1. Introduction
The advent of large language models (LLMs) has transformed how researchers interact with biomedical literature, particularly in high-information domains like healthcare [
1]. In radiology, LLMs show promise as research tools capable of assisting with literature synthesis, protocol development, and hypothesis generation [
2]. However, their adoption within clinical research environments remains limited due to privacy concerns, infrastructure constraints, and lack of domain-specific adaptation.
General-purpose models such as GPT-4-Consensus attempt to mitigate hallucination by citing published sources [
3]. Yet, these models remain externally hosted, making them incompatible with institutional use cases that require strict data privacy and security guarantees. Additionally, broad training corpora result in limited radiology-specific comprehension, leading to outputs that often lack the precision or contextual appropriateness needed in this field [
4].
Prior work has also highlighted the importance of ethical oversight and bias mitigation when deploying artificial intelligence (AI) systems in radiology, where LLM-driven tools may influence clinical decision making [
5]. Despite their impressive capabilities, LLMs are at risk of “stochastic parroting”—regurgitating training data without true understanding [
6]. Public-facing tools like GPT-4-Consensusfurther introduce the risk of overconfidence and misuse by patients who may rely on oversimplified or inaccurate outputs. These challenges necessitate privacy-compliant, auditable, and locally controlled systems.
Retrieval-augmented generation (RAG) system architectures enhance LLM responses by incorporating semantically relevant context from a curated literature corpus rather than relying solely on pre-trained weights [
7]. This improves domain specificity and transparency while also enabling institutions to retain control over input, retrieval, and output. Local, domain-specific RAG systems can serve as viable and secure alternatives to commercial LLMs [
8]. This aspect of RAG systems allows for deployment within an institution without additional external API calls or data transfer.
Prompt engineering, defined as crafting and writing inputs to guide LLMs, increasingly facilitates their skilled use while improving user experience. By tailoring prompts, users can get more precise responses with topic-specific requirements; for example, it can improve the quality and acceptability of LLM-generated responses in a clinical setting [
9,
10].
To determine if RAG models can effectively support radiology research, we developed a secure, radiology-specific RAG system deployed entirely within our institutional infrastructure. Our RAG system leverages a database of 167,028 radiology-related abstracts sourced from PubMed. Prompts answered by both our RAG LLM and GPT-4-Consensus models were evaluated for factual accuracy (FA), citation relevance (CR), and perceived performance (PP). While prior work has used similar scoring frameworks—such as factuality, citation relevance, and readability [
11]—to assess LLM outputs, our study emphasizes not only the objective quality of responses but also the user-perceived utility in the context of radiology research. This paper presents the system design, implementation and evaluation processes, and broader implications of RAG models in radiology clinical research environments.
2. Materials and Methods
2.1. Data Collection via Webscraping
To construct a domain-specific knowledge base, we programmatically extracted radiology-related article metadata and abstracts from PubMed using BeautifulSoup [
12]. PubMed provides public access to abstracts and metadata (titles, authors, affiliations, journals, dates, and keywords), while restricting access to full texts. Due to API limitations and pagination constraints (maximum 10,000 “best matching” results per query), search queries were issued, in compliance with PubMed policies, separately for each calendar year from 2000 through February 2024 using the case-insensitive keyword “radiology.” The year 2000 was chosen as a natural lower bound based on article volume and indexing consistency. For each query, all resulting article URLs were enumerated and asynchronously parsed to extract the title, abstract, authors, affiliations, journal, publication date, keywords, and URL.
Postprocessing involved aggregating raw CSVs and deduplicating entries by title, abstract, and URL. Records without abstracts were excluded. The final dataset comprised 167,028 unique articles spanning 2000–2024, totaling 331 MB, with an average of ~6700 usable entries per year;
Figure 1 shows the exact distribution. Webscraping took approximately 7 days of continuous execution using 4 CPU cores and 8 GB of RAM.
