Abstract
In a worldwide effort to generate clinically useful therapeutic or preventive interventions, harnessing biophysical stimuli for directing cell fate is a powerful strategy. With the vision to control cell function through engineering cell shape, better understanding, measuring, and controlling cell shape for ultimately utilizing cell shape-instructive materials is an emerging “hot” topic in regenerative medicine. This review highlights how quantitation of cellular morphology is useful not only for understanding the effects of different microenvironmental or biophysical stimuli on cells, but also how it could be used as a predictive marker of biological responses, e.g., by predicting future mesenchymal stromal cell differentiation. We introduce how high throughput image analysis, combined with computational tools, are increasingly being used to efficiently and accurately recognize cells. Moreover, we discuss how a panel of quantitative shape descriptors may be useful for measuring specific aspects of cellular and nuclear morphology in cell culture and tissues. This review focuses on the mechano-biological principle(s) through which biophysical cues can affect cellular shape, and recent insights on how specific cellular “baseline shapes” can intentionally be engineered, using biophysical cues. Hence, this review hopes to reveal how measuring and controlling cellular shape may aid in future regenerative medicine applications.
1. Is Shaping the Cell Also Shaping Regenerative Medicine?
Worldwide, regenerative medicine works toward improving practical methods and experimental strategies to generate clinically useful therapeutic or preventive interventions. In this view, mesenchymal stromal cells (MSCs) are recognized as adult, self-renewing, and multipotent stem cells with substantial potential for therapeutic use [1,2] that were forecasted to significantly improve disease outcomes and patient lives [3]. In this review, we chose the term “mesenchymal stromal cells” over “mesenchymal stem cells” to describe a heterogeneous population of cells that can be differentiated in vitro into a range of lineages, and whose self-renewal and subsequent in vivo differentiation remains to be proven [4]. Publication titles such as the “rise of mechano-transduction” [5], “mechano-transduction: use the force” [6], and “mechano-transduction: may the force be with you” [7] illustrate that besides biochemical and genetic factors, biophysical cues may be equally important in controlling cell fate [8], and that harnessing biophysical stimuli is a modern, powerful approach to steering cell function in regenerative medicine. Developmental biology demonstrates that cell shape follows function. Classic examples are skeletal myogenic cells, which exhibit an elongated cylindrical morphology. This morphology aligns multi-nucleated myofibers parallel to the direction of muscle tissue tension and contraction. In the context of biophysically steering cell fate, cellular shape has only recently emerged as potential determinant, because cellular shape influences tissue structure and function [9,10] and determines the lineage of differentiation [11]. Additionally, the physical shape of cells is a fundamental signal for proliferation [12], a potent regulator of cell growth and physiology, and is adapted for specific functions [13]. Thus, in a way, actively shaping cellular morphology by biophysical means contributes to shaping the future of regenerative medicine.
As this invited review was given the task to summarize topic-related advancements presented in the Tissue Engineering and Regenerative Medicine International Society (TERMIS) European Chapter Meeting 2017, this review focuses on cellular morphology as a novel assessment of biological responses and discusses broadly how biophysical cues affect cellular shape, how cellular shape can be measured quantitatively, and introduces the computational tools and approaches necessary for this task. Finally, this review will discuss means of controlling cellular shape, its effects on functional phenotype, and impact it will have on regenerative medicine applications.
2. Cell Morphology as a Novel Tool to Assess Biological Responses in Tissue Engineering
As this review is in included in the TERMIS special issue: selected papers from TERMIS European Chapter Meeting 2017 on “Biomechanics, Morphology and Imaging”, we are highlighting some selected studies that were presented at this meeting that demonstrated the importance of measuring cell morphological features and their implications on tissue engineering. Topics ranged from the effect of exposure to normogravity (or earth gravity force) vs. simulated hypergravity conditions using the large diameter centrifuge (LDC) from the European Space Research and Technology Centre (ESTEC, ESA, The Netherlands) [14] to how different degrees of compressive stiffness in 3D-printed scaffolds containing mesenchymal stromal cells (MSCs) affected cell function and morphology [15]. According to the former group [14], microgravity-induced alterations, found during spaceflight, but more importantly, during bed rest, were comparable to tissue degeneration caused by disuse and ageing. In this study, human tendon-derived cells were exposed to normogravity (earth gravity force) vs. hypergravity. After 16 h, the different g-levels led to cell and cytoskeleton morphology changes (an increase in cell area, actin stress fiber formation and intracellular anisotropy) that correlated with tenogenic differentiation markers, highlighting the importance of measuring cell shape and suggesting that exposure of musculoskeletal tissues to hypergravity may simulate loading and rescue the phenotype of degenerated tendon cells after exposure to near-weightlessness conditions. The latter group’s work demonstrated that 3D printed scaffolds with different mechanical properties (compressive moduli), but with the same 3D microstructure resulted in elongated vs. round and short cell morphologies [15]. A third study by Rocca et al. [16] used a quantitative approach for measuring cell shape parameters and assessed how the effect of mechanical forces through hypergravity stimulation improved differentiation of MSCs into osteoblasts. His group showed that hypergravity resulted in spreading of the cells (i.e., cells exhibiting a decrease in circularity, roundness and solidity), and corresponding upregulation of Ras Homolog Family Member A (RHOA), a transcription factor that regulates the actin cytoskeleton, and increased osteogenesis. A TERMIS 2017 keynote talk highlighted how spatially restricting macrophage spreading through micro-patterning, can prevent the activation of M0 macrophages into pro-inflammatory M1 macrophages and suggested that control of cell shape may be used to regulate the immune response [17]. These recent results demonstrate the importance of measuring cell morphology in assessment of biological responses and in tissue engineering.
