Abstract
The Hainan small-toothed sleeper, Microdous chalmersi, is an endemic species disjunctly distributed in Hainan and Guangxi provinces of China. Morphological differences have been previously observed between these populations. We sequenced the mitochondrial genomes of M. chalmersi from Hainan and Guangxi in order to test whether there is a cryptic species. We reconstructed a phylogenetic tree of samples collected from the two populations along with eight representative species of other odontobutids using thirteen mitochondrial coding genes and two rRNA genes. The results showed that five individuals of M. chalmersi from each population clustered into reciprocal monophyletic clades. Furthermore, genetic distance between individuals of the two populations was much larger than that between individuals from the same population. The genetic distance between the two Microdous populations was comparable to interspecific genetic distance of a closely related genus, Odontobutis. We propose that M. chalmersi from Guangxi and Hainan could belong to two different species, but detailed morphological and genetic studies should be carried out to test this hypothesis.
Key Contribution:
We found large genetic distance between populations of Microdous chalmersi from Guangxi and Hainan provinces using mitochondrial genome data. The limited and disjunct distribution and the large genetic distance between the two populations call attention to conservation of this fish and further taxonomic investigation.
1. Introduction
Microdous chalmersi (Nichols and Pope 1927), synonyms Philypnus chalmersi, Perccottus chalmersi, or Sineleotris chalmersi, is an endemic sleeper fish inhabiting streams, and is distributed disjunctly in Hainan and Guangxi provinces of southern China (Figure 1). Based on the characteristics of its vomerine teeth, pre-operculum bone margin without spines, and large gill openings, M. chalmersi was initially classified as a species of the genus Philypnus under the family Eleotridae [1], but subsequently assigned to the genus Perccottus under the family of Odontobutidae [2,3]. Lately, Chen et al. (2002) considered M. chalmersi as a species of genus Sineleotris (Odontobutidae) [4], but without providing any evidence.
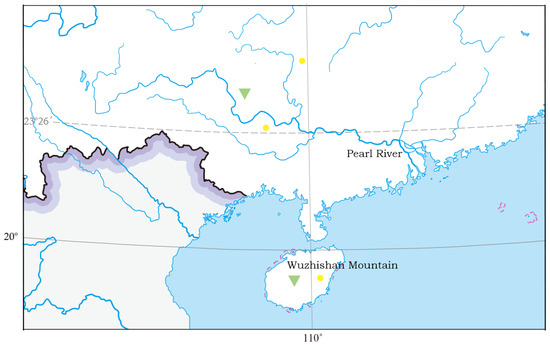
Figure 1.
Sampling sites (green triangles) and additional distribution sites (yellow dots) previously known for Microdous chalmersi (Wu, 2008).
Li et al. (2018) found that M. chalmersi has a synapomorphy of the Odontobutidae, that is, scapula well-developed separating the upper proximal radials of the pectoral fin and the cleithrum [5]. They also reconstructed a phylogenetic tree of the Odontobutidae based on 4434 nuclear gene loci showing that M. chalmersi had a close relationship with the genera Micropercops and Sineleotris. Due to the genetic distance of M. chalmersi to Sineleotris being more divergent than that of Micropercops to Sineleotris, Microdous was proposed as an independent genus instead of being a species of Sineleotris. In their results, the two samples of M. chalmersi from Guangxi were grouped together and then joined to an individual from Hainan with a large distance.
Further to the large genetic distance, we also found some differences in morphological appearance between the specimens collected from the two populations upon closer examination, that is, dense vomerine teeth were found in the individuals from Hainan but sparse in the individuals from Guangxi (Figure 2), and the interorbital width of the individuals from Guangxi was much wider than that of fish from Hainan. Therefore, we suggest that cryptic species may exist in Microdous. In this study, we found ten specimens of Microdous from previous collections, five each from Hainan and Guangxi, respectively. Due to poor preservation status of those specimens, morphological characters could not be further examined. Nonetheless, we sequenced mitochondrial genome of those samples and compared their genetic distance to that between species of a closely related genus, Odontobutis, and performed phylogenetic analyses to test whether there could be cryptic species in Microdous.

Figure 2.
Photos of Microdous chalmersi. M. chalmersi collected from Wuzhishan Mountain, Hainan province (top); M. chalmersi collected from Du’an, Guangxi province (bottom). The red rectangles highlight the region where vomerine teeth can be found.
