Abstract
Inland capture fisheries play a critical role in supporting food security and livelihoods in Africa. Therefore, it is important to evaluate the genetic health of exploited fish populations. The African bonytongue, Heterotis niloticus, supports important commercial and subsistence fisheries in western Africa. However, sharp declines in stocks have been reported. Herein, we estimate contemporary effective population sizes (Ne) of four Heterotis populations in Nigeria, three in Benin, and five in Cameroon using Linkage Disequilibrium methods. Ne estimates were used to assess genetic short-term (i.e., inbreeding depression) and long-term (i.e., loss of evolutionary potential) risks. Ne point estimates obtained with the best estimator (out of 16), as determined by computer simulations, were <50 (range = 5.1–36.2) for nine of the twelve populations examined, which is below the minimum recommended for avoiding the potential deleterious effects of inbreeding depression (original criterion Ne ≥ 50, revised to Ne ≥ 100); and well below the minimum recommended for populations to retain evolutionary potential (original criterion Ne ≥ 500; revised to Ne ≥ 1000). The lower bound of the confidence interval for two of the remaining populations was below the minimum recommended to retain evolutionary potential (with the point estimate of one of them also below this threshold), and for some methods, values were lower than the minimum recommended to avoid inbreeding depression. Accordingly, our results suggest that urgent conservation and management plans are needed to guarantee the persistence and sustainability of the H. niloticus populations examined.
1. Introduction
Inland capture fisheries in Africa comprise 28% of the global total [1] and provide a critical source of protein, micronutrients and income for people in this continent [2,3,4,5]). The sustainability of these fisheries will be crucial for the food security of the rapidly growing human population of sub-Saharan Africa, which is projected to double by 2050 [6]. Multiple threats, however, are imperiling freshwater fish stocks in Africa, including overexploitation, habitat degradation, introduction of exotic species, and aquaculture [7,8,9,10]. Africa’s most populous country, Nigeria (population ~225 million) in West Africa, ranked 9th globally for inland fisheries yield in 2020, accounting for 3% of the global total and ~11% of Africa’s total (Table 1). Nigeria is one of the seven countries driving most of the growth in global inland fisheries [4]. Nigeria is also one of nine countries that collectively will contribute more than half the projected growth of the global human population between 2019 and 2050 [6], and could be one of the world’s largest consumers of fish by that year [11]. Inland fisheries have been historically important in Nigeria’s western neighbor Benin, a country with a much smaller population (13.7 million). During 1960–2013, the inland fishery sector of Benin produced an average 27,000 tons annually, employing 57,500 fishers and 40,000 women active in fish processing and marketing [12]. Benin has ranked 15th globally in inland capture fisheries production per capita, and 17th in inland capture fisheries production per unit area [13]. Nigeria’s eastern neighbor Cameroon (population ~28.6 million), home to several large rivers, also has considerable inland capture production (Table 1). As of 2018, inland fisheries in this country were estimated to employ more than 177,000 fishers [14]. Despite the current and future significance of inland capture fisheries in these countries, and in Africa in general, very little is known about the status of fish stocks, including their effective population size (Ne), one of the most important parameters in evolutionary biology and conservation biology [15,16].

Table 1.
Inland capture (live weight) of all fishes and Heterotis niloticus in Africa, Nigeria and Benin, and for all fishes in Cameroon.
Ne represents a population’s genetic finite sampling size between generations by comparison to a hypothetical idealized population (i.e., the Wright-Fisher model) where all genetic drift is represented by a sample of 2N gametes from one generation to establish the next generation. The Wright-Fisher model is used to predict the genetic consequences of genetic drift as well as to provide a common quantitative metric for drift that can be compared among otherwise heterogeneous populations. Estimates of Ne in an actual population are made using an observable genetic pattern, such as chance disequilibrium between pairs of unlinked loci that varies with the strength of drift. Estimates of Ne in natural populations predict the effects of genetic drift, including the magnitude of random changes in allele frequencies, the rate of random fixation and loss of alleles, and the loss of heterozygosity [15]. Ne predicts the rate of increase in homozygosity as alleles undergo fixation and loss, which increases the potential for inbreeding depression. Ne also predicts the ability of populations to respond to natural selection and undergo adaptation, also known as their evolutionary potential [17].
Franklin [18] and Soulé [19] proposed that Ne ≥ 50 for a single isolated population is required for short-term persistence, because it minimizes the risk of deleterious effects from inbreeding depression. At Ne < 50, an isolated population can enter an “extinction vortex” [20]. Franklin [18] also proposed that Ne ≥ 500 is needed for healthy evolutionary potential and long-term persistence. Known as the “50/500 rule”, these thresholds have been used as a guiding principle in conservation for assessing minimum viable Ne and to determine the extinction risks of populations and species [21]. These minima, however, have been revised upward. Frankham et al. [22] indicate that Ne = 50 is inadequate for preventing inbreeding depression over five generations in the wild, with Ne ≥ 100 being required to limit loss in total fitness to ≤10%. Caballero et al. [23] suggested that Ne ≈ 70 is sufficient to avoid extinction due to inbreeding depression, when considering that deleterious recessive alleles in homozygous genotypes will be purged by natural selection, reducing inbreeding depression over time. Frankham et al. [22] argued that several independent lines of theoretical and empirical evidence indicate that at least Ne = 1000 is required to maintain initial evolutionary potential in perpetuity. Pérez-Pereira et al. [24] indicate Ne ≥ 500 is necessary for long-term persistence for species with moderately high reproductive rates; and Ne ≥ 1000 for species with low reproductive rates. Previously, Lande [25] argued that Ne ≥ 5000 was necessary to retain evolutionary potential; and Lynch and Lande [26] suggested a range between 1000 and 5000 (see [21] for the meaning of these thresholds in conservation). Ne can be estimated from genetic data, which are slowly accruing for inland capture fisheries of Africa. Estimates of the Ne of fish stocks can be used to assess the genetic risk of extinction and to inform conservation and management strategies for sustainable use. The present study focuses on the estimation of Ne for an important inland fisheries species of western Africa.