Another highlight at the TERMIS 2017 meeting was use of different methods for measuring cell morphology parameters. Our group showed how an automated, non-destructive, high-throughput quantification method, using images of calcein-stained cells, combined with binary thresholding, could be used to efficiently and accurately measure a panel of cell morphological parameters on a large number of cells (e.g., 60,000 cells) within a short time [18,19]. This automated high-throughput quantification method was significantly more efficient, precise and reproducible than manual methods. Furthermore, this approach can be applied to other cell types. Another group used a label-free high throughput approach for measuring morphological descriptors during cell culture, providing a non-invasive approach that could be used to assess cells during time-course experiments or in stem cell manufacturing [20]. This group also showed how measuring cell morphological parameters can be used in combination with experimental data in order to predict stem cell phenotypic outcomes at a later time point. Here, stem cell shape was used as predictive marker with the intention to replace measuring later time points [21,22,23]. Hence, studies have begun to increasingly and successfully use quantitative approaches to access morphological parameters and to correlate them to biological parameters and cell fate outcome. For example, our group has used a semi-automated high-throughput method for calculating a specific panel of shape descriptors [18,19,24], which was used to describe MSC shape in correlation to biophysical stimuli such as biomaterial type and nanoscale surface stiffness, as well as in response to externally applied biomechanical forces [19,24] (Figure 1). Therefore, quantitation of cellular morphology is useful for understanding the effects of different microenvironmental or biophysical stimuli on cells and for helping to define a morphological fingerprint for cells [20,25].
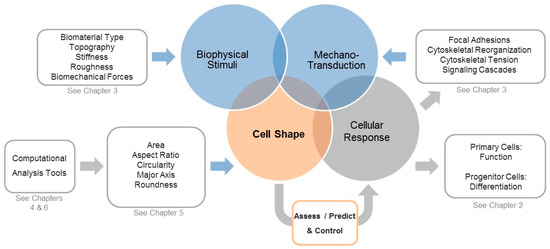
Figure 1.
Overview of how biophysical stimuli are translated into biological responses and the central role of cell shape in assessing, predicting, and controlling these processes. Various static biophysical factors such as biomaterial type, topography, stiffness, roughness, and dynamically applied biomechanical forces (e.g., cyclic stretch) can be used to modulate the cell’s shape and the associated mechano-transduction processes. During such processes, external stimuli initiate changes in cytoskeletal tension, which can modify focal adhesions and cytoskeletal organization and activate integrin-mediated signal pathways. Additionally, these processes affect–and are affected by-cell shape, which highlights why shape can be used to assess, predict, and control the downstream biological responses. Modern computational tools such as automated segmentation and high throughput calculation of quantitative shape descriptors for a large amount of cells, paired with machine learning-assisted analyzes and classifications, will help to associate distinct shape profiles with biological function in order to better understand how these processes are linked and can be controlled.
Given the pertinence of cell shape toward describing a cell’s current phenotype, recent morphological analysis has also developed as a potent tool to effectively predict how cells will respond to a particular stimulus or environment, including differentiation potential. As an example, using high content imaging, Marklein et al. [25] demonstrated that nuclear morphological profiles of MSCs had distinct morphological features that were highly predictive of MSC mineralization capabilities, with >90% accuracy. Kato’s group has proposed a similar theory [21,22,23,26]. Their group demonstrated that cell morphological features from the first three days of differentiation can predict osteogenic differentiation potential (i.e., alkaline phosphatase and calcium deposition). Additionally, quantification of cell morphology is increasingly being used in time-lapse microscopy, combined with non-invasive computational tools, to automatically detect, segment, and analyze unlabeled live cells to assess, track and even predict MSC differentiation phenotypic outcomes [20,26,27]. For example, eccentricity and filopodia were shown to robust classifiers of myogenic differentiation [27]. Pearson et al. combined a unique approach which simultaneously measured cell morphology parameters, migration trajectories, as well as viscoelasticity parameters and cell stiffness for characterizing MSC differentiated phenotypes and naïve vs. stimulated T cells [28].
Quantitative morphological assessment of cell shape is not limited to cells in culture; it has also been used for analyses in tissues. Zanier et al. showed how quantification of cell morphology can be used to define specific population subsets in injured brain tissue following different injurious stimuli [29]. Moreover, they used this technique to demonstrate how different populations of inflammatory cells correlated with different aspects of injury. An image processing approach has also been used to quantify morphology of muscle fibers in tissue by measuring the number of nuclei per cytoplasm to track multinucleated myofiber formation, fiber length and width, and fiber density [30]. This approach could be used to classify abnormalities in muscular diseases or easily applied to track myogenic differentiation of MSCs, satellite cells, or myocytes into myotubes.
Nuclear morphological studies have also showed strong correlations between nuclear shape and changes in cell phenotype in many physiological, as well as pathological conditions [31,32,33,34,35]. Interestingly, even the perinuclear cytoplasmic mechanical properties have been associated with the geometry of the stem cell nucleus; stem cell types can even be discriminated from each other through coupling perinuclear mechanical properties to nuclear shape [32]. Collectively, these findings illustrate that cellular shape is a fundamental element within the multiple complex processes of biophysically modulated cellular function.
Hence, by converting the statistics of quantitative cell morphology into a data-rich morphological fingerprint, computer analysis of high-resolution digitized images of cells and tissues may be used to improve cell culture, tissue engineering techniques, and perhaps even serve as a prognostic marker in which today’s cell shape can be analyzed to deduce a cell cultures state of function prior to implantation.
