Abstract
The Cambeva variegata group (CVG) is endemic to a region situated in the intersection of two endangered biodiversity hotspots, Cerrado and Atlantic Forest, and drained by two important South American river basins, the upper Rio Paraná and upper Rio São Francisco basins. Presently, CVG comprises two nominal species, besides some still undescribed. We first performed a molecular phylogenetic analysis (total of 3368 bp) for five species of the CVG and 30 outgroups, which supported the monophyly of the CVG and its inclusion in Cambeva. Most morphological character states distinguishing the CVG from congeners are also present in Scleronema, possibly consisting of plesiomorphic features. We also performed the first time-calibrated phylogeny of the group, which supported possible relationships between present geographical distribution patterns and palaeogeographical events. The estimated time of origin of CVG in the Middle Miocene is nearly contemporaneous to a past hydrographical configuration when part of the upper Rio Paraná basin was connected to the Rio São Francisco basin. The first CVG lineage split occurring in the Miocene end corresponds to a major break in that palaeo basin. Species diversification between the Pliocene and early Pleistocene is compatible with final drainage rearrangement. This study highlights the urgent need for more detailed studies on the diversity and phylogenetic relationships of still poorly known organisms in this highly diverse and threatened region.
Keywords:
Atlantic Forest; Cerrado; molecular phylogeny; mountain biodiversity; osteology; Rio Paraná; Rio São Francisco Key Contribution:
This paper comprises the first osteological character survey of the CVG, supporting clade diagnoses; the first time-calibrated phylogenetic analysis focusing on Cambeva; and the first attempt to integrate the molecular phylogeny of Cambeva, present distribution patterns, and palaeogeographical scenarios.
1. Introduction
Fast-flowing rivers and streams draining the mountain ranges of southeastern Brazil in the region comprising the upper Rio Paraná basin and the upper Rio São Francisco basin (hereafter Upper Paraná–São Francisco Region, UPSFR), such as Serra da Canastra, Serra do Espinhaço, and Serra da Mantiqueira, among others, shelter a huge diversity of trichomycterine mountain catfishes, mostly revealed in recent years (e.g., [1] and included references). The great majority of trichomycterines endemic to this region belong to the genus Trichomycterus Valenciennes, 1832, whereas species of Cambeva Katz, Barbosa, Mattos, and Costa, 2018, are rare. Cambeva presently includes about 50 species occurring between the tropical area drained by the Rio São Francisco basin, about 16°10′ S, and the subtropical area drained by the rivers connected to the Lagoa dos Patos system, about 29°30′ S [2]. However, only two species of Cambeva occur and are endemic to UPSFR, Cambeva concolor Costa, 1992, and Cambeva variegata Costa, 1992, both endemic to the upper Rio São Francisco basin [3]. These species are easily distinguished from all other species of Cambeva by the presence of a prominent skin crest on the dorsal margin of the caudal peduncle [3], a condition similar to that occurring in Scleronema Eigenmann, 1917 [4,5,6], and to the adipose fin of the Copionodontinae [7]. Our field studies have shown that this group of Cambeva, hereafter the Cambeva variegata group (CVG), also includes undescribed species, some of which are endemic to the Rio Grande drainage, upper Rio Paraná basin, consisting of the first records of CVG for UPSFR.
Whereas the great species diversity of Trichomycterus from UPSFR has been reported in frequent publications (e.g., [2,8] and included references), little has been published about species of the CVG from the same region. Mentions of this group are restricted to a single taxonomic paper published 32 years ago [3] and a few phylogenetic studies inferring the positioning of C. variegata among other congeners (e.g., [2,9,10]). The scarcity of studies on the CVG is probably due to two factors. Firstly, most species of this group are extremely similar when compared only by external morphology, making the identification and recognition of new species inaccessible to biologists not trained in osteological examination or molecular biology. Secondly, there is great difficulty in field collecting, since species in this group have shown to be restricted to small areas and are often rare in their environments.
UPSFR is situated in the intersection of two phytogeographical provinces considered among the main biodiversity hotspots of the world, the savannah-like Cerrado and the Atlantic Forest [11]. On the other hand, rivers and streams of this area are highly impacted by anthropogenic factors, such as large dams for the generation of electricity, destruction of the hydrographic structure for mineral extraction, water pollution by domestic and industrial sewage and by pesticides used in agriculture, and the introduction of exotic species and deforestation of marginal areas (e.g., [1]). The growing process of environmental degradation requires urgency in studies focusing on the diversity of this rare fish group. The objectives of the present study are to present the first phylogenetic analysis involving a large species sample of the CVG, testing the monophyly of the group; to provide a comparative osteological analysis allowing morphological diagnoses of clades supported in molecular analyses; and to present the first time-calibrated phylogeny of the group in order to find evidence of possible relationships between the present geographical distribution patterns and palaeogeographical events.
2. Materials and Methods
2.1. Specimens
Fish collections were performed during daylight with dip nets. Collecting permits were provided by ICMBio (Instituto Chico Mendes de Conservação da Biodiversidade; permit numbers: 38553-13 and 64415-5). Methods for fish collection, euthanasia using a buffered solution of tricaine methane sulphonate (MS-222) at a concentration of 250 mg/L (e.g., AVMA Guidelines for the Euthanasia of Animals) [12], and fixation were approved by CEUA-CCS-UFRJ (Ethics Committee for Animal Use of Federal University of Rio de Janeiro; permit numbers: 065/18 and 084/23). For fixation, specimens were kept in formalin for two weeks, or fixed in absolute ethanol in cases of specimens used in the molecular analysis. Formalin-fixed specimens were preserved in 70% ethanol. Among these specimens, between three and five were prepared for osteological examination, using clearing and staining techniques described by Taylor and Van Dyke [13], and later preserved in glycerine. In the list of specimens below, C&S means specimens cleared and stained for osteological examination, and DNA means specimens directly fixed in absolute ethanol for molecular analysis. Specimens were deposited in the ichthyological collections of Instituto de Biologia, Universidade Federal do Rio de Janeiro (UFRJ), and Centro de Ciências Agrárias e Ambientais, Universidade Federal do Maranhão (CICCAA).