2.2. Embedding and Vector Database Construction
To enable semantic retrieval, each entry in the curated PubMed-derived dataset was embedded into a high-dimensional vector space using the state-of-the-art 7B parameter instruction-tuned model “intfloat/e5-mistral-7b-instruct” [
13], made available through HuggingFace [
14]. This model was selected based on its performance on the Massive Text Embedding Benchmark, where it consistently outperformed other open-weight embedding models of similar size [
15]. It can embed an input size up to 4096 tokens and is optimized for dense retrieval tasks. Abstracts and associated metadata (title, journal, date, etc.) were serialized into string representations and tokenized for embedding generation. Embeddings were normalized and stored in a persistent, disk-backed ChromaDB vector database [
16].
ChromaDB was chosen for its native Python 3.9 support, ease of integration, and efficient handling of up to 1 million entries with low latency. Its support for local filesystem storage aligns with institutional constraints on cloud-based tools. Each record in the ChromaDB collection was stored with an associated unique ID, original metadata, and full document string to support future auditing and traceability.
The embedding process was conducted in batches using 4-bit quantized [
17] inference on an NVIDIA A100 GPU with flash attention enabled [
18], 4 CPU cores, and 32 GB of RAM. Vector generation and ingestion into ChromaDB took approximately 8 h.
The vectorized prompt embedding was compared with stored vector embeddings, and the top similar abstracts were selected using the k-nearest neighbor (kNN) classifier [
19]. Similarity was measured with cosine similarity metrics:
where
and
are two vectors with
elements and
is the cosine similarity [
20].
is for vector transpose.
This semantic retrieval mechanism surfaced conceptually similar documents regardless of vocabulary mismatch, paraphrasing, or surface-level token overlap. By retrieving vectors that are meaningfully similar rather than merely textually similar, the system mitigated irrelevant context and reduced the LLM’s reliance on hallucinated content or unsupported prior assumptions [
21]. In effect, high-quality retrieval shifts the burden of reasoning to grounded, verifiable evidence.