3. The Cellular Components that Sense and Respond to Biophysical Stimuli and Affect Cell Shape
As we have illustrated above that cellular shape is a powerful, emerging parameter for assessing the state of a cell, it is helpful to discuss how biophysical cues can sense the stimuli, transduce the signals, and react to the environment (including but certainly not limited to changes in cell shape), (Figure 1). Cell behavior can be dictated by microenvironmental biophysical cues including (1) mechanical forces such as compression, tension, and shear forces, (2) substrate properties such as biomaterial type, ligand presentation, surface topography, viscoelasticity, and stiffness, or (3) biochemical cues such as autocrine and paracrine signals [19,24,31,36,37,38,39]. Collectively, such cues can ultimately lead to a specific differentiation behavior of cells such as MSCs [38].
One of the most significant aspects of this biomechanical response to external cues is the cytoskeleton. It is a highly dynamic network of interlinked proteins that form the cells’ shape and plays an integral role in utilizing biophysical signals to modulate cell function. Such extracellular biophysical signals exerted on the cell membrane and cytoskeleton change the balance between extracellular and intracellular forces, which, in turn, affects the shape and also the function of a given cell by activating various signaling pathways [31] (Figure 1). Cells transmit extracellular forces by adhering to extracellular matrix proteins or to other cells. The main component of these adhesions are integrins, which sense physical cues in micro- and nanometer ranges through the attachment to specific ECM proteins. By binding to the ECM, integrins build highly dynamic adhesion complexes referred to as focal adhesions (FAs), whose mechanical modulation induces integrin-mediated signal pathway activation. Such activation can lead to the spatio-temporal coordination of multiple downstream events [40] and cause diverse biophysical cues such as mechanical forces or surface-associated properties, such as roughness or elasticity, to affect the cytoskeleton, cell shape and FA assembly and modification [40,41,42]. Integrins can also interact via their cytoplasmic tails via various adaptor and signalling molecules to activate downstream signalling pathways such as tyrosine kinases and phosphatases and regulate gene expression by responding to external biophysical stimuli. Such so-called mechanotransduction pathways establish positive-feedback loops that cause integrin engagement to activate processes such as actin-myosin cytoskeleton contractility, through which FAs are reinforced [40,41,42]. For this reason, the cytoskeleton contractility propagated inside the cell becomes directly proportional to the cells’ adhesion strength and the ECM elastic modulus. In many cases, force-responses are acute and only transiently affect the local FAs, the cytoskeleton, and intracellular signaling messengers. However, long-term phenotypic changes can also transpire in the cell through comparable mechanisms.
4. Computational Tools to Segment Images for Recognizing Cells
Phase contrast microscopy has allowed the visualization of cells for nearly a century, and qualitative assessment of histology has provided valuable insight into effects of culturing conditions of cell morphology. Objective identification and quantification of these changes is the next natural step in the progression of using shape to assess and modulate phenotype. Hence, computational tools for analysis of cell morphology are increasingly becoming necessary to extract and analyze the enormous amount of data from entire cell populations. This technology is continuously being improved to promote a better quantitative and statistical approach for defining specific geometric aspects of cell shape and relationships between cell shape and experimental stimuli or microenvironmental effects. Generally, the process of measuring dimensions and shapes is termed quantitative mathematical morphometry. As a first step, before any morphological parameters can be analyzed from a given cell, an appropriate numerical mathematical representation of a cell’s shape must be generated, which can then be used for more complex calculations of specific shape descriptors mentioned later [43]. In other words, one must first identify and select the cell(s) within a given image in order to calculate specific shape features. In recent years, automated cell segmentation has evolved, which led to the development of diverse approaches for identifying and distinguishing cells from the background of an image [44]. Many different segmentation methods or other approaches for detecting objects and boundaries exist and only a selection can be discussed here. The intensity threshold is the oldest method that is based on measuring the absolute intensity difference between cells and black background, either by global or local adaptive thresholding. Another approach is feature detection, which uses image intensity-derived features that are found using linear image filtering or others such as Gaussian or Laplacian-of-Gaussian filters [45,46,47]. In contrast, the morphological filtering method uses e.g., nonlinear filters to examine geometrical and topological properties of objects within images. Here, combining multiple approaches promises the construction of complicated, but effective, filters possible. In an interactive approach, region accumulation lets the user set selected, labeled areas within the image by connecting points [44]. This model can be implemented in watershed transform, one of the main mathematical segmentation methods [48], in which a given image is seen as a topographic surface, in which so-called watershed lines represent region boundaries. Another approach fits a deformable model to the image data and this particular methodology allows the ability to adapt to topological features and hence could be useful for complex situations such as seen in e.g., cell tracking. Based on these segmentation approaches, one can determine the outline of features such as cells or other objects of interest against a given background. This step is necessary prior to using the outlines for calculating specific shape descriptors. Thus, biologically useful information needs to be extracted from sample images after segmentation. The future of this technology will involve better algorithms for distinguishing cell bodies but also demand modification to account for three-dimensional shape. This will become increasingly important as the field of tissue engineering develops and requires manipulations of cells in complex environments.