The African bonytongue, Heterotis niloticus (Cuvier, 1829), supports significant commercial and subsistence fisheries in western Africa. Nigeria leads wild captures of this fish (Table 1; [27]), totaling 409,784 metric tons between 1990 and 2021 (85.3% of the total catch in Africa during that period), of which 23,875 metric tons were caught in 2021 (86% of the total catch in Africa that year). Benin ranked second for Heterotis yields, with 21,640 metric tons captured between 1987 and 2021 (5% of the total caught in Africa during that period), including 1095 tons in 2021 (Table 1). In Cameroon, this fish naturally occurs in the Sudano-Sahelian zone (Far North and North regions), from where it has been introduced to other regions in the country (i.e., Centre, South, and Littoral), where it supports subsistence fisheries [28,29]. Although FAO does not track capture statistics for H. niloticus in Cameroon, historically, it has been an important capture species in the Logone River in the Far North [30], and Benue River in the North [31] where it occurs naturally. This fish was introduced into the Nyong River (Centre region) in 1958 [32], where it supports an important fishery, with reports of 60 tons landed near Ayos in 1976 [32], ~616 tons/year prior to 1984 in Akonolinga, and ~240 tons/year from the middle Nyong River during 2004–2005 (reviewed in [28]). Overexploitation and declines in Heterotis stocks have been reported for Nigeria [33], Benin [34], and other West African countries [35].
Genetically differentiated populations of the African bonytongue, suggested to represent different conservation/management units, have been identified in Benin [36], Nigeria [37], and Cameroon [38]. These populations provide important food resources for local communities and the sustainability of some stocks appears threatened [36,37]. Herein, we estimate Ne for these populations to assess the risks of inbreeding depression and loss of evolutionary potential.
2. Methods
2.1. Datasets
We used published genotypic datasets of Heterotis populations (Figure 1; Table S1) from Benin [36], Nigeria [37], and Cameroon [38]. The study in Benin identified the following three genetically differentiated populations: Niger River at Malanville; Mono River; and Ouemé-Sô river-floodplain system. The study in Nigeria identified the following four genetically differentiated populations: Kainji Lake; Epe Lagoon; Igbokoda River; and Ethiope River. The study in Cameroon examined the following five populations: Logone River (Far-North region); Benoue River (North region); Nyong River (Centre region); Dja and Lobo Rivers (South region); and Nkam River (Littoral region). Wikondi et al. [38] report no genetic differentiation among Centre, Littoral, and South populations. However, we assessed genetic differentiation among the five populations from Cameroon using the software GenAlEx v. 6.51b2n [39], and found significant differences in all pairwise comparisons (Table S2). Table 2 shows the year of collection for each locality, sample size, number of microsatellites, number of polymorphic microsatellites (only polymorphic microsatellites were used in our analyses), and genetic diversity measures.
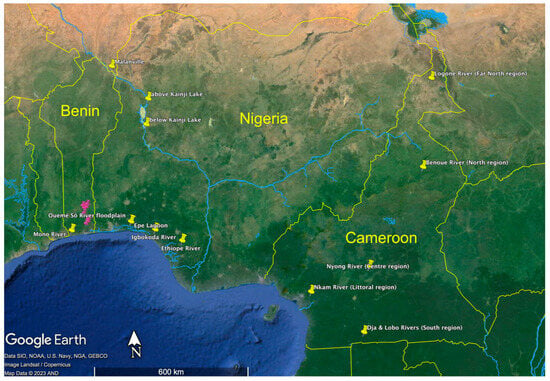
Figure 1.
Map of the study area depicting sampled localities in Benin, Nigeria, and Cameroon. Collectively, the pink pins represent the Ouemé-Sô river-floodplain system.

Table 2.
Information of Benin, Nigeria, and Cameroon datasets.
2.2. Evaluation of Ne Estimators for Genotype Data
Because there are numerous estimators of Ne that employ two-locus disequilibrium (r2), we carried out simulations using SpEED-Ne [40] to evaluate which estimators exhibited the greatest accuracy and precision with sample sizes of individuals, loci, and numbers of alleles per locus that approximated those of sampled populations. Time-forward simulations were carried out following Hamilton et al. [40], generating microsatellite genotype data sets with true Ne equal to 10, 25, 50, 100 and 250, each with 250 independent replicates. Simulated data sets all had eight loci with six initially equally frequent alleles per locus, a 5% rate of null alleles, and a sample size of individuals equal to true Ne.
With multilocus genotype data, two-locus disequilibrium is used to estimate Ne according to 1/[3r2 − (r2 correction)] where the r2 correction is used to subtract disequilibrium caused by mechanisms other than genetic drift, such as finite sample size or within-locus disequilibrium [40]. We compared four categories of Ne estimators based on (1) disequilibrium adjusted for within-locus excess homozygosity () and r2 correction equal to the expected disequilibrium among unlinked loci in a finite sample of individuals, ; (2) disequilibrium estimated with a composite haplotype table () and as the r2 correction; (3) and the sample size of individuals as independent variables in second-order regression equation fits proposed by Waples [41] to adjust for bias and as the r2 correction; and (4) and an r2 correction estimated by median observed for 5000 random permutations of genotypes (pm). Within each of the four categories, estimates of disequilibrium were either allele frequency thresholded (AFT), using AFT = 0.05, or weighted (AFW), to adjust for the disproportionate impact of low-frequency alleles, and either lacked or included (UB) a small sample bias adjustment S/(S − 1) where S is the average of the number of sampled genotypes for each locus pair. Detailed descriptions of these estimators are given in the study by Hamilton et al. [40].
We report Ne point estimates and corresponding confidence intervals (CI) obtained with the estimator(s) that performed best in simulations parameterized with sample sizes of individuals and numbers of loci similar to those for most populations in our study. Reported confidence intervals were obtained using the following three jackknife confidence interval estimation methods implemented in SpEED-Ne: over all locus pairs; over individuals; and over loci. We updated SpEED-Ne to version 2.6 [42] to provide jackknife confidence intervals where the upper and lower bound r2 estimates have an S/(S − 1) adjustment for small sample size bias.
For comparison, Ne was also estimated for all populations using the widely applied second-order regression equation fits using sample size and r2 as independent variables [41,43], as implemented in the program NeEstimator v.2.1 [44]. Ne values were estimated assuming random mating and using a minimum allele frequency (MAF) threshold value of 0.05. Confidence intervals for Ne values were estimated using NeEstimator v.2.1 based on a delete-one-locus jackknife to estimate the variance in r2 to give a parametric confidence interval using the chi-square distribution ([43], see [40]). We used SpEED-Ne to estimate the effective number of independent locus pair comparisons (n′) used for this confidence interval, and compared it with the actual number of unordered pairwise comparisons of loci (npw = [n(n − 1)]/2; where n is the number of loci used) to evaluate how well the assumptions of the chi-square CI were met (i.e., n ≤ n′ ≤ npw; [40]). In our datasets, n varied between 6 and 9, resulting in npw of 15–36 unordered locus pairs.
