ZF-AutoML: An Easy Machine-Learning-Based Method to Detect Anomalies in Fluorescent-Labelled Zebrafish
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethic Approval
2.2. Zebrafish Experiments
2.3. Image Capture
- 4× objective lens with 10× eyepieces (40× total magnification).
- High-resolution mode (1920 × 1440 px).
- Exposure time; brightfield: 1/7500 s, green fluorescent protein
- (GFP: Ex 470/40, Em 525/50): 1.2 s.
- The orientation of zebrafish was random.
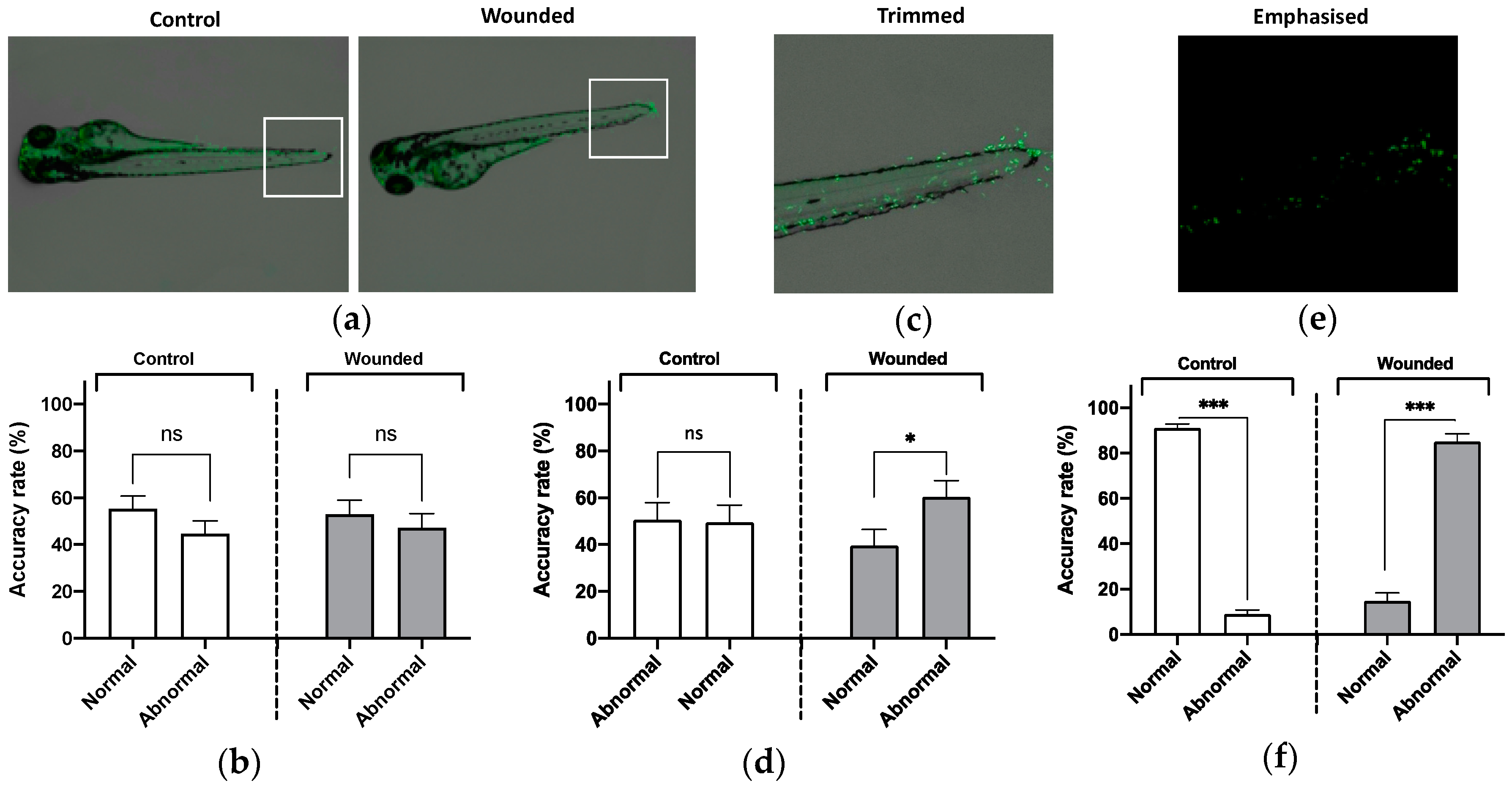
2.4. Image Processing
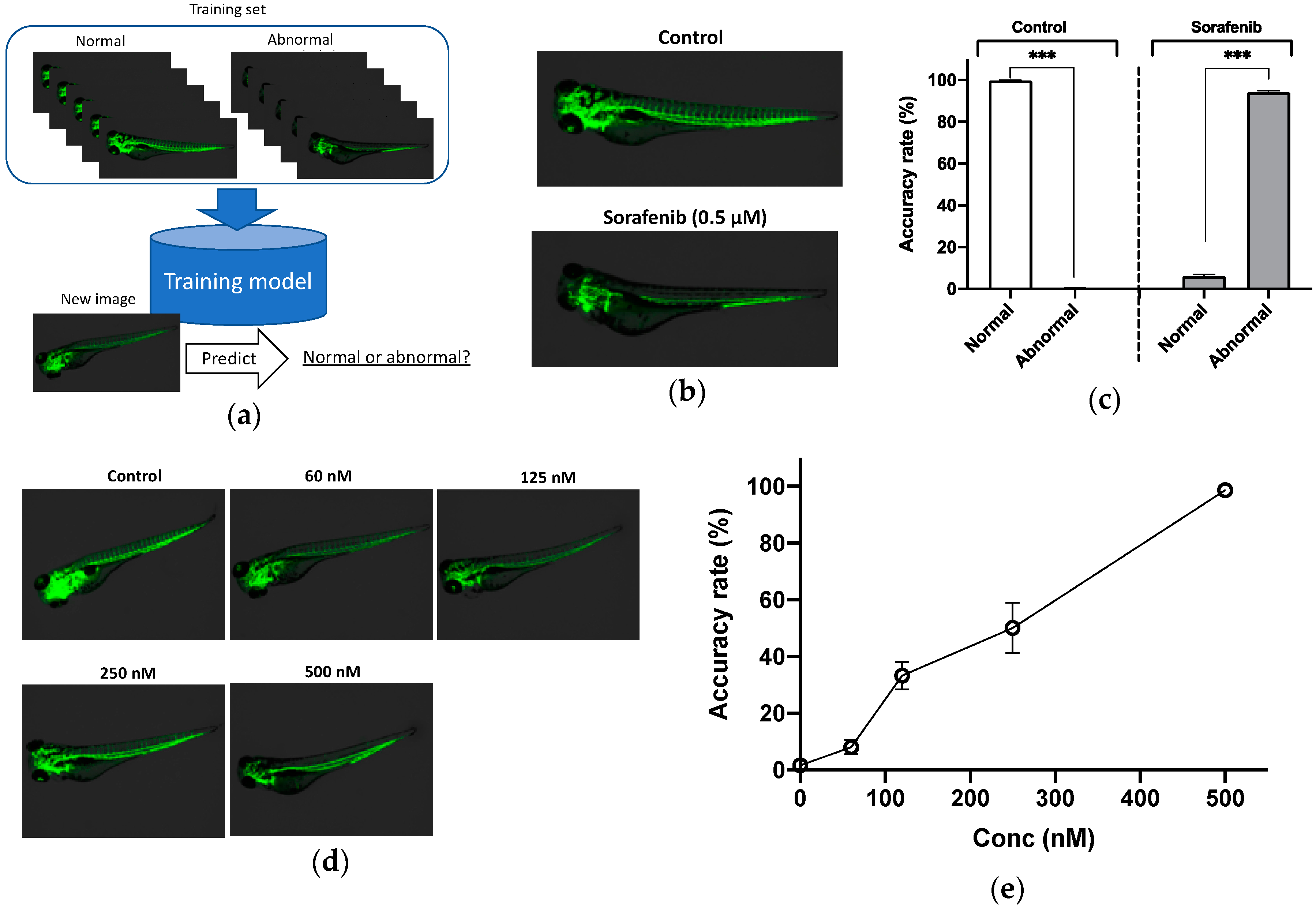
2.5. Machine Learning
2.6. ZF-ImageR
- from google.cloud import automl_v1beta1 .... (1)
- prediction_client = automl_v1beta1.PredictionServiceClient() .... (2)
- prediction_client = prediction_client.from_service_account_json(KEY_FILE) .... (3)
- name = ‘projects/{}/locations/us-central1/models/{}’.format(project_id, model_id) .... (4)
- payload = {‘image’: {‘image_bytes’: content}} .... (5)
- request = prediction_client.predict(name, payload) .... (6)
- print(request) .... (7)
- (1)
- Import Python library for Google Cloud.
- (2)
- Create Instance object of AutoML prediction service client.
- (3)
- Import Google Cloud service account key file which is required for accessing GCP services.
- (4)
- Set AutoML prediction model’s ID which is required for (6).
- (5)
- Set Image data to post to AutoML for prediction which is required for (6).