5. Measuring Cellular Morphology with a Panel of Cell Shape Descriptors
Analysis of cell morphology is increasingly being measured via a more quantitative and high-throughput approach. Although identification of cell morphological features has been used to describe cells for centuries, in pre-digital times, cell shape was assessed qualitatively with the microscope, which of course may have led to imprecise and biased results. Recent developments in image processing technology and computer-aided analyses now allow for high-throughput calculation of quantitative shape descriptors for mathematically describing cellular morphology. Our lab has actively used algorithms in ImageJ, an open-source program for analyzing images [18,19]. In this software, shape descriptors can be calculated based on objective mathematical functions and produce numerical values. Based on the shape descriptor’s respective underlying formula, these numerical values are representative of a specific characteristic of a given cellular shape. Shape descriptors can measure the perimeter, area, length, and roundness of individual cells, or, once an ellipse is fitted over a given cell, they may determine the length of the cell’s (ellipse’s) major and minor axes and, thus, a cell’s length and width. Using major and minor axes, one can determine the cell’s aspect ratio. Whereas length, roundness, and aspect ratio focus on more general shape aspects of the cell, descriptors such as circularity and solidity emphasize the presence of membrane protrusions such as lamellipodia, filopodia, and blebs that are relevant for adhesion, migration, and rigidity sensing [49]. While these shape factors are conceptually less intuitive, they have obvious implications of changes to cell morphology and function. Circularity is a normalized ratio of area to perimeter (with a circle having a circularity of 1 and lines having a circularity of 0) and solidity contrasts convex cell area and concave cell area (an example is the negative area of a crescent moon). Biologically high circularity and solidity values describe fewer of such protrusions. Sickle cells would have low solidity. We refer the biologically interested reader to [19] and the mathematically interested reader to the ImageJ homepage [50,51] and specifically to the “Analyze” menu section “set measurements”, which details the mathematical calculations. In the context of quantifying MSC shape, we advocate using a defined panel of shape descriptors such as those introduced in our previous studies [18,19,24]. Other programs like MetaMorph [52] provide additional, albeit proprietary means for quantifying cell morphology and cell networks. Of course, the choice, mathematical nature and biological meaning of these descriptor values depends on the definition of the shape descriptors but, as specific aspects of morphology can now be quantitatively calculated with high precision and reproducibility [18,21], using different methods discussed below, they are becoming fundamental in quantitative cytology.
6. Computational Approaches to Classify Shape Profiles into Biologically Interpretable Groups
A modern approach for quantifying cellular morphology is to use computer algorithms for the mathematical quantification of shape descriptors that represent specific features of cellular morphology. Scion Image (property of Scion Corporation), ImageJ [50,51] or Fiji [53], an ImageJ-based package with a rich set of tools and plugin focused on scientific image analysis, can potentially be used for such analyses [18,19,24]. Another option is CellProfiler, a Python based system [54]. ImageJ, Fiji, and CellProfiler are open-source software. Hence, the biggest advantage these systems have over commercial systems is that the code can be altered by any individual to fit the personal needs of the user. However, ImageJ and Fiji are not the only software systems for cell morphometry, as MATLAB, Metamoporph [52] and IMARIS are two additional commercial software programs with features for cell analyses. However, commercial systems that have their own codes for image analyses may give the user limited input on how images are segmented prior to calculating shape descriptors.
After selecting cellular shape descriptors that may best represent the phenotypic information in a dataset, a variety of computational strategies can be used to cluster or classify the resultant shape profiles into biologically interpretable groups. Machine learning with novel algorithms are gradually increasing and being used to link cellular morphological features to biomarker measurements and to recognize cell phenotypes [22,26,55,56]. Machine learning uses pattern recognition and computational tools to find functional relationships from the training data with minimal intervention or bias [55]. For example, Logan et al. used cell segmentation combined with pixel-based machine learning to identify hepatocytes vs. fibroblasts in co-culture conditions [57]. While use of cell segmentation accurately identified hepatocytes, use of cell segmentation for identification of fibroblasts was not as accurate [58]. Tuning the software to both cell types using defined regions of interest (ROIs) and pixel-based machine learning enhanced the system and resulted in higher accuracy in identification of both tested cell types [57]. Pixel-based machine learning using was accomplished with the usage of ilastik software [55] succeeded by model-based segmentation of the predefined ROIs using the software CellProfiler [57]. Whereas machine learning or other computational strategies are highly promising approaches, it is not within the scope nor the aim of this review to recommend a particular strategy/system, as almost no comparative data is available. These newly developed systems include fastER, an advanced trainable state-of the art cell segmentation tool for large-scale microscopy-images. This program addresses many of the problems with high through-put microscopy imaging (e.g., high cell density, cell-to-cell heterogeneity, low and variable signal-to-noise ratios, illumination gradients, and changing imaging modalities). Aside from training, the process does not require any further specifications or interventions and is applicable to any given dataset [59]. Another example is the Undecimated Wavelet Transform Multivariate Image Analysis (UWT-MIA) [60], a pattern recognition tool, which can extract textual features from phase contrast images and also extracts shape descriptors such as major and minor axes length, orientation, and roundness. UWT-MIA is an analysis tool, which can simultaneously analyze data relevant for scale and orientation of the cells, which proves to be advantageous at high cell densities [60]. Another approach for addressing the problem of recognizing cells within densely packed groups is the multi-resolution analysis and maximum-likelihood (MAMLE) [61]. The automated image processing toolbox PHANTAST was developed with open-source code for MATLAB and ImageJ and as such may enable a wide range of utilizations for image processing pipelines [62]. PHANTAST was tested on chinese hamster ovary (CHO) cells, human neuroblastoma, and embryonic mouse stem cells and obtains accurate information on culture confluency, cell density, and the morphology of cellular objects. With this impressive list in mind, it is clear that computational systems, including machine learning approaches, are increasingly being used to establish relationships between functional and morphological properties of differing populations of cells. This has allowed to more precisely define relationships and allow predictions between form, such as dynamic cell shape, and function, such as cell phenotype (Figure 1).