3. Results
Simulated genotype data (Figures S1–S3) showed that for true Ne values of 10, 25, and 50, the UB-AFT estimator, followed by the UB-AFW estimator, provide the most accurate estimates among the 16 compared (i.e., median slightly biased, smaller quartile ranges, and markedly fewer outliers). At higher true Ne values (i.e., 100 and 250; Figures S4 and S5), an increasing number of the 16 estimators showed improvement in accuracy and precision. As nine of the Ne point estimates obtained with the empirical data were <50, we report results with the UB estimators (Table 3).

Table 3.
Genetic effective population size (Ne) estimates and 95% confidence intervals (CIs) in parenthesis. Estimates employed a minimum allele frequency threshold of 5% (AFT) or were allele-frequency-weighted (AFW) to adjust for the influence of low-frequency alleles. For the first two columns, Ne estimates were adjusted for small sample bias by S/(S − 1), where S is the average of the number of sampled genotypes for each locus pair, and percentile jackknife CIs are shown in this order: over all locus pairs; over individuals; and over loci. For the third column, Ne estimates were based on second-order regression equation fits using sample size and r2 as independent variables [41], and CIs were based on a jackknife over loci and a parametric chi-square distribution based on an effective number of independent locus comparisons (n′).
Ne point estimates < 50 were obtained for the following nine localities ordered from smallest to largest (estimations using UB with AFT; CIs for jackknifing over all locus pairs): Dja and Lobo rivers 5.1 (4.8–5.9); Malanville 9.6 (7.8–35.1); Mono River 15.0 (13.8–17.2); Epe Lagoon 19.1 (17.5–22.2); Kainji Lake 22.7 (20.8–25.6); Ethiope River 24.4 (23.0–29.2); Logone River 27.8 (24.7–37.3); Igbokoda River 36.1 (30.2–74.6); and Benoue River 36.2 (32.9–42.0). Ne point estimates for the three remaining populations were 171.2 (112.1–2392.1) for Nyong River; ∞ (181.7–∞) for Nkam River; and 1867.8 (1292.1–31,044.5) for the Ouemé-Sô river-floodplain system.
For comparison, we also reported Ne estimated with Waples [41] regression equation fits implemented in NeEstimator v2.1 (Table 3). The chi-square CI often had an infinite upper bound and effective sample of locus pairs (n′) much greater than the actual number of locus pairs (npw = 15–36) used to estimate .
4. Discussion
Ne point estimates for nine of the twelve H. niloticus populations examined are below the minimum recommended to avoid the potential deleterious effects of inbreeding depression (original criterion Ne ≥ 50, revised to Ne ≥ 100; [22]); and, well below the minimum recommended for populations to retain evolutionary potential (original criterion Ne ≥ 500; revised to Ne ≥ 1000 [22]; and Ne ≥ 5000 [25]). These populations are Malanville and Mono River, in Benin; Kainji Lake, Epe Lagoon, Ethiope River and Igbokoda River, in Nigeria; and Logone River, Benoue River, and Dja and Lobo rivers, in Cameroon. The CI lower bounds for Nyong River and Nkam River, in Cameroon, were below the recommended values to retain evolutionary potential, and for some methods, below the recommended values to avoid inbreeding depression. In addition, the Ne point estimate for Nyong River (171.2) was well below the ‘500’ threshold to retain evolutionary potential, whereas the Ne point estimate for Nkam using AFW (Ne = 547) was below the ‘1000’ threshold. Nyong River and Nkam River populations are introduced (see below). The CI lower bounds for the Ouemé-Sô river-floodplain system population were below or closely above the ‘1000’ threshold to retain evolutionary potential. Therefore, our results suggest that urgent conservation and management plans are needed to guarantee the survival and sustainability of most of the H. niloticus populations examined.
The computer simulations we conducted indicated that Ne estimates obtained using r2Δ with AFT and the small sample size bias correction (UB) are expected to be fairly precise for the genotypic data used, especially when Ne 50, which encompasses the Ne estimates we obtained for nine populations. This is consistent with expectations on the performance of the disequilibrium Ne estimation approach, with which precise estimates for relatively small (Ne < 200) isolated populations can be obtained, and small populations are not likely to be mistaken for large ones [45]. Waples (Guest Box 10 in [17]) indicates that if Ne is relatively small (<100), reasonably precise estimates can be obtained using samples of 25–50 individuals with 5–10 moderately variable loci; however, considerably more data are needed to achieve comparable precision if the population is relatively large (Ne ∼ 500–1000 or higher). Gilbert and Whitlock [46] used simulations to compare the performance of seven Ne estimation methods under different scenarios of migration with three different Ne values (50, 500 and 5000), and found that the disequilibrium method (implemented in NeEstimator v2.0 in their study) outperformed the other methods in conditions of isolation for Ne = 50. This method also performed well in scenarios of low migration and small Ne. Accordingly, because the Heterotis populations studied herein exhibit genetic differentiation consistent with isolation [36,37], the small Ne estimates we obtained are likely reflective of the true Ne. Nonetheless, in taxa with overlapping generations, Ne estimates based on mixed-age (i.e., multi-cohort) adult samples, which is likely the case for the Heterotis samples examined herein, tend to be biased downwardly [47]. The simulations of Waples et al. [47], based on random samples of adults from 22 taxa representing different taxonomic groups with different life histories (invertebrates, plants, and major vertebrate groups, including fishes), revealed that for most, estimated Ne was 9–35% below true Ne, with only two outliers (mosquito and primrose) below this range (52% and 45% below true Ne, respectively). The life history of Heterotis likely resembles more closely that of the non-outlier taxa of Waples et al. [47]. However, even if we were to consider a downward bias as extreme as 60%, our Ne < 50 estimates would be upwardly corrected to a range between 13 (for Dja and Lobo rivers, whose Ne point estimate was 5.1) and 90 (for Igbokoda and Benoue rivers, whose Ne point estimates were ~36), and thus fall below the 100 threshold for inbreeding depression.