- (6)
- Send a request to Google Cloud AutoML server for prediction of an image data from an service client which is prepared on (2).
- (7)
- Output prediction result.
3. Results
3.1. Evaluation of Zebrafish after Treatment with Anti-Angiogenesis Drug
3.2. Detection of Macrophage Abnormalities Using Machine Learning.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lessman, C.A.; Taylor, M.R.; Orisme, W.; Carver, E.A. Use of flatbed transparency scanners in zebrafish research: versatile and economical adjuncts to traditional imaging tools for the Danio rerio laboratory. Methods Cell Biol. 2010, 100, 295–322. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Liang, J.; Zheng, X.; Zhang, K.; Zhao, Y. Using a high-throughput zebrafish embryo screening approach to support environmental hazard ranking for cardiovascular agents. Sci. Total Environ. 2019, 702, 134703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shimada, Y.; Kuroyanagi, J.; Umemoto, N.; Nishimura, Y.; Tanaka, T. Quantitative phenotyping-based in vivo chemical screening in a zebrafish model of leukemia stem cell xenotransplantation. PLoS ONE 2014, 9, e85439. [Google Scholar] [CrossRef] [PubMed]
- Sittampalam, G.S.; Grossman, A.; Brimacombe, K.; Arkin, M.; Auld, D.; Austin, C.P.; Baell, J.; Bejcek, B.; Caaveiro, J.M.M.; Chung, T.D.Y.; et al. Assay Guidance Manual; NCBI: Bethesda, MD, USA; p. 2004.