2.3. LLM Integration and Prompt Engineering
For response generation, we integrated the open-weight, instruction-tuned language model mistral-7b-instruct-v0.2, which outperforms several 13B parameter models in instruction-following tasks [
22,
23]. This model, also made available through HuggingFace, was selected for its strong performance in general-purpose language understanding paired with its ability to be run on a single 16 GB VRAM GPU via 4-bit quantization.
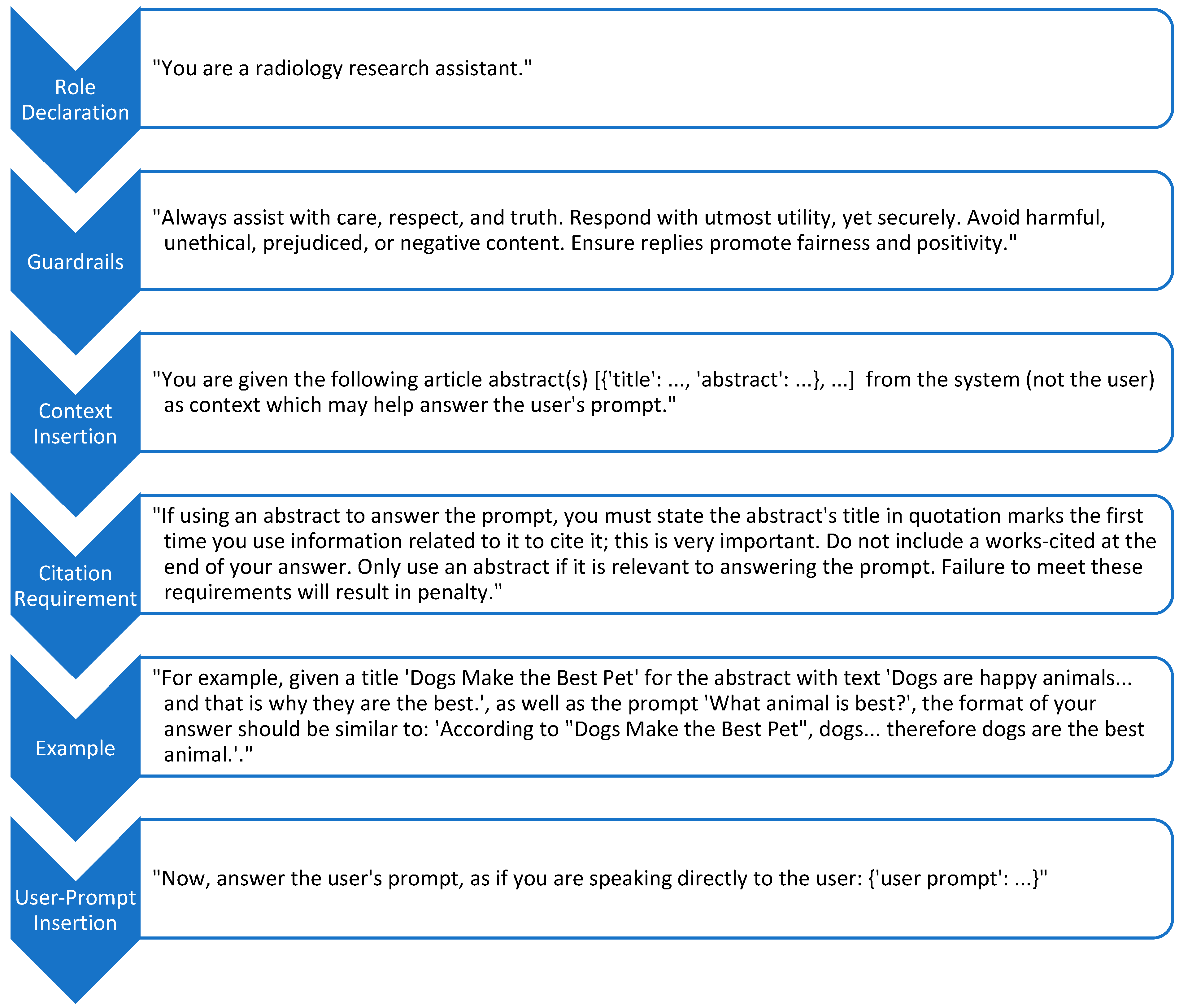
Prompt engineering played a central role in eliciting high-quality responses. The prompt template (
Figure 2) explicitly defined the model’s role as a radiology research assistant, imposed behavioral guardrails (e.g., respectfulness, ethical integrity, fairness), defined citation requirements for the inserted context, and included an example to guide output structure. The user’s question was appended at the end of the prompt to maintain instructional focus.
To provide factual grounding, the prompt was augmented with the top-k most semantically similar articles (k = 5 by default) retrieved from the ChromaDB vector database. We selected k = 5 based on a soft cap of ~2000 words (~3000 tokens) for context to avoid exceeding the 4096-token input limit of the generation model. If the combined length of retrieved abstracts exceeded the limit, fewer than five were used, with priority given to higher-ranked results. This approach ensured consistent retrieval latency and model performance across queries.
Each selected document was serialized to include only the title and abstract, formatted as a list of Python-like dictionaries to discourage hallucinated content. The LLM was instructed to reference source documents by title (in quotes) when incorporating claims, and a structured citation list was explicitly programmed to be appended to the end of each response for reliable transparency.
Inference was executed using 4-bit quantized weights on an NVIDIA 16 GB VRAM GPU with flash attention enabled for memory and throughput optimization. The generation parameters included a temperature of 0.2, top-k of 50, and top-p of 0.95 to balance determinism with creativity. Streaming token generation was implemented to support responsiveness in the user interface (UI).
