Detection and Identification of Fish Skin Health Status Referring to Four Common Diseases Based on Improved YOLOv4 Model
Abstract
1. Introduction
2. Materials and Methods
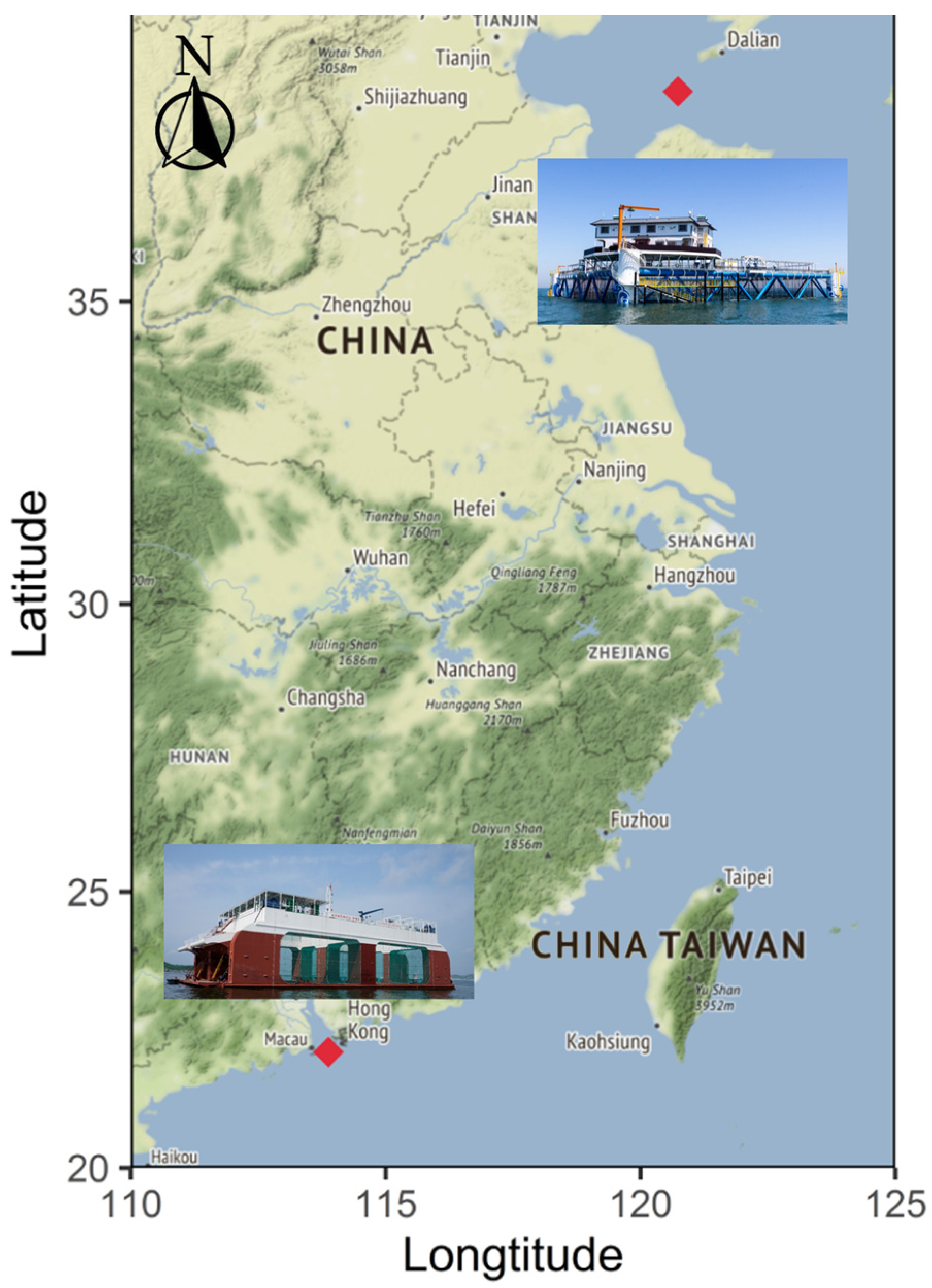
2.1. Dataset
2.2. Detection and Identification Model Based on YOLOv4
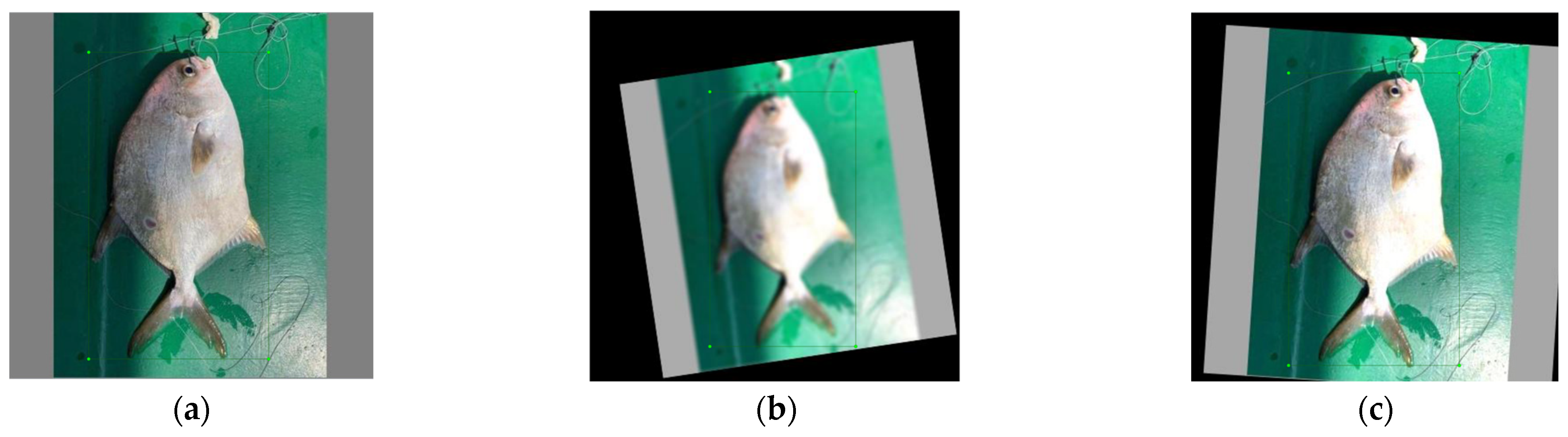
2.2.1. Data Preprocessing
2.2.2. Model Improving
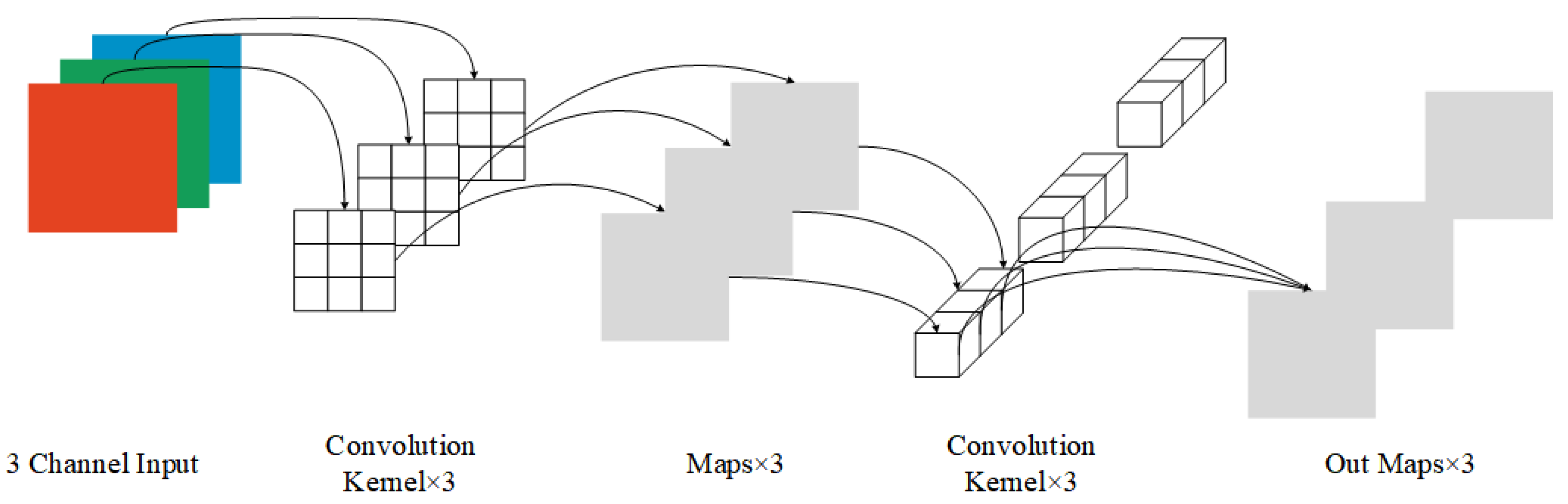
Feature Extraction Network
Activation Function
Depthwise Separable Convolution
2.2.3. Model Training
2.2.4. Performance of Improved YOLOv4 Model
3. Results and Discussion
3.1. Comparison of Different Loss Functions
3.2. Comparison of Changes in the Number of Parameters
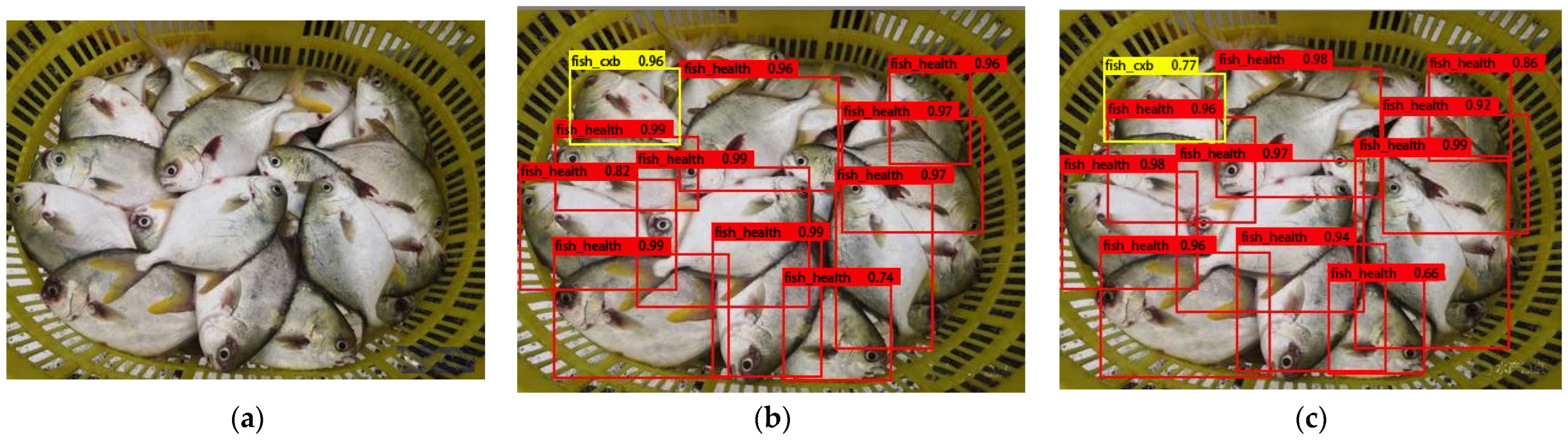
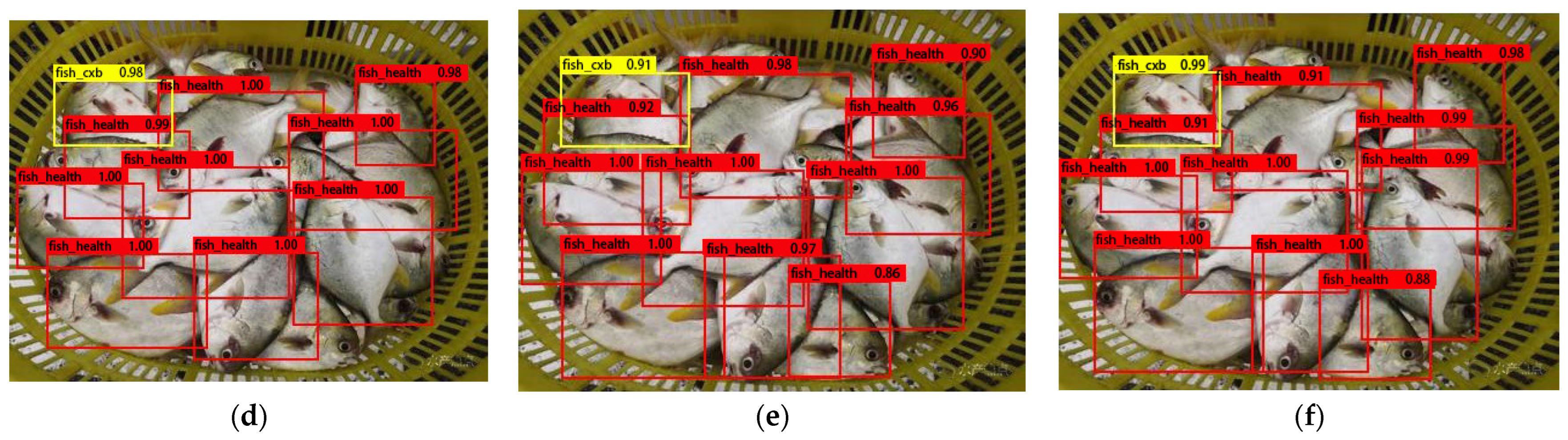
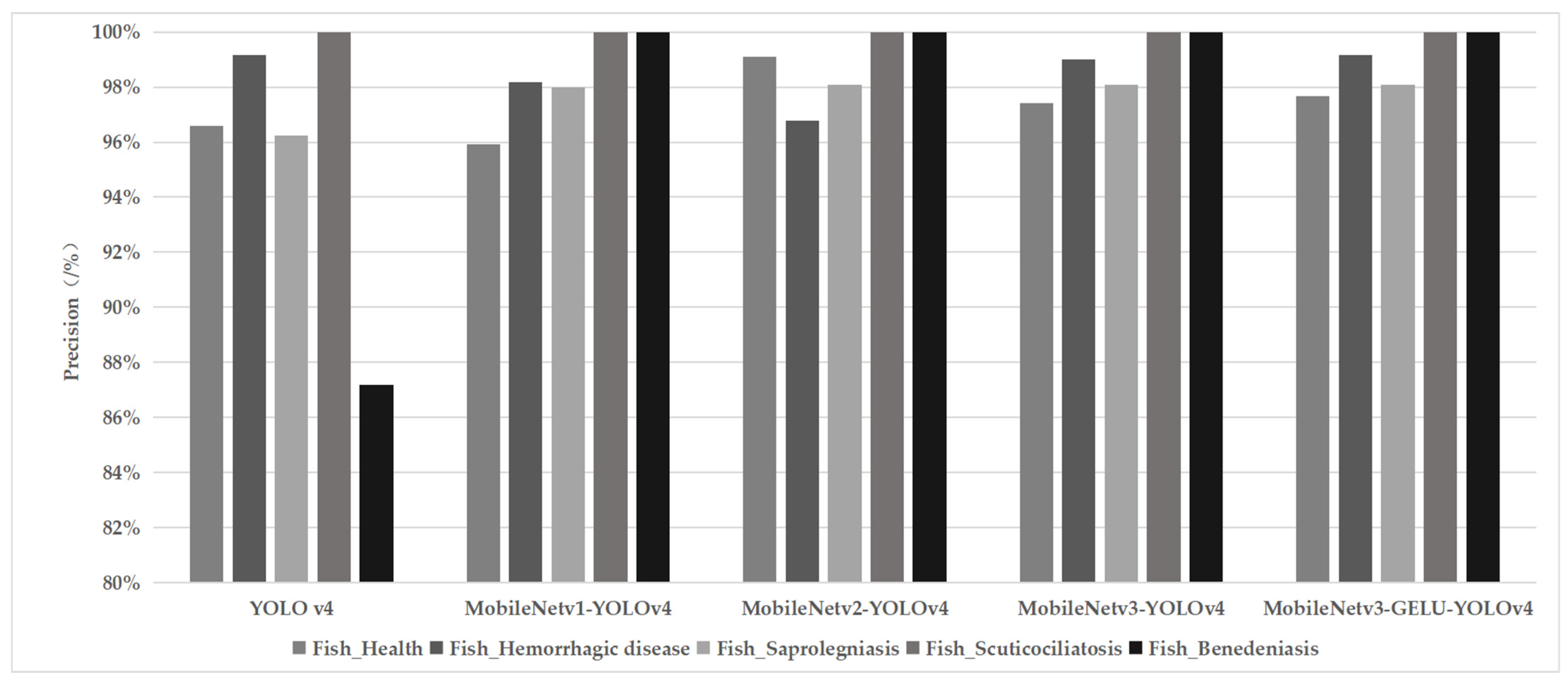
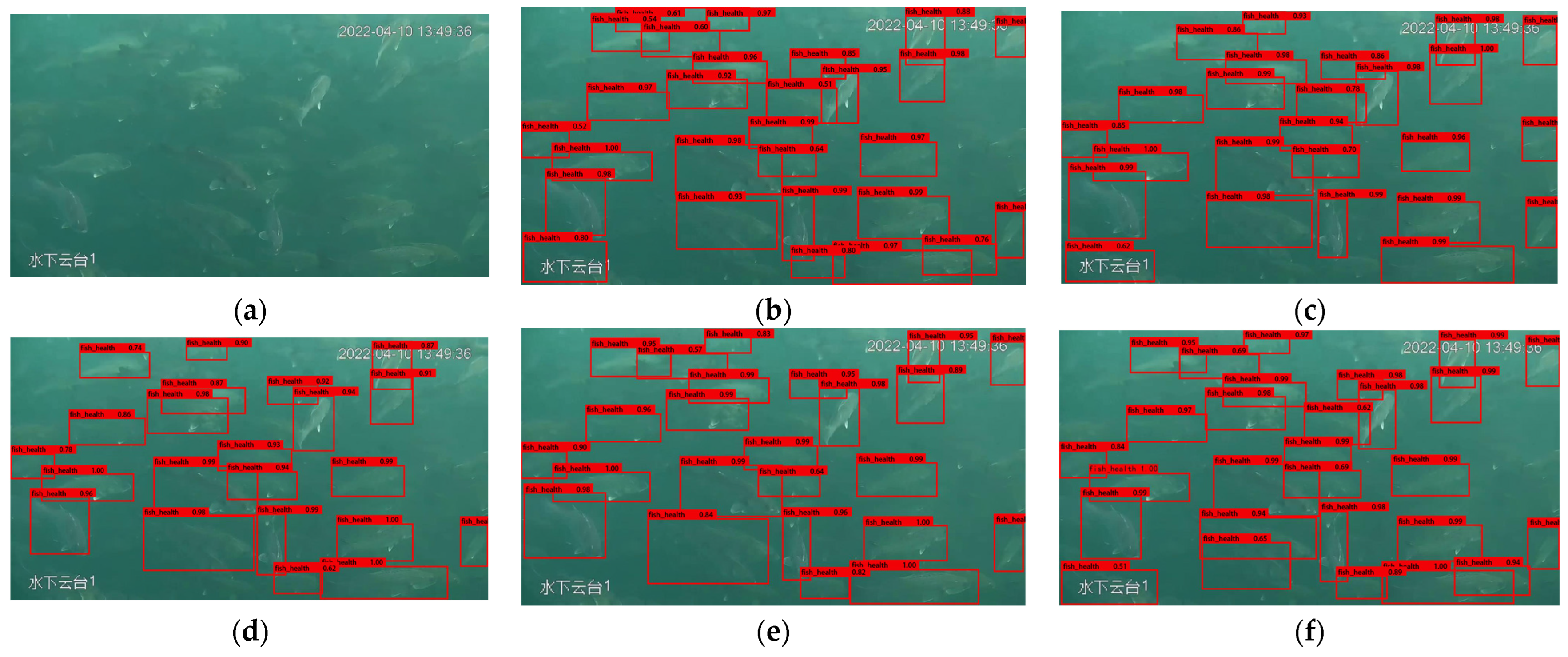
3.3. Model Detection Performance and Precision
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Planning for Aquaculture Diversification: The Importance of Climate Change and Other Drivers; FAO Fisheries and Aquaculture Department: Rome, Italy, 2016. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture 2018; FAO Fisheries and Aquaculture Department: Rome, Italy, 2018. [Google Scholar]
- Yu, J.-K.; Li, Y.-H. Evolution of Marine Spatial Planning Policies for Mariculture in China: Overview, Experience and Prospects. Ocean. Coast. Manag. 2020, 196, 105293. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2020; FAO Fisheries and Aquaculture Department: Rome, Italy, 2022. [Google Scholar]