Specimens examined: Cambeva variegata (UFRJ 8355, 7 ex.; UFRJ 8314, 1 ex. (DNA); UFRJ 8318, 1 ex. (DNA); UFRJ 8319, 1 ex. (DNA); UFRJ 9346, 2 ex. (C&S): topotypes: 20°15′09″ S 46°24′23″ W; UFRJ 585, 2 paratypes, (C&S); UFRJ 12857, 3 ex.; UFRJ 12833, 1 ex. (DNA): 20°15′50″ S 46°20′56″ W; UFRJ 14072, 9 ex.; UFRJ 12928, 3 ex. (DNA): 20°15′50″ S 46°20′56″ W); Cambeva sp. 1 (UFRJ 14061, 12 ex.; UFRJ 14062, 5 ex., C&S; UFRJ 14063, 6 ex., DNA: 22°28′02″ S 45°21′38″ W); Cambeva sp. 2 (UFRJ 14059, 5 ex.; UFRJ 14060, 2 ex., C&S: 20°34′47″ S 46°20′56″ W); Cambeva sp. 3 (UFRJ 14050, 1 ex.; UFRJ 14051, 11 ex; UFRJ 14052, 4 ex. (C&S); UFRJ 14042, 2 ex. (DNA); CICCAA 08193, 4 ex.: 20°36′27″ S 46°26′09″ W); Cambeva sp. 4 (UFRJ 13637, 1 ex.; UFRJ 14054, 1 ex.; UFRJ 13638, 1 ex. (DNA); UFRJ 14055, 9 ex.; UFRJ 14056, 4 ex. (DNA); UFRJ 14057, 5 ex. (C&S): 20°35′19″ S 46°13′41″ W); Cambeva sp. 5: (UFRJ 13653, 1 ex.; UFRJ 12967, 6 ex.; UFRJ 12968, 22 ex.; UFRJ 12983, 9 ex.; UFRJ 13654, 4 ex. (C&S); CICCAA 07950, 10 ex.: 19°58′29″ S 43°51′39″ W; UFRJ 12965, 7 ex.; UFRJ 12966, 15 ex.; UFRJ 12982, 3 ex.: 19°58′26″ S 43°47′47″ W). In addition, we accessed data taken from the type material of C. concolor and C. variegata, deposited in Museu de Zoologia, Universidade de São Paulo, at the time of the original description by one of us (WJEMC). Comparative material appears in Costa et al. [11] and included references.
2.2. Morphological Data
Methods for taking morphological data and terminology followed the methods of our most recent studies on the systematics of eastern South American trichomycterines (e.g., [1]), which were based on Costa [14] and Kubicek [15] for bone nomenclature, Arratia and Huaquin [16] and Bockmann and Sazima [17] for the terminology of pores of the cephalic latero-sensory system, and Bockmann and Sazima [17] for fin-ray formulae. Morphological comparisons were made in the two nominal species, C. concolor and C. variegata, and in five undescribed species, Cambeva sp. 1–5 (see list of specimens above).
2.3. DNA Extraction, Amplification, and Sequencing
Methods for DNA extraction are those described in our recent studies on trichomycterines [1]. Polymerase chain reaction (PCR) was performed with the following primers for mitochondrial encoded genes: Cytb Siluri F and Cytb Siluri R [18] for cytochrome b (CYTB); FISH-F1 and FISH-R1 [19] for cytochrome c oxidase I (COX1); L11935 and H12857 [20] for NADH: ubiquinone oxidoreductase core subunit 4 (ND4), along with t-RNA-His, Ser, and Leu; and for the nuclear gene recombination activating 2 (RAG2), RAG2 TRICHO F and RAG2 TRICHO R [21]. PCR was performed in 45 μL with the following reagent concentrations: 5× GreenGoTaq Reaction Buffer (Promega), 2.0 mM MgCl2, 1 μM of each oligo, 0.2 mM of each dNTP, 1 μL of Promega GoTaq Hot Start polymerase, and 50 ng of total genomic DNA. Negative controls were used to check for contaminants. The thermal profile consisted of initial denaturation at 95 °C for 5 min; followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 45–50 °C for 1 min, and extension at 73 °C for 1–1.5 min; with a final extension at 73 °C for 7 min. PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Sequencing reactions were performed in 20 μL reaction volumes containing 4 μL BigDye, 2 μL sequencing buffer 5× (Applied Biosystems), 2 μL of the PCR products (30–40 ng), 2 μL primer, and 10 μL ultrapure water, and the thermal profile was 35 cycles of 30 s at 95 °C, 30 s at 55 °C, and 1.5 min at 72 °C. MEGA 11 [22] was used for analysing sequencing chromatograms and sequence annotation, and for translating DNA sequences into amino acid residues to confirm the absence of premature stop codons or indels. GenBank accessions are provided in Appendix A.
2.4. Phylogenetic Analysis
Terminal ingroup taxa comprised a total of fifteen species, including five species of the CVG and ten species representing other lineages of Cambeva. The species of the CVG analysed were C. variegata and four undescribed species. Cambeva concolor and Cambeva sp. 2, without available material for DNA sequencing, were not included in the phylogenetic analysis, with their possible relationships inferred from an examination of morphological traits (see list of specimens examined above). Outgroups comprised five species of Scleronema, the sister group of Cambeva, and five species of Trichomycterus s.s., the sister group of Cambeva plus Scleronema [2], two trichomycterines representing another subfamilial lineage, three species representing other trichomycterid subfamilies, four species representing other catfish families, and one representative of another Ostariophysi lineage, order Characiformes. The gene datasets were aligned using the Clustal W algorithm [23] implemented in MEGA 11, not finding gaps or stop codons. The 3368 bp complete dataset (COX1 732 bp, ND4 693 pb, tRNA His Ser Leu 162 pb, CYTB 993 bp, RAG2 788 bp) was analysed using PartitionFinder2.1.1 [24] for optimal partitioning and evolutionary models, using the Corrected Akaike Information (AICc) selection criteria. Partitions and respective models appear in Appendix B.
Bayesian Inference (BI) and Maximum Likelihood (ML) analyses were utilised as independent approaches for phylogenetic reconstruction, aiming to mitigate methodological biases. BI was conducted using Beast 1.10.4 [25] with the following parameters: Birth–Death process as the tree prior [26], and two independent Markov chain Monte Carlo (MCMC) runs with 107 generations with a sampling frequency of 1000 each. The convergence of the MCMC chains and the proper burn-in value were determined by evaluating the achievement of the stationary phase and the effective sample size for all the analysis parameters in both runs using Tracer 1.7.2 [27]. LogCombiner v.1.10.4 and the Tree Annotator version 1.10.4 [25] were employed to combine and calculate the consensus tree, apply the burn-in, and annotate the Bayesian posterior probabilities. The ML analysis utilised IQTREE 2.2.2.6 [28]. Node support was assessed by ultrafast bootstrap (UFBoot) [29] and the Shimodaira–Hasegawa-like approximate likelihood ratio test (SH-aLRT), both with 1000 replicates [30].