2.4. System Deployment
This RAG system leverages only open-weight 7-billion parameter models and is fully runnable on a single 16 GB NVIDIA GPU. Both the embedding model and the generation model require approximately 6 GB of VRAM each, leaving sufficient headroom for runtime memory and intermediate buffers. This compact resource footprint allows the entire RAG system—including embedding, retrieval, and generation—to execute on a single GPU without requiring specialized hardware or distributed infrastructure.
The system, shown in
Figure 3, was deployed on an internal institutional server, ensuring that all computation—including vector embedding, semantic retrieval, and language generation—occurred in a secure environment without reliance on external APIs or data transfer. This design safeguarded patient privacy and institutional data security throughout the inference process.
To facilitate access and usability, the system was exposed to end users via a lightweight Python-based UI built with Gradio [
24], shown in
Figure 4. Gradio enabled rapid prototyping and deployment through a browser-accessible interface while maintaining full local execution. The interface allowed radiology researchers to submit free-text prompts, receive model-generated responses, and inspect the cited supporting abstracts.
2.5. Survey Design and Statistical Analysis
To assess the comparative quality of our RAG-based LLM system, we conducted a single-blinded evaluation study after IRB exemption. Twenty radiology-related prompts were collected from practicing radiologists and clinical researchers to ensure domain relevance and diversity of information needs.
Each prompt was submitted to both our local RAG LLM and GPT-4-Consensus, a publicly available state-of-the-art model. The outputs were randomized in order and anonymized to prevent any indication of the source. Participants—comprising radiology researchers and clinicians—were blinded to the model identity behind each response and instructed to evaluate their pair of answers independently.
Survey responses were collected using a structured three-question rubric. For each response, participants rated the following:
Factual Accuracy (FA): the degree to which the response was correct and free of hallucination;
Citation Relevance (CR): the alignment of the cited literature with the prompt and response content;
Perceived Performance (PP): the overall quality and usefulness of the response as perceived by the participant.