- Sveen, L.; Timmerhaus, G.; Johansen, L.-H.; Ytteborg, E. Deep Neural Network Analysis—A Paradigm Shift for Histological Examination of Health and Welfare of Farmed Fish. Aquaculture 2021, 532, 736024. [Google Scholar] [CrossRef]
- Ina-Salwany, M.Y.; Al-saari, N.; Mohamad, A.; Mursidi, F.-A.; Mohd-Aris, A.; Amal, M.N.A.; Kasai, H.; Mino, S.; Sawabe, T.; Zamri-Saad, M. Vibriosis in Fish: A Review on Disease Development and Prevention. J. Aquat. Anim. Health 2019, 31, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Sorgeloos, P.; Bossier, P. Alternatives to Antibiotics for the Control of Bacterial Disease in Aquaculture. Curr. Opin. Microbiol. 2011, 14, 251–258. [Google Scholar] [CrossRef]
- Labella, A.M.; Arahal, D.R.; Lucena, T.; Manchado, M.; Castro, D.; Borrego, J.J. Photobacterium Toruni Sp. Nov., a Bacterium Isolated from Diseased Farmed Fish. Int. J. Syst. Evol. Microbiol. 2017, 67, 4518–4525. [Google Scholar] [CrossRef] [PubMed]
- Kristoffersen, A.B.; Qviller, L.; Helgesen, K.O.; Vollset, K.W.; Viljugrein, H.; Jansen, P.A. Quantitative Risk Assessment of Salmon Louse-Induced Mortality of Seaward-Migrating Post-Smolt Atlantic Salmon. Epidemics 2018, 23, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Oh, M.