2.5. Divergence Time Estimation
The divergence time analysis was conducted in Beast 1.10.4 using the same dataset, partitions, evolution models, tree priors, and parameters as described above. Additionally, the analysis incorporated a lognormal uncorrelated relaxed clock model. Calibration points were established as follows: the origin of Siluriformes Order with a normal prior distribution (mean = 140 MA, SD = 7) following Lundberg et al. [31], the origin of the Trichomycteridae with a normal prior distribution (mean = 106 MA, SD = 5.0) following the estimative of Betancur-R et al. [32]; and the origin of the genus Corydoras Lacépède, 1803, with a lognormal prior distribution (mean = 55 MA, SD = 2.5), based on the dating of the oldest known Corydoras fossil species, Corydoras revelatus Cockerell, 1925. MCMC chains were assessed to verify convergence by evaluating the effective sample size of the runs in Tracer 1.7.1. The time-scaled tree was obtained using Tree Annotator version 1.10.4 to generate the consensus tree.
3. Results
3.1. Phylogenetic Relationships and Comparative Morphology
The CVG was supported as monophyletic with maximum values, whereas relationships among the more inclusive lineages of Cambeva were weakly supported (Figure 1). Individual tree loci generated compatible results (Supplementary File S1), with the CVG recovered as monophyletic in both trees, but without any internal resolution in the RAG2 tree. In addition to the presence of an adipose crest on the caudal peduncle (Figure 1A,B), CVG species differ from other congeners by the presence of an interrupted supraorbital canal, with an s4 pore (Figure 2; vs. continuous, s4 absent), a pronounced narrowing at the lateral end of the premaxilla (Figure 3A–E; vs. expansion absent), and a ventral expansion in the preopercle (Figure 4; vs. expansion absent). In addition, only in species of the CVG, the foramen of the parurohyal is rudimentary or absent (Figure 5B–E), but a broad foramen is present in a new species of this clade described below (Figure 5A), possibly as a result of a reversal due to its apical position in the phylogenetic tree (Figure 1). The CVG group comprises two main lineages, Cambeva sp. 1 from tributaries of the Rio Grande drainage at the Serra da Mantiqueira and the Cambeva variegata complex, a group of similar species occurring in a broad geographical region encompassing the upper Rio São Francisco basin and an adjacent area of the Rio Grande basin at the Serra da Canastra (Figure 6). A taxonomical revision of the Cambeva variegata complex is in progress by the authors and new species here cited will be described elsewhere.
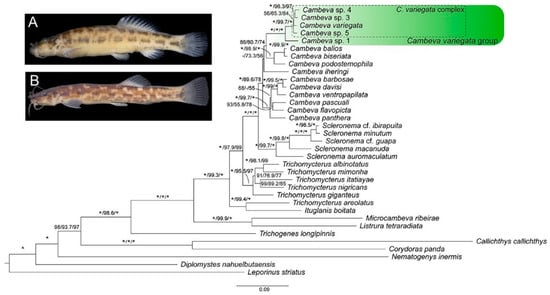
Figure 1.
Phylogenetic tree obtained from the Bayesian analysis in BEAST 1.10.4 for 15 species of Cambeva, including 5 belonging to the C. variegata species group, and 20 outgroup species, using a dataset comprising COI, CYTB, ND4, t-RNA-His, Ser, Leu and RAG2 (total of 3368 bp). Numbers separated by bars (/) above branches indicate posterior probabilities from the Bayesian Inference, followed by ultrafast bootstrap (UFBoot) and the Shimodaira–Hasegawa-like approximate likelihood ratio test (SH-aLRT) from the Maximum Likelihood analysis. Asterisks (*) indicate maximum support values, and dashes (-) indicate support values below 50. (A) Cambeva sp. 1; (B) Cambeva sp. 5.

Figure 2.
Head, dorsal view, showing cephalic pores of the latero-sensory system of Cambeva sp. 5, UFRJ 12967, 46.5 mm SL.
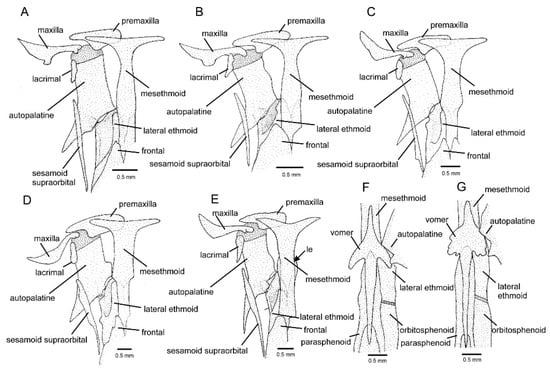
Figure 3.
Mesethmoidal region: (A–E) middle and left portions, dorsal view, (F,G) middle portion, ventral view: (A) Cambeva sp. 3; (B) Cambeva sp. 4; (C,F) Cambeva sp. 5; (D) Cambeva variegata; (E,G) Cambeva sp. 1. Arrow indicates the lateral expansion of the mesethmoid. Larger stippling represents cartilage.
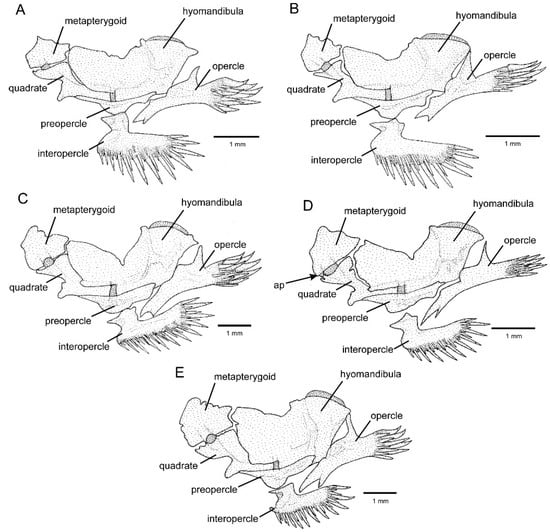
Figure 4.
