 509–521. [Google Scholar] [CrossRef]
- Moeller, R.; Reitz, G.; Li, Z.; Klein, S.; Nicholson, W.L. Multifractional resistance of Bacillus subtilis spores to high-energy proton radiation: Role of spore structural components and the homologous recombination and non-homologous end joining DNA repair pathways. Astrobiology 2012, 12, 1069–1077. [Google Scholar] [CrossRef]
- Zhang, Y.; Huber, N.; Moeller, R.; Stülke, J.; Dubovcova, B.; Akepsimaidis, G.; Meneses, N.; Drissner, D.; Mathys, A. Role of DNA repair in Bacillus subtilis spore resistance to high energy and low energy electron beam treatments. Food Microbiol. 2020, 87, 103353. [Google Scholar] [CrossRef]
- Furusawa, Y.; Fukutsu, K.; Aoki, M.; Itsukaichi, H.; Eguchi-Kasai, K.; Ohara, H.; Yatagai, F.; Kanai, T.; Ando, K. Inactivation of aerobic and hypoxic cells from three different cell lines by accelerated 3He-, 12C- and 20Ne-ion beams. Radiat. Res. 2000, 154, 485–496. [Google Scholar] [CrossRef]
- Jones, B.; Hill, M.A. The Physical separation between the LET associated with the ultimate relative biological effect (RBE) and the maximum LET in a proton or ion beam. Biomed. Phys. Eng. Express 2020, 6, 055001. [Google Scholar] [CrossRef] [PubMed]

| Beam | Surface LET (keV/µm) | Penetration Range (µm) |
|---|---|---|
| 1 MeV H+ | 37.7 | 16.7 |
| 2 MeV H2+ | 37.7 × 2 (75.4) * | 16.7 * |
| 500 keV H+ | 58.9 | 5.8 |
| 340 keV H+ | 75.0 | 3.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hase, Y.; Satoh, K.; Chiba, A.; Hirano, Y.; Moribayashi, K.; Narumi, K. Proton-Cluster-Beam Lethality and Mutagenicity in Bacillus subtilis Spores. Quantum Beam Sci. 2021, 5, 25. https://doi.org/10.3390/qubs5030025
Hase Y, Satoh K, Chiba A, Hirano Y, Moribayashi K, Narumi K. Proton-Cluster-Beam Lethality and Mutagenicity in Bacillus subtilis Spores. Quantum Beam Science. 2021; 5(3):25. https://doi.org/10.3390/qubs5030025
Chicago/Turabian StyleHase, Yoshihiro, Katsuya Satoh, Atsuya Chiba, Yoshimi Hirano, Kengo Moribayashi, and Kazumasa Narumi. 2021. "Proton-Cluster-Beam Lethality and Mutagenicity in Bacillus subtilis Spores" Quantum Beam Science 5, no. 3: 25. https://doi.org/10.3390/qubs5030025
APA StyleHase, Y., Satoh, K., Chiba, A., Hirano, Y., Moribayashi, K., & Narumi, K. (2021). Proton-Cluster-Beam Lethality and Mutagenicity in Bacillus subtilis Spores. Quantum Beam Science, 5(3), 25. https://doi.org/10.3390/qubs5030025