-J.; Han, S. Fish Disease Diagnosis System Based on Image Processing of Pathogens’ Microscopic Images. In Proceedings of the 2007 Frontiers in the Convergence of Bioscience and Information Technologies, Jeju, Republic of Korea, 11–13 October 2007; pp. 878–883. [Google Scholar] [CrossRef]
- Malik, S.; Kumar, T.; Sahoo, A.K. Image Processing Techniques for Identification of Fish Disease. In Proceedings of the 2017 IEEE 2nd International Conference on Signal and Image Processing (ICSIP), Singapore, 4–6 August 2017; pp. 55–59. [Google Scholar] [CrossRef]
- Li, X.; Shang, M.; Qin, H.; Chen, L. Fast Accurate Fish Detection and Recognition of Underwater Images with Fast R-CNN. In Proceedings of the OCEANS 2015—MTS/IEEE Washington, Washington, DC, USA, 19–22 October 2015; pp. 1–5. [Google Scholar] [CrossRef]
- Qin, H.; Li, X.; Liang, J.; Peng, Y.; Zhang, C. DeepFish: Accurate Underwater Live Fish Recognition with a Deep Architecture. Neurocomputing 2016, 187, 49–58. [Google Scholar] [CrossRef]
- Lu, H.; Li, Y.; Uemura, T.; Ge, Z.; Xu, X.; He, L.; Serikawa, S.; Kim, H. FDCNet: Filtering Deep Convolutional Network for Marine Organism Classification. Multimed. Tools Appl. 2018, 77, 21847–21860. [Google Scholar] [CrossRef]
- Christensen, J.H.; Mogensen, L.V.; Galeazzi, R.; Andersen, J.C. Detection, Localization and Classification of Fish and Fish Species in Poor Conditions Using Convolutional Neural Networks. In Proceedings of the 2018 