Left jaw suspensorium and opercular series, lateral view: (A) Cambeva sp. 3; (B) Cambeva sp. 4; (C) Cambeva sp. 5; (D) Cambeva variegata; (E) Cambeva sp. 1. Arrow indicates the anterior articulatory process. Larger stippling represents cartilage.
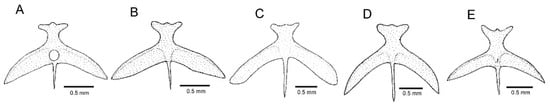
Figure 5.
Parurohyal, ventral view: (A) Cambeva sp. 3; (B) Cambeva sp. 4; (C) Cambeva sp. 5; (D) Cambeva variegata; (E) Cambeva sp. 1.
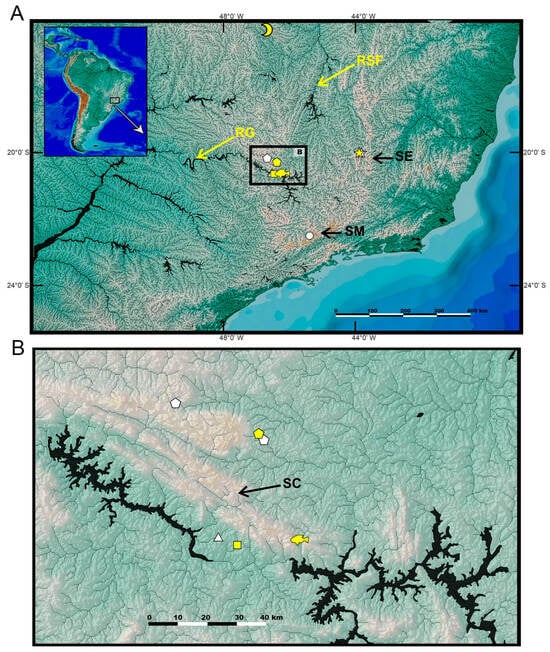
Figure 6.
Geographical distribution of the Cambeva variegata group ((A), general view of the region; (B), detailed view of the area in Serra da Canastra): circle, Cambeva sp. 1; fish, Cambeva sp. 4; half-moon, Cambeva concolor; pentagons, Cambeva variegata; square, Cambeva sp. 3; stars, Cambeva sp. 5; triangle, Cambeva sp. 2. Black arrows indicate mountain ranges: SC, Serra da Canastra; SE, Serra do Espinhaço; SM, Serra da Mantiqueira. Yellow arrows indicate major rivers: RG, Rio Grande; RSF, Rio São Francisco.
Cambeva sp. 1, previously identified as Trichomycteridae sp. by Thereza and Langeani [8], was strongly supported as a member of the CVG here. Externally, it differs from all other congeners by having a long dorsal fin with twelve or thirteen principal rays (vs. nine or ten) (Figure 1A). Seven osteological features distinguish this species from all other congeners: postero-lateral process of the autopalatine posteriorly directed (Figure 3E; vs. postero-laterally directed, Figure 3A–D); posterior extremity of the lateral process of the vomer sinuous (Figure 3G; vs. convex, Figure 3F); six branchiostegal rays (vs. seven or eight); epibranchial 2 elongated, lacking the postero-distal process (Figure 7B; vs. not elongated, with a postero-distal process, Figure 7A); pharyngobranchial 4 relatively large, occupying more than half the surface of the adjacent dentigerous plate (Figure 7B; vs. one-third or less, Figure 7A); dorsal and ventral hypural plates in close proximity, separated by a short posterior interspace (Figure 8B; vs. completely separated, Figure 8A); and pelvic bone relatively broad, with short anterior processes (Figure 8D; vs. narrower, with longer anterior processes, Figure 8C). In addition, the entire anterior portion of the neurocranium is proportionally slenderer (Figure 3E) than in other congeners (Figure 3A–D). This taxon also differs from all other congeners of the CVG by the absence of a pectoral fin filament and the presence of a lateral expansion of the mesethmoid (Figure 3E; vs. absence); the mesethmoid cornua having a relatively short and sharply pointed cornu (Figure 3E; vs. longer, not sharply pointed, Figure 3A–D); and pharyngobranchial 3 with distinctive lateral constriction (Figure 7B; vs. without lateral constriction, Figure 7A). This species is being formally described by F. Langeani and collaborators (Langeani personal communication to WJEMC, 2 January 2024).
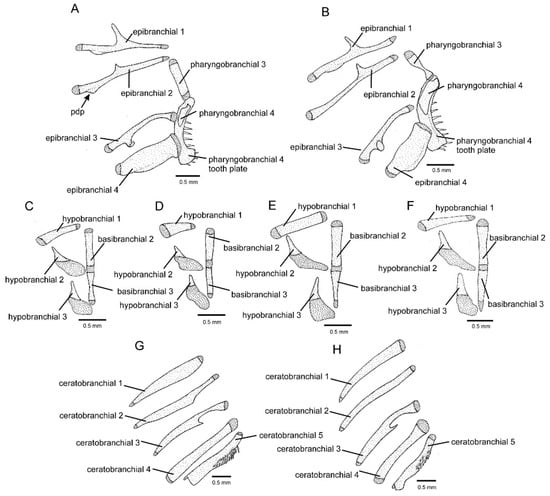
Figure 7.
Branchial bones: (A,B) left dorsal arches, dorsal view, (C–F) basibranchials and left hypobranchials, dorsal view, (G,H) left ceratobranchials, dorsal view: (A) Cambeva variegata; (B,F,H) Cambeva sp. 1; (C) Cambeva sp. 3; (D) Cambeva sp. 4; (E,G) Cambeva sp. 5. Arrow indicates the poster-distal process of the first epibranchial. Larger stippling represents cartilage.
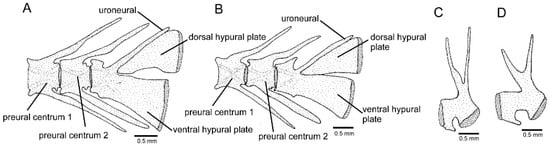
Figure 8.
Post-cranial structures: (A,B) caudal skeleton, left lateral view, (C,D) pelvic bone: (A) Cambeva variegata; (B,D) Cambeva sp. 1; (C) Cambeva sp. 5. Larger stippling represents cartilage.