2. Materials and Methodsfile into a Bam File, and the Parameter
2.1. Sample Collection and DNA Extraction
Ten samples of M. chalmersi were collected, five individuals from Du’an, Guangxi (108°06′25′′ E, 24°23′17′′ N, 5 April 2018) (Supplementary Materials Table S1) and five from Wuzhishan Mountain, Hainan (109°36′13′′ E, 18°52′58′′ N, 24 August 2018). These specimens were stored at the Ichthyological Collection of Shanghai Ocean University (voucher: SOU1801008-2,3,4,5,6 and SOU1801009-1,2,3,5,7; contact person: Dr Ya Zhang, email: zhangya@shou.edu.cn). From each sample, a portion of pectoral fin was taken and stored in 95% alcohol in a −20 °C freezer in the Lab of Molecular Systematics and Ecology, Shanghai Ocean University, Shanghai, China. Twenty-five-milligram tissue was used for DNA extraction according to the Kit instruction (Ezup Column Genomic DNA Purification Kit, Sangon Biotech, Shanghai, China). The integrity of extracted DNA was checked using agarose gel electrophoresis. The concentration of sample DNA was measured using a NanoDropTM 3300 fluorescence spectrophotometer.
2.2. Bait Design, Library Prep, Gene Capture and Sequencing
RNA baits were designed for gene enrichment based on conserved sites of the mitochondrial genome of M. chalmersi (GenBank accession: MH644035; Supplementary Materials Table S2). Three-hundred nanograms of the extracted DNA was used for library preparation. The DNA was sheared into approximately 250 bp in length using Covaris M220 ultrasonic disruptor (Covaris, Inc., Woburn, MA, USA). Library preparation and gene capture were carried out following Meyer and Kircher with some modifications [6,7]. To facilitate multiplex sequencing, each sample was labeled with an 8 bp P7 index adapter. Finally, high-throughput sequencing was performed on an Illumina HiSeq10X platform by Genewiz, Inc. (Shanghai, China).
2.3. Data Assembly
The raw data of each sample were separated by the 8 bp index using a custom perl script [8]. Subsequently, trim_galore v0.6.4 was employed to remove adapter sequences and reads with low-quality bases (Q-score < 20). PCR duplicates were excluded by using “-bfastx_uniques” command in USEARCH v10.0.240 (https://drive5.com/usearch/, accessed on 22 December 2021) [9]. The processed raw reads were assembled using the mitochondrial genome of M. chalmersi (MH644035) as a reference. Briefly, BWA v0.7.16 [10] was used to construct the index of the reference sequence and mem algorithm was used to align the cleaned reads to the reference genome. The ‘fastq’ command of SAMtools v1.10 [11] were used to convert the sam file into a bam file, and the parameter “−F 4” was adopted to retain only the mapped reads. Trinity v2.11.0 [12] were used to reassemble the output reads applying “seqType” command parameter. From the assembly, contigs shorter than 10,000 bp were clipped off. Subsequently, all the contigs and the reference genome were put together in MEGAX [13] for the final alignment. Annotation of the assembly was performed through a web interface platform named ‘Geseq’ that provided an annotation result in the ‘gb’ format (https://chlorobox.mpimp-golm.mpg.de/geseq.html, accessed on 11 January 2022).
2.4. Phylogenetic Analysis
The 13 mitochondrial protein coding genes and 12S and 16S genes of the samples were extracted to reconstruct a phylogenetic tree. Sequences of Micropercops swinhonis (RefSeq accession: NC_021763), Rhyacichthys aspro (RefSeq accession: NC_004414) and 6 species of Odontobutis (O. interrupta (RefSeq accession: NC_027583), O. haifengensis (RefSeq accession: NC_036056), O. potamophilus (RefSeq accession: NC_022706), O. platycephala (RefSeq accession: NC_010199), O. yaluensis, (RefSeq accession: NC_027160), and O. sinensis (RefSeq accession: NC_022818)) were retrieved from NCBI and treated as the outgroups [14,15,16,17,18,19,20]. The respective gene sequences from the above-mentioned species along with M. chalmersi were saved into one fasta file for each gene. The file was imported into MEGAX and DNA sequence of coding genes were converted into amino acid sequence for aligning with MUSCLE algorithm (https://www.ebi.ac.uk/Tools/msa/muscle/, accessed on 11 January 2022), and converted back into DNA sequences afterward. The mitochondrial coding genes were combined into a super-matrix where the 12S and 16S were used as two partitions, and the coding genes were divided into 39 partitions with 1st, 2nd, and 3rd codon position, separately. IQtree v1.7-beta9 [21,22] were used to build the phylogenetic tree. Data partitioning and the best evolution model were estimated through the MFP + MERGE parameters in IQTREE. The final result was viewed through FigTree1.4.2 [23].