Each criterion was scored on a 5-point Likert scale (1 = poor, 5 = excellent) [
25]. Additionally, participants were asked to indicate their preferred response for their submitted prompt. After all prompts and responses were recorded, a board-certified radiologist reviewed the responses, counting the occurrence of hallucination.
Statistical analysis was conducted using the Wilcoxon signed-rank test [
26] to evaluate paired differences in Likert scores between the two systems across all prompts. A
p-value of less than 0.05 was considered statistically significant. Descriptive statistics (mean ± standard deviation [SD]) were calculated for each metric. Preferred response frequencies were also reported to assess overall user preference. The full list of prompts, responses, Likert scores, and output preferences is provided in the
Supplementary Materials.
3. Results
3.1. Performance Metrics
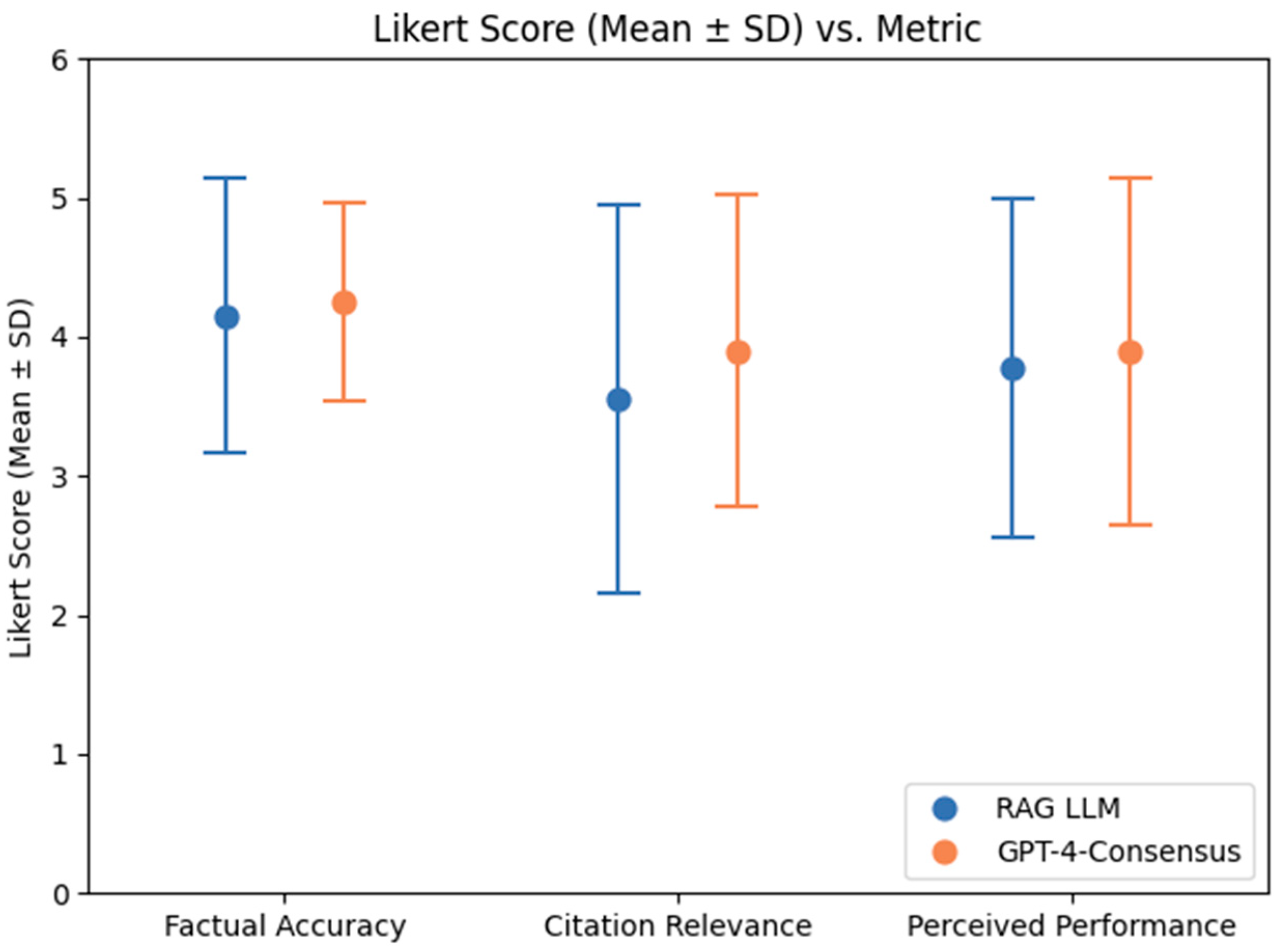
For the RAG system, the means ± SD of the FA, CR, and PP ratings were 4.15 ± 0.99, 3.70 ± 1.17, and 3.55 ± 1.39, respectively. For GPT-4-Consensus, they were 4.25 ± 0.72, 3.85 ± 1.23, and 3.90 ± 1.12, respectively. These Likert scores can be visualized in
Figure 5. No statistically significant differences were found between these ratings (
p = 0.97, 0.65, and 0.42, respectively).
3.2. Output Preference
In addition to scoring individual responses on specific criteria, participants were asked to indicate their overall preference between the two outputs for their prompt. Out of 20 prompts, preferences were evenly divided: 10 participants preferred the output generated by our local RAG LLM system, while the other 10 preferred the GPT-4-Consensus output.
Table 1 showcases the output preference for each prompt. A board-certified radiologist found no occurrence of hallucination in any of the outputs.
4. Discussion
This study demonstrates that a locally deployed RAG-based LLM system, operating entirely on institutional infrastructure and restricted to open-weight 7-billion parameter models, can achieve performance comparable to GPT-4-Consensus—a proprietary state-of-the-art language model. Quantitatively, the two systems received statistically indistinguishable scores across three key evaluation metrics: FA, CR, and PP, with p-values of 0.97, 0.65, and 0.42, respectively, based on 20 prompts. Preferences between outputs were also evenly split (10 vs. 10 prompts), further supporting the interpretation that users perceived the systems as equally viable.