The C. variegata complex, comprising Cambeva variegata, Cambeva sp. 3, Cambeva sp. 4, and Cambeva sp. 5, was corroborated in the phylogenetic analysis with high support values (Figure 1). In addition to the unique character states described for Cambeva sp. 1 that are not present in the C. variegata complex (see above), species of this complex are distinguished from all congeners by a relatively short posterior process of the vomer, which is about 1.5 times longer than the vomer length excluding the posterior process (Figure 3F; vs. about two times longer or more, Figure 3G). Relationships within the C. variegata group were weakly supported, except for the sister group relationships between Cambeva sp. 3 and Cambeva sp. 4, two species occurring in close localities of the upper Rio Paraná basin at Serra da Canastra (Figure 6).
3.2. Time-Calibrated Phylogeny
According to the time-calibrated analysis, the origin of Cambeva would have occurred in the late Oligocene, whereas the origin of CVG occurred in the middle Mioceno, which was followed by the split between Cambeva sp. 1 and the C. variegata complex in the late Miocene, with the diversification within the C. variegata complex occurring between the Pliocene and Pleistocene (Figure 9).
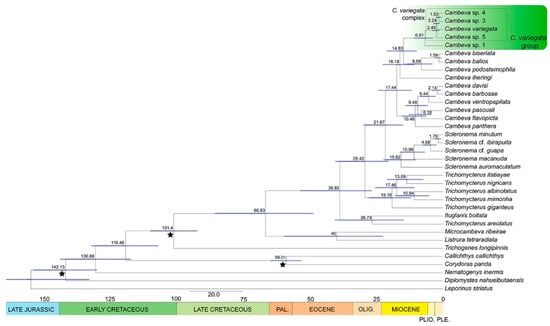
Figure 9.
Time-scaled phylogeny obtained from the Bayesian analysis in BEAST 1.10.4 for 15 species of Cambeva species, including 5 belonging to the C. variegata species group, and 20 outgroup species, using a dataset comprising COI, CYTB, ND4, t-RNA-His, Ser, Leu, and RAG2 (total of 3368 bp). Black stars indicate calibration points, numbers above nodes indicate median age, blue bars on nodes indicate 95% HPD ranges of those ages, and coloured bars below the tree represent geological epochs.
4. Discussion
4.1. Phylogenetic Relationships
Species of the CVG share four morphological character states that are unique among congeners: 1—the presence of an adipose crest on the caudal peduncle; 2—an interrupted supraorbital canal, with an s4 pore (Figure 2); 3—a pronounced narrowing at the lateral end of the premaxilla (Figure 3); 4—a ventral expansion in the preopercle (Figure 4). However, the first three character states are also present in species of the genus Scleronema [4,5,6], which is sister to Cambeva [2,33], but not in Trichomycterus s.s. [14], sister to the clade comprising Cambeva and Scleronema [2].
Considering only the distribution of these morphological character states, the most plausible hypothesis would be to interpret Cambeva as paraphyletic, with the CVG being more related to Scleronema than to the other lineages of Cambeva. However, all molecular phylogenetic analyses available to date point to the monophyly of Cambeva when including the species of the CVG (e.g., [2,9,10]), although the position of this group within Cambeva was different in previous studies (i.e., sister to Cambeva iheringi instead of to the Cambeva gama-clade as in the present study). It is important to note that the topology found in previous studies that used a smaller sample of genetic markers, where the CVG is sister to C. iheringi, is congruent with the unique possession of a long supraorbital sesamoid in these two groups (Figure 3), which does not occur in other species of Cambeva and Scleronema. The possibility of the three conditions shared by species of the C. variegata group and Scleronema consisting of primitive conditions present in the most recent common ancestor of the clade comprising Cambeva plus Scleronema and subsequently lost in other lineages of Cambeva cannot be ruled out. An accurate optimisation of these character states depends on a phylogeny where all Cambeva lineages are positioned with high support values, which is still not available.
Monophyly of the C. variegata group is tentatively supported by the presence of a ventral expansion in the preopercle (character state 4 above), a condition variable within the CST clade. A rudimentary or absent middle foramen of the parurohyal (Figure 5B–E) may be considered further evidence of monophyly if admitting a reversion in Cambeva sp. 3, which is the only species of the group with a broad foramen (Figure 5A) and appearing in an apical position within the CVG topology (Figure 1).
4.2. Distribution Patterns
The geographical distribution of the CVG comprises a wide region of southeastern Brazil encompassing the upper Rio São Francisco basin and the Rio Grande drainage, which is a part of the Rio Paraná basin (Figure 6). As in most groups of Trichomycterinae, the distribution of the CVG is concentrated in mountain ranges, such as the Serra da Canastra, Serra do Espinhaço, and Serra da Mantiqueira. Geological data indicate that the Serra da Mantiqueira region, the area of endemism of Cambeva sp. 1, which is strongly supported as the sister group to all other CVG species, already acted as a watershed between the basins located south of this mountain range and the Rio Grande drainage at least since the Eocene, which happened synchronously with the installation of the Continental Rift of southeastern Brazil [34]. Evidence indicates that the natural flow path of the Rio Grande drainage course headed towards the São Francisco River basin, with which it was connected in the past [35]. Thus, at least during the Paleogene and part of the Neogene, the Rio Grande drainage acted as an upper tributary of the Rio São Francisco basin, not as a main tributary of the Rio Paraná River as presently. Therefore, the origin of the CVG here estimated to have occurred in the Middle Miocene (Figure 9) portrays a palaeogeographic scenario in which the Rio Grande drainage and the São Francisco basin formed a single hydrographic basin.
Geological data indicate that the disruption of the ancient connections between the current Rio Grande drainage and the São Francisco River basin, and the consequent capture of the Rio Grande drainage by the Rio Paraná basin, began in the Middle Miocene, following a process of widespread uplift in the region [35]. This historical process is compatible with the divergence found between Cambeva sp. 1, endemic to the Rio Grande drainage, and the C. variegata complex, endemic to the upper Rio São Francisco basin and a small area of the Rio Grande drainage on the periphery of the upper Rio São Francisco, which would have occurred at the end of the Miocene according to our estimates (Figure 9). Therefore, a major rupture in the Grande-São Francisco palaeo basin would be responsible for a vicariance event involving Cambeva sp. 1 and the Cambeva variegata complex. A similar vicariance event was hypothesised by Vilardo et al. [36] for lineages of the subgenus Paracambeva Costa, 2021, of the genus Trichomycterus inhabiting the same area.