IEEE/OES Autonomous Underwater Vehicle Workshop (AUV), Porto, Portugal, 6–9 November 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Khalifa, N.E.M.; Taha, M.H.N.; Hassanien, A.E. Aquarium Family Fish Species Identification System Using Deep Neural Networks. In Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2018, Cairo, Egypt, 3–5 September 2018; Hassanien, A.E., Tolba, M.F., Shaalan, K., Azar, A.T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 347–356. [Google Scholar] [CrossRef]
- Sun, X.; Shi, J.; Liu, L.; Dong, J.; Plant, C.; Wang, X.; Zhou, H. Transferring Deep Knowledge for Object Recognition in Low-Quality Underwater Videos. Neurocomputing 2018, 275, 897–908. [Google Scholar] [CrossRef]
- Måløy, H.; Aamodt, A.; Misimi, E. A Spatio-Temporal Recurrent Network for Salmon Feeding Action Recognition from Underwater Videos in Aquaculture. Comput. Electron. Agric. 2019, 167, 105087. [Google Scholar] [CrossRef]
- Labao, A.B.; Naval, P.C. Cascaded Deep Network Systems with Linked Ensemble Components for Underwater Fish Detection in the Wild. Ecol. Inform. 2019, 52, 103–121. [Google Scholar] [CrossRef]
- Rauf, H.T.; Lali, M.I.U.; Zahoor, S.; Shah, S.Z.H.; Rehman, A.U.; Bukhari, S.A.C. Visual Features Based Automated Identification of Fish Species Using Deep Convolutional Neural Networks. Comput. Electron. Agric. 2019, 167, 105075. [Google Scholar] [CrossRef]