7. A View into the Future: Controlling Shape for Regenerative Medicine Applications
In the previous sections of this review, we have discussed how morphology can be used quantitatively and computationally as a functional or even predictive marker for assessing the biological response(s) of a given cell type. It is important to stress that this approach has been established on selected cell types, mainly MSCs, but that this approach is not limited to MSCs. In contrast, one would hope that this approach will be extended to a multitude of cell types that are relevant to the various anatomical and functional regions that are of interest in tissue engineering and regenerative medicine. We are shaping the desired cell type’s shape and function and, of course, it is difficult to predict how this shapes the future. However, some studies that have investigated mainly MSCs, but whose punchline should not be limited to MSCs, may help envision future applications. In a classical, well-known study, MSCs plated on polyacrylamide gels, across a wide range elasticities, differentiated into different lineage directions in the absence of exogenous soluble factors. The type of lineage was shown to correspond to specific ranges of the elastic properties of the material. Moreover, through the addition of blebbistatin, which prevents the cell from building intracellular cytoskeleton tension, differentiation by matrix-mechanics was abolished [41]. This study highlighted that matrix elasticity is a crucial factor in determining differentiation lineage and concurrent cell shape. Another study investigated the effect of limiting the adhesion site area for cell growth. Specific growth areas of low and high compactness had considerable effects on cell shape and MSC differentiation behavior [63]. On areas of low compactness, the cells were exposed to less intracellular forces than on areas of high compactness, which directed differentiation into specific phenotypes. Hence, cells restricted to growing on areas of low compactness–reflecting relatively lower intracellular forces-developed into adipose tissue, whereas cells restricted to areas of high compactness–reflecting relatively higher intracellular forces-developed towards cartilage and bone. Another group demonstrated that cells growing on aligned topographical surfaces also influenced cell shape and differentiation [64]. This group used adipose-derived stem cells and cultured them on so-called “random” and “aligned” topographies. They achieved cell differentiation towards a progenitor muscle state on aligned topographies and suggested that an elongated cell shape was due to the confinement of the growth area. Collectively, these studies offer valuable insights into how architectural environments can help steering cell fate.
The well-accepted concept that changes in extracellular forces would prompt a subsequent change in the intracellular forces also suggests that extracellular forces can be harnessed to engineer specific cellular shapes. Indeed, our group showed in a recent study that MSCs exhibit a specific baseline morphology that is associated with the biomaterial type and nanoscale stiffness of a given biomaterial surface [19] and one can intentionally generate a desired baseline shape using this concept. Moreover, our group demonstrated in Walters et al. [24] that defined changes in externally applied biomechanical forces also prompt a subsequent, defined change in MSC shape. By applying various regimens of cyclic tensional forces we were able to engineer distinctly different MSC shapes. Interestingly, cyclic tension not only had time- and amplitude-dependent effects on cell shape, but the key to biomechanically engineering cellular shape was the repetition of a chosen stretch regimen. This demonstrated that the engineered shape and the associated MSC differentiation were complex non-linear processes that is dependent on both active stress input and inactive response time. Indeed, it was shown in [19] that MSCs can be elongated and spreading changes during cyclic stretch, but, after cessation of stretch, the shape reverts back toward the initial baseline shape determined by the biomaterial type and stiffness. It is important to the field that, in the chosen experimental system, dynamic tensile forces were shown to be more significant in defining MSC shape than biomaterial stiffness-related cues [19]. It remains to be seen whether this is a general principle highlighting the potential of harnessing biomechanical forces for controlling cell shape, or whether this result represents a study-specific observation. From these studies, it has become clear that MSC shape can be statically engineered through using e.g., micro-engineered adhesion sites, but MSC shape can also be highly dynamic. Under biomechanically dynamic conditions, such as in a patient’s body, MSC shape can be intentionally designed through biomaterial type and nanoscale stiffness and while also accounting for dynamic forces experienced in vivo.
Overall, insight on how one can manipulate cell shape and cell function is crucial for designing better cell-instructive scaffolds for in situ applications. Using biomaterials with lineage-specific biophysical cues could serve as a starting-point to develop cell-based regenerative therapies [65], which would determine the lineage of differentiation locally through the implant properties. Perhaps, one could reach this goal by integrating the here discussed various shape-controlling biophysical cues into a single biomaterial, e.g., a biomaterial with defined adhesion sites, nano-topography, biomaterial stiffness and viscoelasticity, and specific force-transducing properties that harness the effects of biomechanical forces abundant in our body. Naturally, it would be necessary to extend the here discussed biological and computational concepts to the multitude of cell types and tissues that are the target of worldwide regenerative medicine efforts to generate better and clinically useful therapeutic or preventive interventions. For this task, an impressive range of methods is at our disposal, and it will be interesting to see how these will shape our future. Hence, using advanced segmentation methods and computational approaches to measure cell morphology for classifying shape profiles that correspond to specific biological responses (Table 1) will become ever more important tools for, ultimately, controlling cell shape and fate for regenerative medicine applications.

Table 1.