Our computer simulations show that the Ne estimator used in NeEstimator (defined in [43], Table 1) performed much worse for true Ne values of 10, 25, and 50 than the UB estimators with AFT or AFW, and all other estimates in general (Figures S1–S3). Hamilton et al. [40] made similar observations with simulated data for small Ne but different sample sizes of simulated microsatellite loci with a range of null allele frequencies. Nonetheless, critically small Ne point estimates (range 2.6–61.3) were obtained with NeEstimator for the populations for which Ne was <50 with UB estimators, with the exception of Igbokoda, for which NeEstimator’s Ne = 288 (Table 3). Ne point estimates obtained with NeEstimator were infinity for Nyong River, Nkam River, and the Ouemé-Sô river-floodplain system, the populations for which the largest (and in some cases infinite) UB estimates were obtained. Several observations suggest that Igbokoda is a small (Ne < 50) population (i.e., concordant with SpEED-Ne’s UB point estimates). Firstly, the Igbokoda sample was small (n = 15), and its inbreeding estimate was high (F = 0.18; Table 2), features that could have biased estimates by NeEstimator. Nonetheless, NeEstimator yielded a very small CI lower bound (i.e., 11.6). Secondly, Igbokoda is located halfway between the other two southern Nigeria populations examined (separated by ~100 Km of each), Epe Lagoon and Ethiope River, for which Ne point estimates < 50 were obtained. For these populations, we had larger sample sizes (n = 20 and 19, respectively), and their inbreeding coefficient F was lower (0.06 and −0.01, respectively; Table 2). There is no reason to expect a larger Ne for the Igbokoda population, as fish from this locality face severe threats for their conservation (see discussion below). Finally, the high level of inbreeding that Igbokoda appears to be experiencing is inconsistent with a large Ne.
Compared to the jackknife percentile CIs from SpEED-Ne, NeEstimator yielded much larger upper bound estimates for two populations, and infinite upper CI bounds for the remaining ten populations. A tendency to report infinite upper CI values using the method implemented in NeEstimator has been observed in other fish studies (e.g., [48]). This has also been observed in very small endangered populations (i.e., several tens of individuals), for which the Ne point estimates and CI lower bounds were finite and consistent with historical census size estimations, and comparable to those obtained with other Ne estimation methods [36]. Disequilibrium methods can produce negative estimates, which are reported as infinite. This occurs because to estimate Ne, the expected contribution of disequilibrium from sources other than drift is subtracted from the total estimated disequilibrium or r2. Accordingly, if the sample of individuals and loci is relatively small given the true Ne, mean disequilibrium (r2) can be smaller than the correction for disequilibrium due to sources such as a finite sample size, leading to a negative Ne estimate [45]. Nonetheless, even in cases where the point estimate is negative, the CI lower bound generally will be finite and can provide useful information about plausible limits of Ne [45]. The CI method implemented by NeEstimator is considered a ‘pseudo-jackknife’, in that the variance of r2 is estimated by resampling but then used to construct a parametric confidence interval [40,49]. In the present study, the effective number of unordered locus pairs (n′; Table 3) was much greater than the actual number (npw = 15–36), showing that the assumptions of the chi-square CIs were not met. The standard delete-one jackknife confidence intervals in SpEED-Ne provided alternatives to estimate confidence intervals with fewer distributional assumptions. Hamilton et al. (2018) showed that percentile CIs jackknifing over loci and jackknifing over individuals for the estimators provided >97% confidence interval coverage for Ne from 10 to 250. For this study, we expanded SpEED-Ne to compute jackknife CIs for disequilibrium estimates with a simple adjustment for small sample sizes. The SpEED-Ne jackknifing methods should therefore be suitable for datasets with the characteristics (i.e., sample size, number of loci, number of alleles) similar to those of this study.
It is possible that Ne of the Nyong River and Nkam River populations are critically small, as suggested by the CI lower bounds, but limited data prevented accurate point estimates. These populations, along with the population of Dja and Lobo rivers (South region), are introduced, probably from wild populations in northern Cameroon. According to Depierre and Vivien [32], Heterotis was introduced from northern Cameroon populations to a fish farming station in Melen, a suburb of Yaounde, in 1955, and from there, ~20 fry were released into the Nyong River in 1958. Subsequently, in 1961, an accidental spill from a fishpond released several hundred fry into the Nyong River. These authors also indicate that Heterotis appeared in the Lower Sanaga River (Littoral region) around 1968, probably from the lower course of the Nyong, as the two rivers are connected at the mouth of the Sanaga through mangrove channels during periods of high water. It is possible that from the Sanaga, Heterotis colonized other rivers in the Littoral region, such as the Nkam, and that the Dja and Lobo rivers were also colonized by individuals related to the Nyong population. Indeed, STRUCTURE analyses of the five Cameroon populations found two main clusters, one corresponding to the native populations (Benoue and Logone rivers), and the other to the introduced populations, suggesting a common origin for the introduced populations [38]. Small Ne point estimates were obtained for Benoue River (36.2) and Logone River (27.8); thus, it is reasonable to expect the Ne of the introduced populations to be smaller, due to expected founder events typically associated with introductions. The three Cameroon introduced populations have notably lower allelic diversity and heterozygosity (Na = 3.4–3.7; He = 0.47–0.56; Table 2) than their northern Cameroon native counterparts (Na = 5.9–6.0; He = 0.64–0.69; Table 2); consistent with founder events. In addition, for Nkam River, one of the seven loci used by Wikondi et al. [38] was monomorphic, and therefore was discarded for Ne estimations. The negative (infinity) point estimate obtained for Nkam River may stem from its limited genetic information, and more individuals and/or microsatellites may be needed to obtain a reliable estimate. Nonetheless, some of the CI lower bounds of Nkam River and Nyong River indicate that it is plausible these populations also may be at risk of inbreeding depression. Congruent with a history of introductions and genetic diversity reduction, a very small Ne estimate, the smallest in this study, was obtained for Dja and Lobo rivers (5.1).
The Ouemé-Sô river-floodplain appears to have the largest Ne (1868) of all populations examined. The area sampled is vast (~1680 Km2; with sampling localities separated by up to 75 Km) with many interconnected water bodies, including the two major channels, five permanent lakes, and numerous secondary channels and seasonal floodplain pools. Nonetheless, CI lower bounds were close to the threshold for maintaining evolutionary potential. We note that the Benin samples used to estimate Ne were collected between 2008 and 2009, and the ones from Nigeria were collected in 2018. Thus, it is possible that the present-day Ne of these populations is even smaller than our estimates, especially for the Benin stocks, which used samples collected over a decade ago, and considering that fishing pressure and habitat impacts have continued or increased since then.