- Liawatimena, S.; Atmadja, W.; Abbas, B.S.; Trisetyarso, A.; Wibowo, A.; Barlian, E.; Hardanto, L.T.; Triany, N.A.; Faisal; Sulistiawan, J.; et al. Drones Computer Vision Using Deep Learning to Support Fishing Management in Indonesia. IOP Conf. Ser. Earth Environ. Sci. 2020, 426, 012155. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, X.; Wang, Y.; Zhao, Z.; Liu, J.; Liu, Y.; Sun, C.; Zhou, C. Automatic Fish Population Counting by Machine Vision and a Hybrid Deep Neural Network Model. Animals 2020, 10, 364. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, S.; Liu, J.; Gao, Q.; Dong, S.; Zhou, C. Deep Learning for Smart Fish Farming: Applications, Opportunities and Challenges. Rev. Aquac. 2021, 13, 66–90. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Aurpa, T.T.; Azad, M.A.K. Fish Disease Detection Using Image Based Machine Learning Technique in Aquaculture. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 5170–5182. [Google Scholar] [CrossRef]
- Waleed, A.; Medhat, H.; Esmail, M.; Osama, K.; Samy, R.; Ghanim, T.M. Automatic Recognition of Fish Diseases in Fish Farms. In Proceedings of the 2019 14th International Conference on Computer Engineering and Systems (ICCES), Cairo, Egypt, 17–18 December 2019; pp. 201–206. [Google Scholar] [CrossRef]