Overview of quantitatively measured shape descriptors for assessing cell morphology, the stimuli used for controlling shape, and the correlated biological functions. Various cell types such as mesenchymal stromal cells (MSCs) or primary myoblasts can be actively shaped by biophysical and biomechanical forces. The morphological phenotype can be visualized by either staining the cells and subsequently taking fluorescence images or using phase-contrast images for non-labeled cells. Image processing can then be performed using computational tools, e.g., ImageJ, MATLAB, or CellProfiler, calculating shape descriptors. The measured shape descriptors are listed in alphabetical order. For detailed definitions of listed shape descriptors, refer to each reference. Quantitatively obtained cell and nuclear shape features can be used to predict biological responses such as MSC differentiation into specific lineages, physiology and health of primary cells, and even tissue injury. Due to the broad application range and their correlation to biological functions, quantitative shape descriptors contribute vastly to progress in regenerative medicine. Species (Sp.) abbreviations: Human (hu), mouse/murine (mu). Cell type abbreviations: Adipose-derived stem cells (APCs), embryonic stem cells (ESCs), human umbilical vein endothelial cells (HUVECs), induced pluripotent stem cells (iPSCs), oligodendrocyte precursor cells (OPCs), smooth muscle cell (SMC).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bianco, P. “Mesenchymal” stem cells. Annu. Rev. Cell. Dev. Biol. 2014, 30, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Hart, M.L.; Brinchmann, J.E.; Rolauffs, B.; Stenzl, A.; Sievert, K.-D.; Aicher, W.K. Mesenchymal stromal cells for sphincter regeneration. Adv. Drug Deliv. Rev. 2015, 82–83C, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Caplan, A.I.; Mason, C.; Reeve, B. The 3Rs of Cell Therapy. Stem Cells Transl. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.; Cao, X.; Frenette, P.S.; Mao, J.J.; Robey, P.G.; Simmons, P.J.; Wang, C.Y. The meaning, the sense and the significance: Translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013, 19, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Iskratsch, T.; Wolfenson, H.; Sheetz, M.P. Appreciating force and shape-the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 2014, 15, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.K.; Nelson, C.M.; Biais, N.; Fabry, B.; Moeller, J.; Pruitt, B.L.; Wollnik, C.; Kudryasheva, G.; Rehfeldt, F.; Federle, W. Mechanotransduction: Use the force(s). BMC Biol. 2015, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Strzyz, P. Mechanotransduction: May the force be with you. Nat. Rev. Mol. Cell Biol. 2016, 17, 533. [Google Scholar] [CrossRef] [PubMed]
- Albert, P.J.; Schwarz, U.S. Modeling cell shape and dynamics on micropatterns. Cell Adhes. Migr. 2016, 10, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Relan, N.K.; Przywara, D.A.; Schuger, L. Embryonic mesenchymal cells share the potential for smooth muscle differentiation: Myogenesis is controlled by the cell’s shape. Development 1999, 126, 3027–3033. [Google Scholar] [PubMed]
- Manasek, F.J.; Burnside, M.B.; Waterman, R.E. Myocardial cell shape change as a mechanism of embryonic heart looping. Dev. Biol. 1972, 29, 349–371. [Google Scholar] [CrossRef]
- Gao, L.; McBeath, R.; Chen, C.S. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells 2010, 28, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, R.; Kumar, A.; Lopez, G.P.; Stephanopoulos, G.N.; Wang, D.I.; Whitesides, G.M.; Ingber, D.E. Engineering cell shape and function. Science 1994, 264, 696–698. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J.; Moscona, A. Role of cell shape in growth control. Nature 1978, 273, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Costa-Almeida, R.; Carvalho, D.T.O.; Ferreira, M.J.S.; Pesqueira, T.; Monici, M.; Van Loon, J.J.W.A.; Granja, P.L.; Gomes, M.E. Simulated hypergravity induces changes in human tendon-derived cells: From cell morphology to gene expression. In eCells & Materials (eCM) Meeting Abstracts 2017, Collection 2. Personalised Therapies for Regenerative Medicine, TERMIS-EU 2017 Conference, Davos, Switzerland, 26–30 June 2017; AO Research Institute Davos: Davos, Switzerland, 2017; p. 429. [Google Scholar]
- Zhang, J.; Rubert, M.; Baumgartner, J.; Wehrle, E.; Paul, G.; Müller, R. The influence of mechanical properties of 3D bioprinted hMSCs-laden alginate composite scaffolds on cell viability and morphology. In eCells & Materials (eCM) Meeting Abstracts 2017, Collection 2. Personalised Therapies for Regenerative Medicine, TERMIS-EU 2017 Conference, Davos, Switzerland, 26–30 June 2017; AO Research Institute Davos: Davos, Switzerland, 2017; p. 608. [Google Scholar]
- Rocca, A.; Marino, A.; Rocca, V.; Moscato, S.; De Vito, G.; Piazza, V.; Mazzolai, B.; Mattoli, V.; Ngo-Anh, T.J.; Ciofani, G. Barium titanate nanoparticles and hypergravity stimulation improve differentiation of mesenchymal stem cells into osteoblasts. Int. J. Nanomed. 2015, 10, 433–445. [Google Scholar]
- Jain, N.; Vogel, V. Cell geometry regulates temporal regulation of gene expression during macrophage activation. In eCells & Materials (eCM) Meeting Abstracts 2017, Collection 2. Personalised Therapies for Regenerative Medicine TERMIS-EU 2017 Conference, Davos, Switzerland 26–30 June 2017; AO Research Institute Davos: Davos, Switzerland, 2017. [Google Scholar]