Additionally, geological data suggest that the drainage rearrangement was a gradual process, reaching the current hydrographic configuration only in the Pliocene or early Pleistocene [35]. Precisely, the area of the Serra da Canastra region that separates the distribution of C. variegata, endemic to the São Francisco River basin, from the area inhabited by the clade comprising Cambeva sp. 3 and Cambeva sp. 4 (Figure 6) is one of the points of altimetric anomalies, characterising an area of probable past connection between the drainage of the Rio Grande and the Rio São Francisco [35]. Therefore, despite the low support values found at internal nodes of the C. variegata complex, making the relationship hypothesis weak, the estimated split between those two lineages in the Pliocene may indicate a past rupture event in this particular area, artificially restored with transposition works during construction of the Furnas hydroelectric power dam carried out in the 1960s [37].
5. Conclusions
This study reports the occurrence of the genus Cambeva in a vast area of southeastern Brazil, including the first records of the genus in two mountain ranges considered among the most important centres of biodiversity in the world, with high rates of species endemism: the Serra do Espinhaço and Serra da Mantiqueira [38,39]. Over 40 trichomycterines of the genus Trichomycterus s.s. have been continuously reported from these two mountain ranges since the nineteenth century [40,41] and early twentieth century [42,43] to the present (e.g., [2] and included references), where they are usually common in every river drainage, contrasting with only two species of the CVG reported here for the first time. Therefore, this study demonstrates the relative rarity of species of the CVG and its important role in recovering ancient biogeographic patterns, highlighting the urgent need for more detailed studies on the diversity and phylogenetic relationships of still poorly known organisms in this highly diverse and threatened region.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9040116/s1, Supplementary File S1: Gene loci trees.
Author Contributions
Conceptualisation, W.J.E.M.C.; data obtaining, W.J.E.M.C., J.L.O.M., V.M.A.-S., C.R.M.F., P.F.A., F.P.O., P.J.V. and A.M.K.; formal analysis, W.J.E.M.C. and J.L.O.M.; investigation and data curation, W.J.E.M.C., J.L.O.M., V.M.A.-S., C.R.M.F., P.F.A. and A.M.K.; writing—original draft preparation, W.J.E.M.C.; writing—final version, W.J.E.M.C. and J.L.O.M.; visualisation, W.J.E.M.C., J.L.O.M. and A.M.K.; supervision, W.J.E.M.C.; project administration, W.J.E.M.C.; funding acquisition, W.J.E.M.C., J.L.O.M., F.P.O. and A.M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; grant 304755/2020-6 to WJEMC, 307974/2021-9 to FPO, 140689/2022-2 to PJV), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ; grant E-26/201.213/2021 to WJEMC, E-26/203.524/2023 to JLOM, and E-26/202.005/2020 to AMK), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; grant 88887.466724/2019-00 to PFA), and Fundação de Amparo à Pesquisa e ao Desenvolvimento Científico e Tecnológico do Maranhão (FAPEMA; grant BPD-03083/20 to FPO). This study was also supported by CAPES (Finance Code 001) through the Programa de Pós-Graduação em: Biodiversidade e Biologia Evolutiva/UFRJ and Genética/UFRJ.
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee for Animal Use of the Federal University of Rio de Janeiro (protocol code: 065/18, approved on August 2018, and 084/23, approved on October 2023).
Informed Consent Statement
Not applicable.
Data Availability Statement
DNA sequences used in this study are deposited in GenBank.
Acknowledgments
We are grateful to D. G. A. Lopes, G. Correia-Silva, M. F. Vieira, M. E. Molina, and R. V. Demez, all from Eduvale, for help during field collections in the Serra da Canastra region, and G. L. Canella and R. dos Santos-Junior in Serra da Mantiqueira. We also thank D. Deliberali, I. Loyola and S. A. Domingues for logistical support during field studies in Serra do Espinhaço. Thanks are also due to M. Petrungaro, L. I. Chaves, L. Santos, and L. Neves for technical assistance in the laboratory. The manuscript benefitted from the criticisms made by two anonymous reviewers.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A

Table A1.
Terminal taxa for molecular phylogeny and respective GenBank accession numbers.
Table A1.
Terminal taxa for molecular phylogeny and respective GenBank accession numbers.
| COI | ND4 | tRNA Ser His Leu | CYTB | RAG2 | |
|---|---|---|---|---|---|
| Leporinus striatus | JN989019.1 | — | — | EU183020.1 | AY804096.1 |
| Diplomystes nahuelbutaensis | AP012011.1 | NC015823.1 | NC015823.1 | MN640590 | DQ492317 |
| Nematogenys inermis | EU359428 | AY307250.1 | AY307250.1 | — | KY858182.1 |
| Corydoras panda | NC049097.1 | GU210065.1 | GU210065.1 | NC049097.1 | KP960362.1 |
| Callichthys callichthys | MZ051783.1 | AY307241.1 | AY307241.1 | KP960058 | DQ492324 |
| Trichogenes longipinnis | OQ810037 | MN389484 | MN389484 | MK123704 | MF431117 |
| Microcambeva ribeirae | MN385807.1 | MN389502.1 | MN389502.1 | OK334290 | MN385832 |
| Listrura tetraradiata | JQ231083 | MN389497 | MN389497.1 | JQ231088.1 | MN385826.1 |
| Trichomycterus areolatus | AP012026.1 | AP012026.1 | AP012026.1 | FJ772214 | KY858188 |
| Ituglanis boitata | OQ810038 | MN389485.1 | MN389485.1 | MK123706 | MK123758 |
| Trichomycterus itatiayae | MW671552 | OR948809 | — | MW679291 | OL779233 |
| Trichomycterus nigricans | MN813005 | MN389488.1 | MN389488.1 | MK123723 | MK123765 |