2.5. Calculation of Genetic Distance
Genetic distance between individuals from Hainan and Guangxi and between different individuals of the same population was calculated for 12S, 16S, Cytb, COI, and ND2 genes used for phylogenetic reconstruction. Pairwise distance was calculated using MEGAX, with the option of ‘compute pairwise distances’. The resulting genetic distance was ranked and shown in a histogram. Moreover, sequences of the 12S, 16S, Cytb, COI, and ND2 genes of O. interrupta, O. haifengensis, O. potamophilus, O. platycephala, O. yaluensis, and O. sinensis were used to calculate the genetic distance between these species. The average genetic distance and its standard deviation between different populations of M. chalmersi were compared with the genetic distances between different species of the genus Odontobutis.
3. Results
3.1. Data Assembly and Mitochondrial Genomes
On average, 13,326,503 raw reads per sample were obtained. After trimming the adapter sequence and low-quality bases, there were 12,056,126 reads per sample. In the end, 6,513,391 reads remained after removing the PCR duplicates. On average, 48.8% raw reads were useful for mitochondrial genome assembly after data cleanup.
The mitochondrial genomes of samples from both Guangxi and Hainan consisted of 13 protein coding genes, 22 tRNA, 2 rRNA, and a D-loop region. A mitochondrial genome of an individual from Guangxi had 16,485 bp with 27.3% A, 26.2% T, 29.3% C, and 17.2% G (Accession number: ON312089). A mitochondrial genome of an individual from Hainan had 16,482 bp with 27.4% A, 26.1% T, 29.1% C, and 17.5% G (Accession number: ON312088). Mitochondrial genome of other individuals can be found at NCBI under Accession number: OQ852467-OQ852470.
3.2. Phylogenetic Analysis
Sequences of 12S (948 bp), 16S (1678 bp), Cytb (1141 bp), COI (1551 bp), COII (691 bp), COIII (786 bp), ND1 (977 bp), ND2 (1045 bp), ND3 (349 bp), ND4 (1386 bp), ND4L (297 bp), ND5 (1872 bp), ND6 (522 bp), ATP6 (686 bp), and ATP8 (168 bp) were extracted from Microdous samples along with outgroups and aligned for phylogenetic analysis (doi:10.17632/n4j9zdb9wk.1). The resulting phylogenetic tree showed that M. chalmersi collected from Hainan and Guangxi were clustered together, but separated into two monophyletic clades (Figure 3). Microdous chalmersi formed a sister group to M. swinhonis first, but with a low bootstrap support, and then clustered with the Odontobutis spp. It can be seen clearly from Figure 3 that the genetic distance of Microdous between Hainan and Guangxi was comparable to the genetic distance between different species of the Odontobutis.
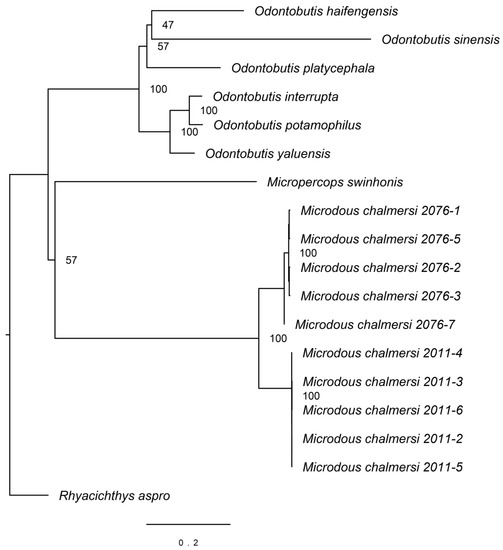
Figure 3.