- Yamamoto, D.; Sato, D.; Nakayama, H.; Nakagawa, Y.; Shimada, Y. ZF-Mapper: Simple and Complete Freeware for Fluorescence Quantification in Zebrafish Images. Zebrafish 2019. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, X.; Xu, M.; Peng, J.R.; Setiono, R. A hybrid SOM-SVM approach for the zebrafish gene expression analysis. Genom. Proteom. Bioinform. 2005, 3, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, W.; Xie, Y.; Wang, W.J.; Li, L.L.; Yang, S.Y. Rapid and accurate assessment of seizure liability of drugs by using an optimal support vector machine method. Toxicol. In Vitro 2011, 25, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Mirat, O.; Sternberg, J.R.; Severi, K.E.; Wyart, C. ZebraZoom: an automated program for high-throughput behavioral analysis and categorization. Front. Neural Circuits 2013, 7, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikut, R.; Dickmeis, T.; Driever, W.; Geurts, P.; Hamprecht, F.A.; Kausler, B.X.; Ledesma-Carbayo, M.J.; Marée, R.; Mikula, K.; Pantazis, P.; et al. Automated processing of zebrafish imaging data: a survey. Zebrafish 2013, 10, 401–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronneberger, O.; Liu, K.; Rath, M.; Rueβ, D.; Mueller, T.; Skibbe, H.; Drayer, B.; Schmidt, T.; Filippi, A.; Nitschke, R.; et al. ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat. Methods 2012, 9, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, O.; Sadanandan, S.K.; Wahlby, C. Deep Fish: Deep Learning-Based Classification of Zebrafish Deformation for High-Throughput Screening. SLAS Discov. 2017, 22, 102–107. [Google Scholar] [CrossRef] [Green Version]
- Cordero-Maldonado, M.L.; Perathoner, S.; van der Kolk, K.J.; Boland, R.; Heins-Marroquin, U.; Spaink, H.P.; Meijer, A.H.; Crawford, A.D.; de Sonneville, J. Deep learning image recognition enables efficient genome editing in zebrafish by automated injections. PLoS ONE 2019, 14, e0202377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussmann, J.; Bakkers, J.; Schulte-Merker, S. Early endocardial morphogenesis requires Scl/Tal1. PLoS Genet. 2007, 3, e140. [Google Scholar] [CrossRef] [PubMed]
- Chimote, G.; Sreenivasan, J.; Pawar, N.; Subramanian, J.; Sivaramakrishnan, H.; Sharma, S. Comparison of effects of anti-angiogenic agents in the zebrafish efficacy-toxicity model for translational anti-angiogenic drug discovery. Drug Des. Dev. Ther. 2014, 8, 1107–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen-Chi, M.; Laplace-Builhe, B.; Travnickova, J.; Luz-Crawford, P.; Tejedor, G.; Lutfalla, G.; Kissa, K.; Jorgensen, C.; Djouad, F. TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell Death Dis. 2017, 8, 12. [Google Scholar] [CrossRef] [PubMed]
| Experiment | Normal Phenotypes | Abnormal Phenotypes |
|---|---|---|
| Angiogenesis 1 | 47 | 65 |
| Macrophage 2 | 104 | 156 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawaki, R.; Sato, D.; Nakayama, H.; Nakagawa, Y.; Shimada, Y. ZF-AutoML: An Easy Machine-Learning-Based Method to Detect Anomalies in Fluorescent-Labelled Zebrafish. Inventions 2019, 4, 72. https://doi.org/10.3390/inventions4040072
Sawaki R, Sato D, Nakayama H, Nakagawa Y, Shimada Y. ZF-AutoML: An Easy Machine-Learning-Based Method to Detect Anomalies in Fluorescent-Labelled Zebrafish. Inventions. 2019; 4(4):72. https://doi.org/10.3390/inventions4040072
Chicago/Turabian StyleSawaki, Ryota, Daisuke Sato, Hiroko Nakayama, Yuki Nakagawa, and Yasuhito Shimada. 2019. "ZF-AutoML: An Easy Machine-Learning-Based Method to Detect Anomalies in Fluorescent-Labelled Zebrafish" Inventions 4, no. 4: 72. https://doi.org/10.3390/inventions4040072
APA StyleSawaki, R., Sato, D., Nakayama, H., Nakagawa, Y., & Shimada, Y. (2019). ZF-AutoML: An Easy Machine-Learning-Based Method to Detect Anomalies in Fluorescent-Labelled Zebrafish. Inventions, 4(4), 72. https://doi.org/10.3390/inventions4040072