These similarities in performance demonstrate promise for implementing RAG LLM systems in the clinical space. Unlike general-purpose LLMs, our approach leverages domain-specific knowledge bases, allowing control over source content and subsequently increasing model interpretability, which are important in clinical settings. The relatively high CR scores suggest that the retrieved citations closely matched the prompts, indicating effective semantic retrieval. Future work should include controlled experiments to measure how much this context improves response quality.
Both systems received consistently high FA scores, and a formal review of all outputs confirmed that no hallucinations occurred. While GPT-4-Consensus generates responses using full-text articles, our RAG LLM system relies solely on abstracts. Papers—particularly their abstracts—often emphasize positive or novel findings, which could lead to overrepresentation of certain conclusions relative to their actual frequency or clinical relevance. For instance, a response citing a study on lymphatic imaging described spin labeling MRI techniques as a key modality, a claim technically supported by the paper but overstated in terms of real-world clinical usage. Future work should assess whether using full texts helps mitigate such biases and yields more balanced, practice-aligned outputs.
Security and data privacy were also central to our design. Because all computation occurred on a local GPU server without API calls or external dependencies, the system inherently avoided exposing patient health information to third-party services.
An additional strength of our system lies in its modularity and adaptability. The knowledge base was built from PubMed abstracts related to radiology, but the same system could be easily extended to other medical specialties—such as cardiology or oncology—by altering the embedded corpus. Furthermore, the generation model (mistral-7b-instruct-v0.2) performed well without fine-tuning, suggesting that domain-specific retrieval combined with robust prompting can remove the need for costly model customization. Nonetheless, fine-tuning remains a promising direction for future optimization.
Despite its success, the current system has limitations. The UI, developed in Gradio, was suitable for prototyping but lacks the robustness required for clinical deployment. A production-ready system would require secure authentication, access controls, logging mechanisms, and audit trails. Additionally, while participants were given consistent instructions, consistency in prompt formulation was not assessed in this study. Future work should incorporate a larger number of prompts and examine how variations in prompt formulation affect system output.
Overall, our findings highlight the potential of localized RAG LLM systems as scalable, privacy-preserving research tools. By enabling transparent, verifiable, and adaptable AI-assisted literature synthesis, this approach offers a compelling model for responsible LLM integration in medical research environments. Such systems could serve as foundational tools across academic medical centers, enabling secure, high-quality, and explainable AI assistance at scale.
5. Conclusions
There is a growing institutional demand for LLM-based tools that can support medical research while preserving data privacy, offering output transparency, and enabling domain-specific customization. Existing public-facing LLM models, while powerful, are constrained by privacy risks and general-purpose design. To address this gap, we developed and validated a secure, locally deployable RAG LLM system tailored for radiology research. Our system enabled semantic retrieval via a vector database of 167,028 PubMed-derived abstracts using e5-mistral-7b-instruct and generated grounded responses using mistral-7b-instruct-v0.2, all while running efficiently on a single 16 GB GPU within institutional infrastructure.
In a blinded evaluation comparing our system to GPT-4-Consensus, participants rated responses from both systems on FA, CR, and PP. The results revealed statistically indistinguishable scores across all three metrics, where user preferences were evenly split, and no hallucination occurred. These findings indicate that a well-designed local RAG LLM system can match the perceived quality of proprietary state-of-the-art systems while offering substantial advantages in terms of privacy, interpretability, and adaptability across medical domains.
Nonetheless, challenges remain. The prototype UI, while functional, requires further development to meet clinical-grade deployment standards, including authentication, logging, and auditability. Future improvements will focus on refining the UI, expanding to other specialties, enabling user feedback loops, and exploring fine-tuning strategies to further enhance generation quality and alignment.