- Hart, M.L.; Schmid, L.; Walters, B.; Rolauffs, B. Implementation and verification of an efficient, precise, reproducible and accurate automatic quantification method for measuring mesenchymal stromal cell morphological parameters. In eCells & Materials (eCM) Meeting Abstracts 2017, Collection 2. Personalised Therapies for Regenerative Medicine TERMIS-EU 2017 Conference, Davos, Switzerland, 26–30 June 2017; AO Research Institute Davos: Davos, Switzerland, 2017; p. 567. [Google Scholar]
- Uynuk-Ool, T.; Rothdiener, M.; Walters, B.; Hegemann, M.; Palm, J.; Nguyen, P.; Seeger, T.; Stöckle, U.; Stegemann, J.P.; Aicher, W.K.; et al. The geometrical shape of mesenchymal stromal cells measured by quantitative shape descriptors is determined by the stiffness of the biomaterial and by cyclic tensile forces. J. Tissue Eng. Regen. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kato, R. Application and understanding of morphological data for non-invasive quality control of stem cell manufacturing. In eCells & Materials (eCM) Meeting Abstracts 2017, Collection 2. Personalised Therapies for Regenerative Medicine TERMIS-EU 2017 Conference, 26–30 June 2017, Davos, Switzerland; AO Research Institute Davos: Davos, Switzerland, 2017; p. 798. [Google Scholar]
- Fujitani, M.; Kawai, S.; Kanie, K.R.K. Morphology-based real-time evaluation of culture condition. In eCells & Materials (eCM) Meeting Abstracts 2017, Collection 2. Personalised Therapies for Regenerative Medicine TERMIS-EU 2017 Conference, Davos, Switzerland, 26–30 June 2017; AO Research Institute Davos: Davos, Switzerland, 2017; p. 797. [Google Scholar]
- Kato, R.; Matsumoto, M.; Sasaki, H.; Joto, R.; Okada, M.; Ikeda, Y.; Kanie, K.; Suga, M.; Kinehara, M.; Yanagihara, K.; et al. Parametric analysis of colony morphology of non-labelled live human pluripotent stem cells for cell quality control. Sci. Rep. 2016, 6, 34009. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Takeuchi, I.; Okada, M.; Sawada, R.; Kanie, K.; Kiyota, Y.; Honda, H.; Kato, R. Label-free morphology-based prediction of multiple differentiation potentials of human mesenchymal stem cells for early evaluation of intact cells. PLoS ONE 2014, 9, e93952. [Google Scholar] [CrossRef] [PubMed]
- Walters, B.; Uynuk-Ool, T.; Rothdiener, M.; Palm, J.; Hart, M.L.; Stegemann, J.P.; Rolauffs, B. Engineering the geometrical shape of mesenchymal stromal cells through defined cyclic stretch regimens. Sci. Rep. 2017, 7, 6640. [Google Scholar] [CrossRef] [PubMed]
- Marklein, R.A.; Lo Surdo, J.L.; Bellayr, I.H.; Godil, S.A.; Puri, R.K.; Bauer, S.R. High Content Imaging of Early Morphological Signatures Predicts Long Term Mineralization Capacity of Human Mesenchymal Stem Cells upon Osteogenic Induction. Stem Cells 2016, 34, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, F.; Takeuchi, I.; Agata, H.; Kagami, H.; Shiono, H.; Kiyota, Y.; Honda, H.; Kato, R. Morphology-based prediction of osteogenic differentiation potential of human mesenchymal stem cells. PLoS ONE 2013, 8, e55082. [Google Scholar] [CrossRef] [PubMed]
- Seiler, C.; Gazdhar, A.; Reyes, M.; Benneker, L.M.; Geiser, T.; Siebenrock, K.A.; Gantenbein-Ritter, B. Time-lapse microscopy and classification of 2D human mesenchymal stem cells based on cell shape picks up myogenic from osteogenic and adipogenic differentiation. J. Tissue Eng. Regen. Med. 2014, 8, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Pearson, Y.E.; Lund, A.W.; Lin, A.W.; Ng, C.P.; Alsuwaidi, A.; Azzeh, S.; Gater, D.L.; Teo, J.C. Non-invasive single-cell biomechanical analysis using live-imaging datasets. J. Cell Sci. 2016, 129, 3351–3364. [Google Scholar] [CrossRef] [PubMed]
- Zanier, E.R.; Fumagalli, S.; Perego, C.; Pischiutta, F.; De Simoni, M.G. Shape descriptors of the “never resting” microglia in three different acute brain injury models in mice. Intensive Care Med. Exp. 2015, 3, 39. [Google Scholar] [CrossRef] [PubMed]
- Comin, C.H.; Xu, X.; Wang, Y.; Costa Lda, F.; Yang, Z. An image processing approach to analyze morphological features of microscopic images of muscle fibers. Comput. Med. Imaging Graph. 2014, 38, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Dahl, K.N.; Ribeiro, A.J.; Lammerding, J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008, 102, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Lozoya, O.A.; Gilchrist, C.L.; Guilak, F. Universally Conserved Relationships between Nuclear Shape and Cytoplasmic Mechanical Properties in Human Stem Cells. Sci. Rep. 2016, 6, 23047. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Gordonov, S.; Treiser, M.D.; Moghe, P.V. Parsing the early cytoskeletal and nuclear organizational cues that demarcate stem cell lineages. Cell Cycle 2010, 9, 2108–2117. [Google Scholar] [CrossRef] [PubMed]
- Vega, S.L.; Liu, E.; Arvind, V.; Bushman, J.; Sung, H.J.; Becker, M.L.; Lelievre, S.; Kohn, J.; Vidi, P.A.; Moghe, P.V. High-content image informatics of the structural nuclear protein NuMA parses trajectories for stem/progenitor cell lineages and oncogenic transformation. Exp. Cell Res. 2017, 351, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Vega, S.L.; Liu, E.; Patel, P.J.; Kulesa, A.B.; Carlson, A.L.; Ma, Y.; Becker, M.L.; Moghe, P.V. High-content imaging-based screening of microenvironment-induced changes to stem cells. J. Biomol. Screen 2012, 17, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Nikkhah, M.; Edalat, F.; Manoucheri, S.; Khademhosseini, A. Engineering microscale topographies to control the cell-substrate interface. Biomaterials 2012, 33, 5230–5246. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, C.S.; Fu, J. Forcing stem cells to behave: A biophysical perspective of the cellular microenvironment. Annu. Rev. Biophys. 2012, 41, 519–542. [Google Scholar] [CrossRef] [PubMed]