Maximum likelihood tree of Microdous chalmersi with other odontobutids as outgroups. Samples labeled as “Microdous chalmersi 2076” were collected from Hainan and “Microdous chalmersi 2011” were from Guangxi. The following sequences were used for comparison: Rhyacichthys aspro, NC_004414, Micropercops swinhonis, NC_021763, and 6 species of Odontobutis (O. interrupta, NC_027583; O. haifengensis, NC_036056; O. platycephala, NC_010199; O. yaluensis, NC_027160; O. potamophilus, NC_022706; and O. sinensis, NC_022818).
3.3. Genetic Distance and Comparison
The genetic distance between samples from the two disjunct populations based on the 12S gene ranged from 0.052 to 0.06, whereas the genetic distance between samples within each location ranged from 0 to 0.011 (Supplementary Materials Table S3). The genetic distance for 16S gene was 0.068–0.078 between populations, and 0–0.013 within populations. The number for the COI gene was 0.095–0.102 between populations and 0–0.009 within populations; for the Cytb gene, it was 0.131–0.134 between populations and 0–0.004 within populations; for the ND2 gene, it was 0.146–0.163 between populations and 0–0.018 within populations (Supplementary Materials Table S3). The results of these genes showed that genetic distance between samples from Guangxi and Hainan was much greater than the genetic distance between different individuals in the same location and there was no overlap between the within-population and between-population genetic distances (Figure 4).
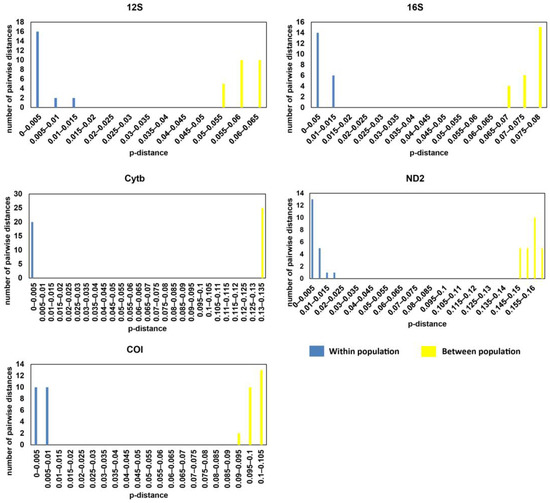
Figure 4.
Genetic distance (p-distance) of mitochondrial genes (12S, 16S, COI, Cytb, and ND2) of Microdous chalmersi between populations of Guangxi and Hainan and between individuals within populations.
The genetic distance between Odontobutis species ranged from 0.028 to 0.128 for 12S, 0.026 to 0.171 for 16S, 0.05 to 0.257 for Cytb, 0.062 to 0.27 for ND2, and 0.041 to 0.175 for COI (Supplementary Materials Table S4). For a comparison, the genetic distance between individuals of M. chalmersi from Guangxi and Hainan ranged from 0.095 to 0.102 for COI, with a mean of 0.096 and a standard deviation of 0.003 (Figure 5).
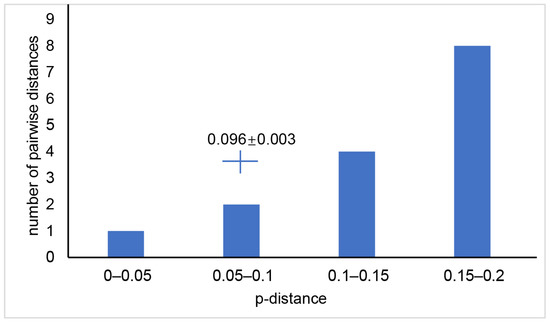
Figure 5.
Genetic distance between different species of the genus, Odontobutis based on COI gene. The cross shows average genetic distance and standard deviation (0.096 ± 0.003) of Microdous chalmersi between Guangxi and Hainan.
4. Discussion
Our results are similar to those of Li et al. [6], in that individuals of M. chalmersi from Guangxi and Hainan were placed as sister groups close to each other apart from Micropercops swinhonis and the six species of the Odontobutis. (Figure 3). It was also observed that Microdous from the two locations were reciprocally monophyletic, which suggested that these populations have been diverged from each other, probably due to their disjunct distribution. In addition, the genetic distance between the two populations is larger than interspecific distance between many Odontobutis species (Figure 3), suggesting that the degree of differentiation between the two populations is at interspecific level.