Ne estimates of Heterotis populations in West Africa are, in general, much lower than those reported for the region’s wild populations of Nile tilapia (Oreochromis niloticus), an important species supporting capture fisheries and aquaculture production [50]. For Nile tilapia examined across West Africa, including eight countries representing the major catchments of the Volta, Niger, Senegal, and Gambia river basins, Ne was below the threshold for long-term genetic risks (range: 56–352 individuals), whereas 10 stocks (43.5%) were below the revised threshold of 100 for short-term genetic risk; and 14 (60.9%) had CI lower bounds below 100 (the lowest was 30.3). The Ne point estimate for Nile tilapia at Malanville was much larger (Ne = 236) than that for Heterotis from the same location (Ne = 9.6). Nile tilapia at Malanville might be part of a more widely distributed metapopulation, whereas Heterotis at this locality might correspond to a more isolated population. Nile tilapia from Malanville and Mopti (Mali), another locality in the Niger River located ~1400 Km upstream, show high genetic similarity, suggesting high levels of gene flow. Nile tilapia Ne estimated at Mopti is very similar (Ne = 289) to that in Malanville; thus, it is possible that such Ne estimates reflect the Ne of a broader metapopulation. According to Waples and England [51], “LD estimates of Ne accurately reflect local (subpopulation) Ne unless m ≥ 5–10%. With higher m, Ne converges on the global (metapopulation) Ne”. For Heterotis, high genetic differentiation appears to occur at comparatively shorter distances within the Niger River, i.e., between Malanville and Kainji Lake (~230 km), and between Kainji Lake and the lower Niger portion (~700 km).
We note that heterozygosity and allelic diversity are poor predictors of short- and long-term genetic risks, underscoring the importance of Ne estimates. Heterozygosity in Kainji Lake was the highest among all populations (uHe = 0.72), with the second highest allelic diversity (Na = 8.33), yet Ne was only 23. Similarly, for Nile tilapia, the locality with the highest estimated Ne had comparatively low heterozygosity and allelic diversity among the Nile tilapia populations examined. Severe declines in Ne can occur without a significant loss of genetic diversity [52].
Critically small values of Ne have also been reported for populations of Arapaima gigas, the African bonytongue’s closest living relative, and the only other member of Arapaiminae, which is distributed throughout the Amazon River basin in South America. Farias et al. [53] estimated Ne for 19 populations of this fish along the Amazon River basin and three locations in the Araguaia-Tocantins basin. For 19 populations, Ne ranged between 0.8 and 48.8 (average = 15.6); for an additional population, Ne was 95; and for the remaining two, Ne was undetermined (infinity), probably due to small sample sizes and limited genetic diversity. Similar to Heterotis, high levels of genetic population differentiation were detected among these Arapaima populations; 226 of 231 pairwise FST comparisons were significant (range = 0.02–0.57; average = 0.22). An effect of floodplain connectivity and geographic scale on the patterns of genetic differentiation is observed for both species [36,37,53,54]. Genetic homogeneity occurs for both species at a fine scale (e.g., within the same floodplain, such as in the Ouemé-Sô river-floodplain system for Heterotis); at a meso-scale (e.g., in separate floodplain systems, such as in the southern Nigerian locations for Heterotis), they exhibit low but significant values of genetic differentiation; and the highest levels of genetic differentiation occur at the largest geographic scale (e.g., >1300 km in Arapaima and >510 km in Heterotis). These patterns suggest limited migration, with individuals of both species probably confined mainly to a single floodplain [34,55]. High isolation, overexploitation and critically small Ne of populations is concerning for both species, the only members of Arapaiminae.
Implications for Conservation and Management
Overfishing and habitat destruction likely pose the greatest threats for Heterotis stocks. This species is highly sought because of its high protein content and firm flesh [56]. Overfishing has been documented in Kainji Lake [57,58], Malanville [59], Ouemé-Sô river-floodplain system [60], Mono River [61], and Epe Lagoon [62]. Illegal fishing activities exacerbate this problem. In Kainji Lake, illegal practices include the use of prohibited gear (e.g., small mesh size nets and destructive fishing gear), fish poisoning, and explosives [63]. In the Ouemé-Sô river-floodplain system, fish traps are placed near active Heterotis nests to capture brooding adults [34]. Pollution also impacts Heterotis stocks in some regions [64,65,66,67].
Our results indicate that Ne values for most populations of Heterotis niloticus in Nigeria, Benin, and Cameroon are below the recommended thresholds to avoid short- and long-term genetic risks. In addition, high inbreeding (FIS) was reported for the following four local stocks [36,37]: Igbokoda (FIS = 0.18; the highest inbreeding among Nigerian populations); Malanville (FIS = 0.20); Mono River (FIS = 0.13); and Ouemé-Sô river-floodplain (FIS = 0.13). Ne estimates of Heterotis are comparable to those of endangered species [68,69]. For example, Ne, heterozygosity, and FIS values estimated for populations of an endangered snake [70] were similar to estimates for African Heterotis. Small Ne estimates of genetically differentiated Heterotis populations indicate that conservation and management actions are urgently needed, with special consideration of risks associated with inbreeding depression (but see [71]).
Heterotis niloticus is currently listed as a species of Least Concern by the International Union for Conservation of Nature (IUCN) [72], a designation that applies to the species as a whole. It is important that future assessments of this species consider (1) that this species is comprised of multiple genetically differentiated populations with small ranges relative to the species’ global distribution, many of which face multiple threats (i.e., overexploitation, habitat destruction, pollution); (2) most populations examined to date reveal Ne point estimates below the critical long-term threshold of 500, revised to 1000, for retaining evolutionary potential, and most are also below the critical short-term threshold of 50 and 100 for preventing inbreeding depression; and (3) when considering the Ne CI lower bounds of most populations, their vulnerability to inbreeding depression appears more serious.