- Brun, J.; Abruzzese, T.; Rolauffs, B.; Aicher, W.K.; Hart, M.L. Choice of xenogenic-free expansion media significantly influences the myogenic differentiation potential of human bone marrow-derived mesenchymal stromal cells. Cytotherapy 2016, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Brun, J.; Lutz, K.A.; Neumayer, K.M.; Klein, G.; Seeger, T.; Uynuk-Ool, T.; Worgotter, K.; Schmid, S.; Kraushaar, U.; Guenther, E.; et al. Smooth Muscle-Like Cells Generated from Human Mesenchymal Stromal Cells Display Marker Gene Expression and Electrophysiological Competence Comparable to Bladder Smooth Muscle Cells. PLoS ONE 2015, 10, e0145153. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Khalili, A.A.; Ahmad, M.R. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. Int. J. Mol. Sci. 2015, 16, 18149–18184. [Google Scholar] [CrossRef] [PubMed]
- Pincus, Z.; Theriot, J.A. Comparison of quantitative methods for cell-shape analysis. J. Microsc. 2007, 227, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Dzyubachyk, O.; Smal, I.; Van Cappellen, W.A. Tracking in cell and developmental biology. Semin. Cell Dev. Biol. 2009, 20, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yu, S.; Yang, Y.; Xie, Y. Feature and contrast enhancement of mammographic image based on multiscale analysis and morphology. Comput. Math. Methods Med. 2013, 2013, 716948. [Google Scholar] [CrossRef] [PubMed]
- Jirapatnakul, A.C.; Fotin, S.V.; Reeves, A.P.; Biancardi, A.M.; Yankelevitz, D.F.; Henschke, C.I. Automated nodule location and size estimation using a multi-scale Laplacian of Gaussian filtering approach. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009, 2009, 1028–1031. [Google Scholar] [PubMed]
- Kong, H.; Akakin, H.C.; Sarma, S.E. A generalized Laplacian of Gaussian filter for blob detection and its applications. IEEE Trans. Cybern. 2013, 43, 1719–1733. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E. Cell Segmentation: 50 Years Down the Road. IEEE Signal Process. Mag. 2012, 29, 140–145. [Google Scholar] [CrossRef]
- Krause, M.; Gautreau, A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, W.L.; Anglin, L.W. Metamorph: Computer Support for Qualitative Research. Mid-West. Educ. Res. 1993, 6, 30–34. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Soliman, K. CellProfiler: Novel Automated Image Segmentation Procedure for Super-Resolution Microscopy. Biol. Proced. Online 2015, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Sommer, C.; Gerlich, D.W. Machine learning in cell biology—Teaching computers to recognize phenotypes. J. Cell Sci. 2013, 126, 5529–5539. [Google Scholar] [CrossRef] [PubMed]
- Grys, B.T.; Lo, D.S.; Sahin, N.; Kraus, O.Z.; Morris, Q.; Boone, C.; Andrews, B.J. Machine learning and computer vision approaches for phenotypic profiling. J. Cell Biol. 2017, 216, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.J.; Shan, J.; Bhatia, S.N.; Carpenter, A.E. Quantifying co-cultured cell phenotypes in high-throughput using pixel-based classification. Methods 2016, 96, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Schwartz, R.E.; Ross, N.T.; Logan, D.J.; Thomas, D.; Duncan, S.A.; North, T.E.; Goessling, W.; Carpenter, A.E.; Bhatia, S.N. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013, 9, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Hilsenbeck, O.; Schwarzfischer, M.; Loeffler, D.; Dimopoulos, S.; Hastreiter, S.; Marr, C.; Theis, F.J.; Schroeder, T. fastER: A user-friendly tool for ultrafast and robust cell segmentation in large-scale microscopy. Bioinformatics 2017, 33, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Juneau, P.M.; Garnier, A.; Duchesne, C. Monitoring of adherent live cells morphology using the undecimated wavelet transform multivariate image analysis (UWT-MIA). Biotechnol. Bioeng. 2017, 114, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Kandhavelu, M.; Yli-Harja, O.; Ribeiro, A.S. Cell segmentation by multi-resolution analysis and maximum likelihood estimation (MAMLE). BMC Bioinform. 2013, 14, S8. [Google Scholar]
- Jaccard, N.; Griffin, L.D.; Keser, A.; Macown, R.J.; Super, A.; Veraitch, F.S.; Szita, N. Automated method for the rapid and precise estimation of adherent cell culture characteristics from phase contrast microscopy images. Biotechnol. Bioeng. 2014, 111, 504–517. [Google Scholar] [CrossRef] [PubMed]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef]
- Newman, P.; Galenano-Nino, J.L.; Graney, P.; Razal, J.M.; Minett, A.I.; Ribas, J.; Ovalle-Robles, R.; Biro, M.; Zreiqat, H. Relationship between nanotopographical alignment and stem cell fate with live imaging and shape analysis. Sci. Rep. 2016, 6, 37909. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Abdeen, A.A.; Kilian, K.A. Rewiring mesenchymal stem cell lineage specification by switching the biophysical microenvironment. Sci. Rep. 2014, 4, 5188. [Google Scholar] [CrossRef] [PubMed]
- Costa-Almeida, R.; Carvalho, D.T.; Ferreira, M.J.; Aresta, G.; Gomes, M.E.; van Loon, J.J.W.A.; Van der Heiden, K.; Granja, P.L. Effects of hypergravity on the angiogenic potential of endothelial cells. J. R. Soc. Interface 2016, 13. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).