- Sung, M.; Yu, S.-C.; Girdhar, Y. Vision Based Real-Time Fish Detection Using Convolutional Neural Network. In Proceedings of the OCEANS 2017—Aberdeen, Aberdeen, UK, 19–22 June 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Cai, K.; Miao, X.; Wang, W.; Pang, H.; Liu, Y.; Song, J. A Modified YOLOv3 Model for Fish Detection Based on MobileNetv1 as Backbone. Aquac. Eng. 2020, 91, 102117. [Google Scholar] [CrossRef]
- Hu, X.; Liu, Y.; Zhao, Z.; Liu, J.; Yang, X.; Sun, C.; Chen, S.; Li, B.; Zhou, C. Real-Time Detection of Uneaten Feed Pellets in Underwater Images for Aquaculture Using an Improved YOLO-V4 Network. Comput. Electron. Agric. 2021, 185, 106135. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Liu, F.; Xu, X.; Qing, C.; Jin, J. Probability Matrix SVM+ Learning for Complex Action Recognition. In Proceedings of the Internet Multimedia Computing and Service, Qingdao, China, 23–25 August 2017; Springer: Singapore; pp. 403–410. [Google Scholar] [CrossRef]
- Meng, L.; Hirayama, T.; Oyanagi, S. Underwater-Drone with Panoramic Camera for Automatic Fish Recognition Based on Deep Learning. IEEE Access 2018, 6, 17880–17886. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, D.; Zhang, Y.; Zhou, C.; Chen, W. Real-Time Nondestructive Fish Behavior Detecting in Mixed Polyculture System Using Deep-Learning and Low-Cost Devices. Expert Syst. Appl. 2021, 178, 115051. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU Loss: Faster and Better Learning for Bounding Box Regression. In Proceedings of the AAAI Conference on Artificial Intelligence 2020, 34, 12993–13000. [Google Scholar] [CrossRef]