Growth of isolated populations may allow their Ne to increase [73], and fishing restrictions may be required to allow stocks to recover [74]. Gene flow between populations may also boost Ne [75]; however, given that stocks are genetically differentiated, gene flow could result in outbreeding depression [17] and erosion of locally adapted variation. Furthermore, translocation of stocks erodes distinct population genetic structures, which runs counter to a goal of the Convention on Biological Diversity’s post-2020 global biodiversity framework [76]. Results of the present study provide baseline information for continued monitoring of Ne. Future research should also examine genetic diversity of other Heterotis populations, as well as monitor Ne of genetically distinct populations. Heterotis is just one of several fishes important for subsistence and commercial fisheries in Africa, and the genetic diversity of other fish species also needs to be investigated.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9060196/s1, Figures S1–S5: Ne estimates from simulated microsatellite genotype data with true Ne = 10, 25, 50, 100, and 250, respectively; Table S1: Individual genotypes and allele frequencies per population per country; Table S2: Pairwise genetic differentiation among populations from Cameroon.
Author Contributions
L.A.H.: conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing—original draft, conducted revision, supervision; M.M.: conceptualization, formal analysis, resources, writing—original draft; I.C.C.: formal analysis, writing—review and editing; T.E.O.: resources, writing—review and editing; A.A.: resources, writing—review and editing; M.O.A.: resources; K.O.W.: resources, writing—review and editing; M.B.H.: formal analysis, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This article uses genotypic datasets of already published research. Therefore, approval is not necessary.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAO. The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022; 226p. [Google Scholar] [CrossRef]
- Béné, C.; Heck, S. Fish and food security in Africa. NAGA, WorldFish Cent. Q. 2005, 28, 8–13. [Google Scholar]
- De Graaf, G.; Garibaldi, L. The value of African fisheries. FAO Fish. Aquac. Circ. 2015, 1093, 1–76. [Google Scholar]
- Funge-Smith, S.J. Review of the State of World Fishery Resources: Inland Fisheries; FAO Fisheries and Aquaculture Circular; FAO: Rome, Italy, 2018. [Google Scholar]
- Funge-Smith, S.; Bennett, A. A fresh look at inland fisheries and their role in food security and livelihoods. Fish Fish. 2019, 20, 1176–1195. [Google Scholar] [CrossRef]
- United Nations-Department of Economic and Social Affairs-Population Division. World Population Prospects 2019: Highlights, New York, USA (st/esa/ser. A/423). 2019. Available online: https://population.un.org/wpp/Publications/Files/WPP2019_Highlights.pdf (accessed on 18 May 2024).
- Abban, E.K. Considerations for the conservation of African fish genetic resources for their sustainable exploitation. Towards policies for conservation and sustainable use of aquatic genetic resources. ICLARM Conf. Proc. 1999, 59, 95–100. [Google Scholar]
- Lind, C.E.; Brummett, R.E.; Ponzoni, R.W. Exploitation and conservation of fish genetic resources in Africa: Issues and priorities for aquaculture development and research. Rev. Aquac. 2012, 4, 125–141. [Google Scholar] [CrossRef]
- Marshall, B.E. Inland fisheries of tropical Africa. In Freshwater Fisheries Ecology; Graig, J.F., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 249–362. [Google Scholar] [CrossRef]
- Olaosebikan, B.D.; Bankole, N.O. An analysis of Nigerian freshwater fishes: Those under threat and conservation options. In Proceedings of the 19th Annual Conference of the Fisheries Society of Nigeria (FISON), Ilorin, Nigeria, 29 November–3 December 2004; pp. 754–762. [Google Scholar]
- Chan, C.Y.; Tran, N.; Pethiyagoda, S.; Crissman, C.C.; Sulser, T.B.; Phillips, M.J. Prospects and challenges of fish for food security in Africa. Glob. Food Secur. 2019, 20, 17–25. [Google Scholar] [CrossRef]
- Sonneveld, B.; Thoto, F.; Houessou, D.; Wesenbeeck, L. Tragedy of the inland lakes. Int. J. Commons 2019, 13, 609–636. [Google Scholar] [CrossRef]
- FAO. Review of the State of World Fishery Resources: Inland Fisheries; FAO: Rome, Italy, 2003. [Google Scholar]
- FAO. Fishery and Aquaculture Country Profiles. Cameroun, 2022. Country Profile Fact Sheets. In Fisheries and Aquaculture; Rome, Italy, Updated Feb 1, 2022. Available online: https://www.fao.org/fishery/en/facp/cmr?lang=fr (accessed on 17 May 2024).
- Husemann, M.; Zachos, F.E.; Paxton, R.J.; Habel, J.C. Effective population size in ecology and evolution. Heredity 2016, 117, 191–192. [Google Scholar] [CrossRef]
- Waples, R.S. What Is Ne, Anyway? J. Hered. 2022, 113, 371–379. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations; John Wiley & Sons: Chichester, UK, 2013. [Google Scholar]
- Franklin, I.R. Conservation biology: An evolutionary-ecological perspective. In Conservation Biology: An Evolutionary-Ecological Perspective; Soulé, M.E., Wilcox, B.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1980; pp. 135–149. [Google Scholar]
- Soulé, M.E. Thresholds for survival: Maintaining fitness and evolutionary potential. In Conservation Biology: An Evolutionary-Ecological Perspective; Soulé, M.E., Wilcox, B.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1980; pp. 151–169. [Google Scholar]
- Gilpin, M.E.; Soulé, M.E. Minimum viable populations: Processes of species extinction. In Conservation Biology: The Science of Scarcity and Diversity; Soulé, M.E., Ed.; Sinauer Associates: Sunderland, MA, USA, 1986; pp. 19–34. [Google Scholar]
- Jamieson, I.G.; Allendorf, F.W. How does the 50/500 rule apply to MVPs? Trends Ecol. Evol. 2012, 27, 578–584. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.A.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Caballero, A.; Bravo, I.; Wang, J. Inbreeding load and purging: Implications for the short-term survival and the conservation management of small populations. Heredity 2017, 118, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pereira, N.; Wang, J.; Quesada, H.; Caballero, A. Prediction of the minimum effective size of a population viable in the long term. Biodivers. Conserv. 2022, 31, 2763–2780. [Google Scholar] [CrossRef]
- Lande, R. Mutation and Conservation. Conserv. Biol. 1995, 9, 782–791. [Google Scholar] [CrossRef]
- Lynch, M.; Lande, R. The critical effective size for a genetically secure population. Anim. Conserv. 1998, 1, 70–72. [Google Scholar] [CrossRef]
- FAO. Global Capture Production Quantity (1950–2021). 2021. Available online: https://www.fao.org/fishery/statistics-query/en/capture/capture_quantity (accessed on 5 November 2023).