| Trichomycterus albinotatus | MN813007 | OM324337.1 | OM324337.1 | MK123716 | MN812990 |
| Trichomycterus mimonha | MW196749 | OM324343.1 | OM324343.1 | MW196758 | MW196783 |
| Trichomycterus giganteus | MT470413.1 | PP333226 | PP336672 | MK123720.1 | MT446426.1 |
| Scleronema minutum | MK123685 | MN389486.1 | MN389486.1 | MK123707 | MK123759.1 |
| Scleronema cf. guapa | PP319012 | PP333227 | PP336673 | MK123709.1 | MF431118.1 |
| Scleronema cf. ibirapuita | MK123688.1 | PP333228 | PP336674 | MK123710.1 | MK123761.1 |
| Scleronema macanuda | MK123686.1 | PP333229 | PP336675 | MK123708.1 | MK123760.1 |
| Scleronema auromaculatum | OM037445.1 | — | — | OM037134.1 | OM037136.1 |
| Cambeva barbosae | MK123689.1 | MN389487.1 | MN389487.1 | OQ110808 | OQ110815.1 |
| Cambeva balios | OQ810040 | PP333230 | PP336676 | OQ814186 | OQ814193 |
| Cambeva pascuali | MF034463 | PP333231 | PP336677 | OQ110811 | OQ110820 |
| Cambeva panthera | OQ810041 | PP333232 | PP336678 | OQ814187 | OQ814194 |
| Cambeva flavopicta | OQ810042 | PP333233 | PP336679 | OQ814188 | OQ814195 |
| Cambeva davisi | PP319014 | PP333234 | PP336680 | MK123714 | MK123762 |
| Cambeva biseriata | PP319015 | PP333235 | PP336681 | OQ110806 | OQ110817 |
| Camveba ventropapilata | PP319016 | PP333236 | PP336682 | OQ110807 | OQ110818 |
| Cambeva iheringi | GU701893 | — | — | KY858074 | KY858223 |
| Cambeva sp. 1 | PP319017 | PP333237 | PP336683 | PP328532 | PP333215 |
| Cambeva sp. 5 | PP319018 | PP333238 | PP336684 | PP328533 | PP333216 |
| Cambeva variegata | PP319019 | PP333239 | PP336685 | PP328534 | PP333217 |
| Cambeva sp. 3 | PP319020 | PP333240 | PP336686 | PP328535 | PP333218 |
| Cambeva sp. 4 | PP319021 | PP333241 | PP336687 | PP328536 | PP333219 |
| Cambeva podostemophila | OQ810043 | — | — | OQ814189 | OQ814196 |
Appendix B

Table A2.
Best-fitting partition schemes and evolutive models.
Table A2.
Best-fitting partition schemes and evolutive models.
| Partition | Base Pairs | Evolutive Model |
|---|---|---|
| COI 1st | 244 | TRN+I+G |
| COI 2nd | 244 | HKY+I |
| COI 3rd | 244 | TRN+I+G |
| ND4 1st | 231 | GTR+I+G |
| ND4 2nd, CYTB 1nd | 562 | TRN+I+G |
| ND4 3rd | 231 | GTR+G |
| tRNA His Ser Leu | 162 | GTR+G |
| CYTB 1st | 331 | TRN+I+G |
| CYTB 3rd | 331 | GTR+I+G |
| RAG2 1st | 263 | HKY+G |
| RAG2 2nd | 263 | GTR+I |
| RAG2 3rd | 263 | K80+I |
References
- Costa, W.J.E.M.; Azevedo-Santos, V.M.; Mattos, J.L.O.; Katz, A.M. Molecular phylogeny, taxonomy and distribution patterns of trichomycterine catfishes in the middle Rio Grande drainage, south-eastern Brazil (Siluriformes: Trichomycteridae). Fishes 2023, 8, 206. [Google Scholar] [CrossRef]
- Katz, A.M.; Barbosa, M.A.; Mattos, J.L.O.; Costa, W.J.E.M. Multigene analysis of the catfish genus Trichomycterus and description of a new South American trichomycterine genus (Siluriformes, Trichomycteridae). Zoosyst. Evol. 2018, 94, 557–566. [Google Scholar] [CrossRef]
- Costa, W.J.E.M. Description de huit nouvelles espèces du genre Trichomycterus (Siluriformes: Trichomycteridae), du Brésil oriental. Rev. Franç. d’aquariol. Herpetol. 1992, 18, 101–110. [Google Scholar]
- Ferrer, J.; Malabarba, L.R. Systematic revision of the Neotropical catfish genus Scleronema (Siluriformes: Trichomycteridae), with descriptions of six new species from Pampa grasslands. Neotrop. Ichthyol. 2020, 18, e190081. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Sampaio, W.M.S.; Giongo, P.; de Almeida, F.B.; Azevedo-Santos, V.M.; Katz, A.M. An enigmatic interstitial trichomycterine catfish from south-eastern Brazil found at about 1000 km away from its sister group (Siluriformes: Trichomycteridae). Zool. Anz. 2022, 297, 85–96. [Google Scholar] [CrossRef]
- Bockmann, F.A.; Ferrer, J.; Rizzato, P.P.; Esguícero, A.L.H.; Duboc, L.F.; Ingenito, L.F.S. Anatomy, ecology, and behavior of a new species of Scleronema Eigenmann, 1917 (Siluriformes: Trichomycteridae) from coastal drainages in the southern Brazilian Atlantic Rainforest, with comments on the monophyly and phylogeny of the genus. Zootaxa 2023, 5297, 1–47. [Google Scholar] [CrossRef] [PubMed]
- de Pinna, M.C.C. A new subfamily of Trichomycteridae (Teleostei, Siluriformes), lower locarioid relationships and a discussion on the impact of additional taxa for phylogenetic analysis. Zool. J. Linn. Soc. 1992, 106, 175–229. [Google Scholar] [CrossRef]
- Thereza, M.R.; Langeani, F. Bagres e Cascudos do Rio Grande, Alto Rio Paraná; Editora CRV: Curitiba, Brazil, 2019; 122p. [Google Scholar]
- Ochoa, L.E.; Roxo, F.F.; DoNascimiento, C.; Sabaj, M.H.; Datovo, A.; Alfaro, M.; Oliveira, C. Multilocus analysis of the catfish family Trichomycteridae (Teleostei: Ostariophysi: Siluriformes) supporting a monophyletic Trichomycterinae. Mol. Phyl. Evol. 2017, 115, 71–81. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Feltrin, C.R.M.; Mattos, J.L.M.; Amorim, P.F.A.; Katz, A.M. Phylogenetic relationships of new taxa support repeated pelvic fin loss in mountain catfishes from southern Brazil (Siluriformes: Trichomycteridae). Zool. Anz. 2023, 305, 82–90. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeir, R.A.; Mittermeir, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Leary, S.; Underwood, W.; Anthony, R.; Cartner, S.; Corey, D.; Grandin, T.; Greenacre, C.; Gwaltney-Brant, S.; McCrackin, M.; Meyer, R.; et al. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. 2020. Available online: http://www.avma.org/sites/default/files/2020-02/Guidelines-on-Euthanasia-2020.pdf (accessed on 3 December 2022).