- Huang, Z.; Sui, B.; Wen, J.; Jiang, G. An Intelligent Ship Image/Video Detection and Classification Method with Improved Regressive Deep Convolutional Neural Network. Complexity 2020, 2020, 1520872. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the International Conference on Machine Learning, Proceedings of Machine Learning Research, Lille, France, 1 June 2015; pp. 448–456. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Nguyen, A.; Pham, K.; Ngo, D.; Ngo, T.; Pham, L. An Analysis of State-of-the-Art Activation Functions for Supervised Deep Neural Network. In Proceedings of the 2021 International Conference on System Science and Engineering (ICSSE), Ho Chi Minh City, Vietnam, 26–28 August 2021; pp. 215–220. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Połap, D.; Wawrzyniak, N.; Włodarczyk-Sielicka, M. Side-Scan Sonar Analysis Using ROI Analysis and Deep Neural Networks. IEEE Trans. Geosci. Remote Sens. 2022, 60, 4206108. [Google Scholar] [CrossRef]
- Galusha, A.; Dale, J.; Keller, J.M.; Zare, A. Deep Convolutional Neural Network Target Classification for Underwater Synthetic Aperture Sonar Imagery. In Proceedings of the Detection and Sensing of Mines, Explosive Objects, and Obscured Targets XXIV., SPIE, Baltimore, MD, USA, 10 May 2019; Volume 11012, pp. 18–28. [Google Scholar] [CrossRef]