The genetic distances of Microdous between the two locations were significantly greater than the genetic distances between different individuals in the same location, and there was no overlap between the intra- and inter-population distance (Figure 4; Supplementary Materials Table S3). Interspecific distance greater than intraspecific distance and no overlap between them often are used as the criteria for delimiting species [24,25]. Therefore, our results strongly support that the genetic difference between the populations in Guangxi and Hainan is above species level. In addition, comparing the genetic distances between the two populations to the interspecific distance between O. interrupta, O. haifengensis, O. potamophilus, O. platycephala, O. yaluensis, and O. sinensis corroborates that the divergence between M. chalmersi in Hainan and Guangxi should be at interspecific level (Supplementary Materials Table S4; Figure 5). The average distance of COI between the two populations was 9.6%, which is much higher than 4%, the threshold commonly considered as interspecific [26]. Moreover, Hainan Island has been adjacent to northern Vietnam and Guangxi in Eocene [27]. The ichthyological fauna of freshwater in Hainan show distinct patterns, and many species have diverged into separate subspecies or species, such as Vanmanenia hainanensis and Squalidus minor [28]. Microdous chalmersi of Guangxi and Hainan showed a similar divergence pattern, probably due to the same historic geological events. Nonetheless, we hesitate to make a conclusion on whether the two populations are two different species or subspecies without collecting more morphological data. The samples used in this study were fixed in ethanol from the beginning, so they were too deformed to be used for further morphological examination. Meanwhile, limited samples from only two populations also restrained us from making the conclusion. More samples from the distribution areas of M. chalmersi should be collected and combined molecular and morphological analysis should be carried out to test the hypothesis about cryptic species of M. chalmersi.
Microdous chalmersi previously was known from the Changhua River and the Wanquan River of Hainan [29], but we only found it in Tropic Rainforest Park of Wuzhishan Mountain of the Changhua River after two years of sampling. In Pearl River drainage, M. chalmersi is distributed in the upper reaches of the Pearl River in Guangxi Province, but cannot be found downstream in Guangdong Province. Although M. chalmersi was listed only as “least concern and needs updating” in The IUCN Red List of Threatened Species in 2010, its limited and disjunct distribution imply its threatened status. Therefore, conservation of M. chalmersi is urgently needed, and two management units probably should be considered due to the disjunct distribution of the two populations and larger genetic distance between them.
5. Conclusions
The results of the phylogenetic reconstruction and genetic distance analysis based on mitochondrial genome data indicated that M. chalmersi distributed in Guangxi and Hainan may have been differentiated and could be different species due to long-term geographical isolation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8050228/s1, Table S1: Voucher numbers of samples and accession numbers of various loci; Table S2: Eleven target sequences of Microdous; Table S3: Pairwise p-distance within populations and between populations of Microdous; Table S4: Pairwise distance between Odontobutis spp.
Author Contributions
L.J. and C.L. designed the project and drafted the manuscript. L.J. and M.Z. collected and prepared the samples. J.H. and W.C. assembled and analyzed the data. K.K.S. revised the manuscript and provided critical suggestions for designing the experiment and data analyses. All authors reviewed and approved the final version to be published, and all authors agree to be accountable for all aspects of the work. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by “Science and Technology Commission of Shanghai Municipality (19410740500)”.
Institutional Review Board Statement
This study was conducted in accordance with guidelines of Animal Ethics Committee of Shanghai Ocean University, China (2020), with the approval of all experimental procedures including specimen handling. There was no permission required for collecting the fish.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Materials. The genome sequence data that support the findings of this study are available in GenBank of the NCBI at (https://www.ncbi.nlm.nih.gov/, accessed on 21 April 2023) under the accession nos. ON312088, ON312089, and OQ852467-OQ852470. The associated “BioProject” number is PRJNA857570. Other numbers are listed in Supplementary Materials Table S1.