- Taylor, W.R.; Van Dyke, G.C. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 1985, 9, 107–119. [Google Scholar]
- Costa, W.J.E.M. Comparative osteology, phylogeny and classification of the eastern South American catfish genus Trichomycterus (Siluriformes: Trichomycteridae). Taxonomy 2021, 1, 160–191. [Google Scholar] [CrossRef]
- Kubicek, K.M. Developmental osteology of Ictalurus punctatus and Noturus gyrinus (Siluriformes: Ictaluridae) with a discussion of siluriform bone homologies. Verteb. Zool. 2022, 72, 661–727. [Google Scholar] [CrossRef]
- Arratia, G.; Huaquin, L. Morphology of the lateral line system and of the skin of diplomystid and certain primitive loricarioid catfishes and systematic and ecological considerations. Bonn. Zool. Monogr. 1995, 36, 1–110. [Google Scholar]
- Bockmann, F.A.; Sazima, I. Trichomycterus maracaya, a new catfish from the upper rio Paraná, southeastern Brazil (Siluriformes: Trichomycteridae), with notes on the T. brasiliensis species-complex. Neotrop. Ichthyol. 2004, 2, 61–74. [Google Scholar] [CrossRef]
- Villa-Verde, L.; Lazzarotto, H.; Lima, S.Q.M. A new glanapterygine catfish of the genus Listrura (Siluriformes: Trichomycteridae) from southeastern Brazil, corroborated by morphological and molecular data. Neotrop. Ichthyol. 2012, 10, 527–538. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Shimabukuro-Dias, C.K.; Oliveira, C.; Reis, R.E.; Foresti, F. Molecular phylogeny of the armored catfish family Callichthyidae (Ostariophysi, Siluriformes). Mol. Phylog. Evol. 2004, 32, 152–163. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Henschel, E.; Katz, A.M. Multigene phylogeny reveals convergent evolution in small interstitial catfishes from the Amazon and Atlantic forests (Siluriformes: Trichomycteridae). Zool. Scr. 2020, 49, 159–173. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Vir. Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Gernhard, T. The conditioned reconstruction process. J. Theor. Biol. 2008, 253, 769–778. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefor, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, J.G.; Sullivan, J.P.; Rodiles-Hernández, R.; Hendrickson, D.A. Discovery of African roots for the Mesoamerican Chiapas catfish, Lacantunia enigmatica, requires an ancient intercontinental passage. Proc. Acad. Nat. Sci. Philad. 2007, 156, 39–53. [Google Scholar] [CrossRef]
- Betancur, R.-R.; Órti, G.; Pyron, R.A. Fossil-based comparative analyses reveal ancient marine ancestry erased by extinction in ray-finned fishes. Ecol. Lett. 2015, 18, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, L.E.; Datovo, A.; DoNascimiento, C.; Roxo, F.F.; Sabaj, M.H.; Chang, J.; Melo, B.F.; Silva, G.S.C.; Foresti, F.; Alfaro, M.; et al. Phylogenomic analysis of trichomycterid catfshes (Teleostei: Siluriformes) inferred from ultraconserved elements. Sci. Rep. 2020, 10, 2697. [Google Scholar] [CrossRef]
- Riccomini, C.; Sant’anna, L.G.; Ferrari, A.L. Evolução geológica do Rift Continental do Sudeste do Brasil. In Geologia do Continente Sul-Americano: Evolução da obra de Fernando Flávio Marques de Almeida; Mantesso-Neto, V., Bartorelli, A., Carneiro, C.D.R., Brito-Neves, B.B., Eds.; Beca: São Paulo, Brazil, 2004; pp. 383–405. [Google Scholar]
- Rezende, E.A.; Salgado, A.A.R.; Castro, P.T.A. Evolução da rede de drenagem e evidências de antigas conexões entre as bacias dos rios Grande e São Francisco no sudeste brasileiro. Ver. Brasil. Geomorfol. 2018, 19, 483–501. [Google Scholar] [CrossRef]
- Vilardo, P.J.; Katz, A.M.; Costa, W.J.E.M. Phylogeny and historical biogeography of neotropical catfishes Trichomycterus (Siluriformes: Trichomycteridae) from eastern Brazil. Mol. Phylog. Evol. 2023, 186, 107836. [Google Scholar] [CrossRef]
- Moreira Filho, O.; Buckup, P.A. A poorly known case of watershed transposition between the São Francisco and upper Paraná river basins. Neotrop. Ichthyol. 2005, 3, 449–452. [Google Scholar] [CrossRef]
- Giulietti, A.M.; Pirani, J.R.; Harley, R.M. Espinhaço Range region, eastern Brazil. In Centres of Plant Diversity: A Guide and Strategy for Their Conservation; Davis, S.D., Heywood, V.H., Herrera-Macbryde, O., Villa-Lobos, J., Hamilton, A.C., Eds.; IUCN Publication Unit: Cambridge, UK, 1997; pp. 397–404. [Google Scholar]
- Gonzaga, D.R.; Peixoto, A.L.; Neto, L.M. Patterns of richness and distribution of Cactaceae in the Serra da Mantiqueira, Southeast Brazil, and implications for its conservation. Acta Bot. Bras. 2018, 33, 1–9. [Google Scholar] [CrossRef]
- Lütken, C.F. Siluridae novae Brasiliae centralis a clarissimo J. Reinhardt in provincia Minas-geraës circa oppidulum Lagoa Santa, praecipue in flumine Rio das Velhas et affluentibus collectae, secundum characteres essentiales breviter descriptae. Overs. Kongel. Danske Vidensk. Selsk. Forhandl. Kjobenhavn 1874, 1873, 29–36. [Google Scholar]
- Eigenmann, C.H.; Eigenmann, R.S. Preliminary notes on South American Nematognathi, II. Proc. Calif. Acad. Sci. 1889, 2, 28–56. [Google Scholar] [CrossRef]
- Miranda Ribeiro, A. Fauna brasiliense, peixes, tomo 4. Arquiv. Mus. Nac. 1912, 16, 1–504. [Google Scholar]
- Eigenmann, C.H. The Pygidiidae, a family of South American catfishes. Mem. Carnegie Mus. 1918, 7, 259–398. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).