- Brummett, R.; Nguenga, D.; Tiotsop, F.; Abina, J.-C. The Commercial Fishery of the Middle Nyong River, Cameroon: Productivity and Environmental Threats. Smithiana Bull. 2010, 11, 3–16. [Google Scholar]
- Moreau, J. Expose Synoptique des Donnees Biologiques sur Heterotis niloticus (Cuvier, 1829); FAO Synopsis sur les Pèches: Rome, Italy, 1982; pp. 1–45. Available online: https://openknowledge.fao.org/items/ab20fb70-bc00-42c4-a497-cb78936c05da (accessed on 18 May 2024).
- Blache, J.; Miton, F. Première contribution à la connaissance de la pêche dans le bassin hydrographique Logone-Chari-Lac Tchad: Aspect général des activités de la pêche et de la commercialisation des produits. Description des engins de pêche et leur emploi. Mémoires Off. Sci. Tech. Res. Overseas (ORSTOM) 1962, 4, 1–143. [Google Scholar]
- Stauch, A. Le bassin Camerounais de la Benoue et sa Peche; IRD Editions; Office of Scientific and Technical Research Overseas (ORSTOM): Paris, France, 1966; pp. 1–152. Available online: https://horizon.documentation.ird.fr/exl-doc/pleins_textes/pleins_textes_2/memoires/10593.pdf (accessed on 18 May 2024).
- Depierre, D.; Vivien, J. Une réussite du Service Forestier du Cameroun: L’introduction d’Heterotis niloticus dans le Nyong. Rev. Bois Et Forêts Des Trop. 1977, 173, 59–68. [Google Scholar]
- Mustapha, M.K. Heterotis niloticus (Cuvier, 1829) a threatened fish species in Oyun reservoir, Offa, Nigeria; the need for its conservation. Asian J. Exp. Biol. Sci. 2010, 1, 1–7. [Google Scholar]
- Adite, A.; Winemiller, K.O.; Fiogbe, E.D. Population structure and reproduction of the African bonytongue Heterotis niloticus in the Sô River-floodplain system (West Africa): Implications for management. Ecol. Freshw. Fish 2006, 15, 30–39. [Google Scholar] [CrossRef]
- Laë, R. Climatic and anthropogenic effects on fish diversity and fish yields in the Central Delta of the Niger River. Aquat. Living Resour. 1995, 8, 43–58. [Google Scholar] [CrossRef]
- Hurtado, L.A.; Carrera, E.; Adite, A.; Winemiller, K.O. Genetic differentiation of a primitive teleost, the African bonytongue Heterotis niloticus, among river basins and within a floodplain river system in Benin, West Africa. J. Fish Biol. 2013, 83, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Oladimeji, T.E.; Caballero, I.C.; Mateos, M.; Awodiran, M.O.; Winemiller, K.O.; Adite, A.; Hurtado, L.A. Genetic identification and diversity of stocks of the African bonytongue, Heterotis niloticus (Osteoglossiformes: Arapaiminae), in Nigeria, West Africa. Sci. Rep. 2022, 12, 8417. [Google Scholar] [CrossRef]
- Wikondi, J.; Ngono, E.P.J.; Nana, A.T.; Meutchieye, F.; Tomedi, M.E.T. Farming Features of African Bonytongue Fish Heterotis niloticus in Cameroon, Central Africa. Open J. Anim. Sci. 2023, 13, 232–248. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, M.B.; Tartakovsky, M.; Battocletti, A. speed-ne: Software to simulate and estimate genetic effective population size (Ne) from linkage disequilibrium observed in single samples. Mol. Ecol. Resour. 2018, 18, 714–728. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv. Genet. 2006, 7, 167–184. [Google Scholar] [CrossRef]
- Simulation and Estimation Gametic Disequilibrium Genetic Effective Population Size (Ne). Available online: https://github.com/mbhamilton/SpEED-Ne (accessed on 16 May 2024).
- Waples, R.S.; Do, C. LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 2008, 8, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C.H.I. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef]
- Gilbert, K.J.; Whitlock, M.C. Evaluating methods for estimating local effective population size with and without migration. Evolution 2015, 69, 2154–2166. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Antao, T.; Luikart, G. Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 2014, 197, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Almodóvar, A.; Nicola, G.G.; Ayllón, D.; Leal, S.; Marchán, D.F.; Elvira, B. A Benchmark for Atlantic Salmon Conservation: Genetic Diversity and Structure in a Southern European Glacial Refuge before the Climate Changed. Fishes 2023, 8, 321. [Google Scholar] [CrossRef]
- Jones, A.T.; Ovenden, J.R.; Wang, Y.-G. Improved confidence intervals for the linkage disequilibrium method for estimating effective population size. Heredity 2016, 117, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Lind, C.E.; Agyakwah, S.K.; Attipoe, F.Y.; Nugent, C.; Crooijmans, R.P.; Toguyeni, A. Genetic diversity of Nile tilapia (Oreochromis niloticus) throughout West Africa. Sci. Rep. 2019, 9, 16767. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; England, P.R. Estimating contemporary effective population size on the basis of linkage disequilibrium in the face of migration. Genetics 2011, 189, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Lonsinger, R.C.; Adams, J.R.; Waits, L.P. Evaluating effective population size and genetic diversity of a declining kit fox population using contemporary and historical specimens. Ecol. Evol. 2018, 8, 12011–12021. [Google Scholar] [CrossRef] [PubMed]
- Farias, I.P.; Willis, S.; Leao, A.; Verba, J.T.; Crossa, M.; Foresti, F.; Porto-Foresti, F.; Sampaio, I.; Hrbek, T. The largest fish in the world’s biggest river: Genetic connectivity and conservation of Arapaima gigas in the Amazon and Araguaia-Tocantins drainages. PLoS ONE 2019, 14, e0220882. [Google Scholar] [CrossRef] [PubMed]
- Araripe, J.; Rêgo, P.S.D.; Queiroz, H.; Sampaio, I.; Schneider, H. Dispersal capacity and genetic structure of Arapaima gigas on different geographic scales using microsatellite markers. PLoS ONE 2013, 8, e54470. [Google Scholar] [CrossRef]
- Gurdak, D.J.; Stewart, D.J.; Klimley, A.P.; Thomas, M. Local fisheries conservation and management works: Implications of migrations and site fidelity of Arapaima in the Lower Amazon. Environ. Biol. Fishes 2022, 105, 2119–2132. [Google Scholar] [CrossRef]
- Agbugui, M.O.; Egbo, H.O.; Abhulimen, F.E. The Biology of the African Bonytongue Heterotis niloticus (Cuvier, 1829) from the Lower Niger River at Agenebode in Edo State, Nigeria. Int. J. Zool. 2021, 2021, 1748736. [Google Scholar] [CrossRef]
- Du-Feu, T.A.; Abiodun, J.A. Fisheries statistics of Kainji Lake, northern Nigeria, Nov. 1994 - Dec. 1998. 128p. Nigerian-German Kainji Lake Fisheries Promotion Project Technical Report Series 13. 1999. Available online: https://aquadocs.org/handle/1834/21520 (accessed on 18 May 2024).