Acknowledgments
We are grateful to Shuli Song, Linxi Pan, and Jiahu Lan for helping with sampling.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nichols, J.T.; Pope, C.H. The Fishes of Hainan. Bull. Am. Mus. Nat. Hist. 1927, 54, 321–394. [Google Scholar]
- Fowler, H.W. A Synopsis of the Fishes of China; Antiquariaat Junk: Amsterdam, The Netherlands, 1972. [Google Scholar]
- Hoese, D.F.; Gill, A.C. Phylogenetic Relationships of Eleotridid Fishes (Perciformes: Gobioidei). Bull. Mar. Sci. 1993, 52, 415–440. [Google Scholar]
- Chen, I.S.; Kottelat, M.; Wu, H. A New Genus of Freshwater Sleeper (Teleostei: Odontobutididae) from Southern China and Mainland Southeast Asia. J. Aquac. Soc. Taiwan 2002, 29, 229–235. [Google Scholar]
- Li, H.; He, Y.; Jiang, J.; Liu, Z.; Li, C. Molecular Systematics and Phylogenetic Analysis of the Asian Endemic Freshwater Sleepers (Gobiiformes: Odontobutidae). Mol. Phylogenet. Evol. 2018, 121, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hofreiter, M.; Straube, N.; Corrigan, S.; Naylor, G.J. Capturing Protein-Coding Genes across Highly Divergent Species. Biotechniques 2013, 54, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Kircher, M. Illumina Sequencing Library Preparation for Highly Multiplexed Target Capture and Sequencing. Cold Spring Harb. Protoc. 2010, 6, pdb-prot5448. [Google Scholar] [CrossRef]
- Yuan, H.; Atta, C.; Tornabene, L.; Li, C. Assexon: Assembling Exon Using Gene Capture Data. Evol. Bioinform. Online 2019, 15, 1176934319874792. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and Samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from Rna-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Takeshima, H.; Endo, H.; Ishiguro, N.B.; Inoue, J.G.; Mukai, T.; Satoh, T.P.; Yamaguchi, M.; Kawaguchi, A.; Mabuchi, K.; et al. Major Patterns of Higher Teleostean Phylogenies: A New Perspective Based on 100 Complete Mitochondrial DNA Sequences. Mol. Phylogenet. Evol. 2003, 26, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Huang, Z.; Zhang, S.; Xie, L.; Yang, X.; Zhang, X.; Yang, R. The Complete Mitochondrial Genome of Micropercops swinhonis (Perciformes: Gobioidei: Odontobutidae). Mitochondrial DNA 2015, 26, 116–117. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.; Choi, S.H.; Kum, J.D. Complete Mitochondrial Genome of the Endemic South Korean Species Odontobutis interrupta (Perciformes, Odontobutidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 2957–2959. [Google Scholar] [CrossRef]
- Zhong, L.; Wang, M.; Li, D.; Tang, S.; Zhang, T.; Bian, W.; Chen, X. Complete Mitochondrial Genome of Odontobutis haifengensis (Perciformes, Odontobutiae): A Unique Rearrangement of Trnas and Additional Non-Coding Regions Identified in the Genus Odontobutis. Genomics 2018, 110, 382–388. [Google Scholar] [CrossRef]
- Ki, J.S.; Jung, S.O.; Hwang, D.; Lee, Y.M.; Lee, J.S. Unusual Mitochondrial Genome Structure of the Freshwater Goby Odontobutis platycephala: Rearrangement of Trnas and an Additional Non-Coding Region. J. Fish Biol. 2008, 73, 414–428. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, X.; Zhang, X.; Yang, R. Complete Mitochondrial Genome of the Freshwater Goby Odontobutis potamophila (Perciformes: Odontobutidae). Mitochondrial DNA 2015, 26, 299–300. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, X.; Zhang, X.; Yang, R.; Qiu, P. Organization of the Mitochondrial Genome of Odontobutis sinensis (Perciformes: Odontobutidae): Rearrangement of Trnas and Additional Non-Coding Regions. Mitochondrial DNA 2015, 26, 327–328. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. Iq-Tree: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree v1.4.2. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 March 2022).
- Collins, R.A.; Cruickshank, R.H. Known Knowns, Known Unknowns, Unknown Unknowns and Unknown Knowns in DNA Barcoding: A Comment on Dowton Et Al. Syst. Biol. 2014, 63, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Meiklejohn, K.; Cameron, S.L.; Wallman, J. A Preliminary Framework for DNA Barcoding, Incorporating the Multispecies Coalescent. Syst. Biol. 2014, 63, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H. Biogeographical Evidences Help Revealing the Origin of Hainan Island. PLoS ONE 2016, 11, e0151941. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C. Freshwater Fish—Fauna of the Guangdong Province. J. Jinan Univ. (Nat. Sci. Med. Ed.) 1989, 1989, 68–73. [Google Scholar]
- Wu, H. Fauna Sinica Ostichthyes Perciformes (V) Gobioidei; Science Press: Beijing, China, 2008. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).