Supplementary Materials
The following supporting information can be downloaded at
https://www.mdpi.com/article/10.3390/inventions10040055/s1: Table S1: List of 20 radiology-related prompts used in the survey and the corresponding responses generated by the two evaluated systems: our RAG-based LLM system (Output 1) and GPT-4-Consensus (Output 2); Table S2: Survey participant ratings for each of the 20 radiology-related prompts. Scores are based on a 1–5 scale (1 = very low, 5 = very high).
Author Contributions
Conceptualization, M.W., J.L.-R., D.A., V.K., X.W. and S.S.; methodology, M.W., J.L.-R., D.A., V.K., X.W. and S.S.; software, M.W., V.K., X.W. and S.S.; validation, M.W., J.L.-R., D.A., V.K., M.S.-Y. and S.S.; formal analysis, M.W., J.L.-R., D.A., V.K., M.S.-Y. and S.S.; investigation, M.W., J.L.-R., D.A., V.K. and S.S.; resources, M.W., J.L.-R., D.A. and S.S.; data curation, M.W., J.L.-R., D.A. and S.S.; writing—original draft preparation, M.W., J.L.-R., D.A., V.K., M.S.-Y. and S.S.; writing—review and editing, M.W., J.L.-R., D.A., V.K., X.W., M.S.-Y. and S.S.; visualization, M.W., V.K. and S.S.; supervision, M.W. and S.S.; project administration, M.W. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Restrictions apply to the availability of these data. The dataset used in this study was derived from PubMed abstracts, which are publicly accessible but may be subject to third-party copyright. As such, the data cannot be redistributed by the authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| CR | Citation Relevance |
| FA | Factual Accuracy |
| GPU | Graphics Processing Unit |
| LLM | Large Language Model |
| NLP | Natural Language Processing |
| PP | Perceived Performance |
| RAG | Retrieval-Augmented Generation |
| RAM | Random Access Memory |
| SD | Standard Deviation |
| UI | User Interface |
| VRAM | Video Random Access Memory |
References
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large Language Models Encode Clinical Knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; Kim, J.; Kamineni, M.; Pang, M.; Lie, W.; Succi, M.D. Evaluating ChatGPT as an Adjunct for Radiologic Decision-Making. medRxiv 2023. [Google Scholar] [CrossRef]
- Consensus. Introducing: GPT-4-Powered, Scientific Summaries. 2023. Available online: https://consensus.app/home/blog/introducing-gpt-4-powered-scientific-summaries/ (accessed on 30 May 2025).
- Shen, Y.; Heacock, L.; Elias, J.; Hentel, K.D.; Reig, B.; Shih, G.; Moy, L. ChatGPT and Other Large Language Models Are Double-Edged Swords. Radiology 2023, 307, e230163. [Google Scholar] [CrossRef] [PubMed]
- Weinert, D.A.; Rauschecker, A.M. Enhancing Large Language Models with Retrieval-Augmented Generation: A Radiology-Specific Approach. Radiol. Artif. Intell. 2025, 7, e240313. [Google Scholar] [CrossRef] [PubMed]
- Akinci D’Antonoli, T.; Stanzione, A.; Bluethgen, C.; Vernuccio, F.; Ugga, L.; Klontzas, M.E.; Cuocolo, R.; Cannella, R.; Koçak, B. Large Language Models in Radiology: Fundamentals, Applications, Ethical Considerations, Risks, and Future Directions. Diagn Interv Radiol. 2024, 30, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.; Perez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Küttler, H.; Lewis, M.; Yih, W.; Rocktäschel, T.; et al. Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 9459–9474. [Google Scholar]