- Yem, I.Y.; Sani, A.O.; Bankole, N.O.; Onimisi, H.U.; Musa, Y.M. Over fishing as a factor responsible for declined in fish species diversity of Kainji, Nigeria. In Proceedings of the 21st Annual Conference of the Fisheries Society of Nigeria (FISON), Calabar, Nigeria, 13–17 November 2007; pp. 79–85. Available online: http://hdl.handle.net/1834/37723 (accessed on 18 May 2024).
- Hauber, M.E.; Bierbach, D.; Linsenmair, K.E. A description of teleost fish diversity in floodplain pools (‘Whedos’) and the Middle-Niger at Malanville (north-eastern Benin). J. Appl. Ichthyol. 2011, 27, 1095–1099. [Google Scholar] [CrossRef]
- Allan, J.D.; Abell, R.; Hogan, Z.E.B.; Revenga, C.; Taylor, B.W.; Welcomme, R.L.; Winemiller, K. Overfishing of inland waters. Bioscience 2005, 55, 1041–1051. [Google Scholar] [CrossRef]
- Lederoun, D.; Lalèyè, K.R.; Boni, A.R.; Amoussou, G.; Vodougnon, H.; Adjibogoun, H.; Lalèyè, P.A. Length–weight and length–length relationships of some of the most abundant species in the fish catches of Lake Nokoué and Porto-Novo Lagoon (Benin, West Africa). Lakes Reserv. Res. Manag. 2018, 23, 351–357. [Google Scholar] [CrossRef]
- Olukolajo, S.O.; Hillary, E.C. Species diversity and growth pattern of the fish fauna of Epe Lagoon, Nigeria. J. Fish. Aquat. Sci. 2012, 7, 392–401. [Google Scholar] [CrossRef][Green Version]
- Mshelia, M.B.; Okaeme, A.N.; Dantoro, N.O.; Abiodun, J.A.; Olowosegun, O.M.; Yemi, I.Y. Responsible fisheries enhancing poverty alleviation of fishing communities of Lake Kainji. In Proceedings of the 19th Annual Conference of the Fisheries Society of Nigeria (FISON), Ilorin, Nigeria, 29 November–3 December 2004; pp. 597–604. Available online: https://aquadocs.org/handle/1834/21737 (accessed on 18 May 2024).
- Akinsanya, B.; Ayanda, I.O.; Fadipe, A.O.; Onwuka, B.; Saliu, J.K. Heavy metals, parasitologic and oxidative stress biomarker investigations in Heterotis niloticus from Lekki Lagoon, Lagos, Nigeria. Toxicol. Rep. 2020, 7, 1075–1082. [Google Scholar] [CrossRef]
- Akinsanya, B.; Ayanda, I.O.; Onwuka, B.; Saliu, J.K. Bioaccumulation of BTEX and PAHs in Heterotis niloticus (Actinopterygii) from the Epe Lagoon, Lagos, Nigeria. Heliyon 2020, 6, e03272. [Google Scholar] [CrossRef] [PubMed]
- Arojojoye, O.A.; Oyagbemi, A.A.; Ola-Davies, O.E.; Asaolu, R.O.; Shittu, Z.O.; Hassan, B.A. Assessment of water quality of selected rivers in the Niger Delta region of Nigeria using biomarkers in Clarias gariepinus. Environ. Sci. Pollut. Res. 2021, 28, 22936–22943. [Google Scholar] [CrossRef] [PubMed]
- Ikomi, R.B.; Arimoro, F.O. Effects of recreational activities on the littoral macroinvertebrates of Ethiope River, Niger Delta, Nigeria. J. Aquat. Sci. 2014, 29, 155–170. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Palstra, F.P.; Ruzzante, D.E. Genetic estimates of contemporary effective population size: What can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 2008, 17, 3428–3447. [Google Scholar] [CrossRef]
- Sovic, M.; Fries, A.; Martin, S.A.; Gibbs, H.L. Genetic signatures of small effective population sizes and demographic declines in an endangered rattlesnake, Sistrurus catenatus. Evol. Appl. 2019, 12, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.L.A.; Yates, M.C.; Fraser, D.J. Are heritability and selection related to population size in nature? Meta-analysis and conservation implications. Evol. Appl. 2016, 9, 640–657. [Google Scholar] [CrossRef] [PubMed]
- Diouf, K.; Akinyi, E.; Azeroual, A.; Entsua-Mensah, M.; Getahun, A.; Lalèyè, P.; Moelants, T.H. Heterotis niloticus. 2020. Available online: https://www.iucnredlist.org/species/182580/134764025 (accessed on 5 November 2023).
- Kamath, P.L.; Haroldson, M.A.; Luikart, G.; Paetkau, D.; Whitman, C.; Van Manen, F.T. Multiple estimates of effective population size for monitoring a long-lived vertebrate: An application to Yellowstone grizzly bears. Mol. Ecol. 2015, 24, 5507–5521. [Google Scholar] [CrossRef] [PubMed]
- Pita, A.; Pérez, M.; Velasco, F.; Presa, P. Trends of the genetic effective population size in the Southern stock of the European hake. Fish. Res. 2017, 191, 108–119. [Google Scholar] [CrossRef]
- Frankham, R. Evaluation of proposed genetic goals and targets for the Convention on Biological Diversity. Conserv. Genet. 2022, 23, 865–870. [Google Scholar] [CrossRef]
- Hoban, S.; Bruford, M.W.; da Silva, J.M.; Funk, W.C.; Frankham, R.; Gill, M.J.; Grueber, C.E.; Heuertz, M.; Hunter, M.E.; Kershaw, F. Genetic diversity goals and targets have improved, but remain insufficient for clear implementation of the post-2020 global biodiversity framework. Conserv. Genet. 2023, 24, 181–191. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).