- Savage, C.H.; Kanhere, A.; Parekh, V.; Langlotz, C.P.; Joshi, A.; Huang, H.; Doo, F.X. Open-Source Large Language Models in Radiology: A Review and Tutorial for Practical Research and Clinical Deployment. Radiology 2025, 314, e241073. [Google Scholar] [CrossRef] [PubMed]
- Bluethgen, C.; Van Veen, D.; Zakka, C.; Link, K.E.; Fanous, A.H.; Daneshjou, R.; Frauenfelder, T.; Langlotz, C.P.; Gatidis, S.; Chaudhari, A. Best Practices for Large Language Models in Radiology. Radiology 2025, 315, e240528. [Google Scholar] [CrossRef] [PubMed]
- Meskó, B. Prompt Engineering as an Important Emerging Skill for Medical Professionals: Tutorial. J. Med. Internet Res. 2023, 25, e50638. [Google Scholar] [CrossRef] [PubMed]
- Soong, D.; Sridhar, S.; Si, H.; Wagner, J.-S.; Sá, A.C.C.; Yu, C.Y.; Karagoz, K.; Guan, M.; Kumar, S.; Hamadeh, H.; et al. Improving Accuracy of GPT-3/4 Results on Biomedical Data Using a Retrieval-Augmented Language Model. PLOS Digit. Health 2024, 3, e0000568. [Google Scholar] [CrossRef] [PubMed]
- Beautifulsoup4. Available online: https://beautiful-soup-4.readthedocs.io/en/latest/ (accessed on 30 May 2025).
- Wang, L.; Yang, N.; Huang, X.; Yang, L.; Majumder, R.; Wei, F. Improving Text Embeddings with Large Language Models. arXiv 2024, arXiv:2401.003638. [Google Scholar]
- Hugging Face—The AI Community Building the Future. Available online: https://huggingface.co/ (accessed on 30 May 2025).
- MTEB Leaderboard—A Hugging Face Space by Mteb. Available online: https://huggingface.co/spaces/mteb/leaderboard (accessed on 30 May 2025).
- Chroma. Available online: https://trychroma.com (accessed on 30 May 2025).
- Liu, S.; Liu, Z.; Huang, X.; Dong, P.; Cheng, K.-T. LLM-FP4: 4-Bit Floating-Point Quantized Transformers. In Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing, Vancouver, BC, Canada, 6–12 December 2023; pp. 592–605. [Google Scholar]
- Dao, T.; Fu, D.; Ermon, S.; Rudra, A.; Ré, C. FlashAttention: Fast and Memory-Efficient Exact Attention with IO-Awareness. Adv. Neural Inf. Process. Syst. 2022, 35, 16344–16359. [Google Scholar]
- Peterson, L.E. K-Nearest Neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Cosine Similarity. Wikipedia. Available online: https://en.wikipedia.org/w/index.php?title=Cosine_similarity (accessed on 30 May 2025).
- Shuster, K.; Poff, S.; Chen, M.; Kiela, D.; Weston, J. Retrieval Augmentation Reduces Hallucination in Conversation. arXiv 2021, arXiv:2104.07567. [Google Scholar]
- Mistralai/Mistral-7B-Instruct-v0.2 Hugging Face. Available online: https://huggingface.co/mistralai/Mistral-7B-Instruct-v0.2 (accessed on 30 May 2025).
- Jiang, A.Q.; Sablayrolles, A.; Mensch, A.; Bamford, C.; Chaplot, D.S.; de las Casas, D.; Bressand, F.; Lengyel, G.; Lample, G.; Saulnier, L.; et al. Mistral 7B. arXiv 2023, arXiv:2310.06825. [Google Scholar]
- Team, G. Gradio. Available online: https://gradio.app (accessed on 30 May 2025).
- Batterton, K.A.; Hale, K.N. The Likert Scale What It Is and How To Use It. Phalanx 2017, 50, 32–39. [Google Scholar]
- Wilcoxon Signed-Rank Test-Woolson-2005-Major Reference Works—Wiley Online Library. Available online: https://onlinelibrary-wiley-com.proxy.library.upenn.edu/doi/full/10.1002/0470011815.b2a15177 (accessed on 30 May 2025).
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).