- Akgül, T.; Çalik, N.; Töreyın, B.U. Deep Learning-Based Fish Detection in Turbid Underwater Images. In Proceedings of the 2020 28th Signal Processing and Communications Applications Conference (SIU), Istanbul, Turkey, 5–7 October 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Ayob, A.F.; Khairuddin, K.; Mustafah, Y.M.; Salisa, A.R.; Kadir, K. Analysis of Pruned Neural Networks (MobileNetV2-YOLO v2) for Underwater Object Detection. In Proceedings of the 11th National Technical Seminar on Unmanned System Technology 2019, Kuantan, Malaysia, 2–3 December 2019; Md Zain, Z., Ahmad, H., Pebrianti, D., Mustafa, M., Abdullah, N.R.H., Samad, R., Mat Noh, M., Eds.; Springer Nature: Singapore, 2021; pp. 87–98. [Google Scholar] [CrossRef]
- Zheng, Z.; Liang, G.; Luo, H.; Yin, H. Attention Assessment Based on Multi-View Classroom Behaviour Recognition. IET Comput. Vis. 2022; early view. [Google Scholar] [CrossRef]
- Yu, W.; Ren, P. Vehicle and Pedestrian Target Detection in Auto Driving Scene. J. Phys. Conf. Ser. 2021, 2132, 012013. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; Wang, Q.; Hao, Y. Advanced Techniques for the Intelligent Diagnosis of Fish Diseases: A Review. Animals 2022, 12, 2938. [Google Scholar] [CrossRef]
- Gupta, A.; Bringsdal, E.; Knausgård, K.M.; Goodwin, M. Accurate Wound and Lice Detection in Atlantic Salmon Fish Using a Convolutional Neural Network. Fishes 2022, 7, 345. [Google Scholar] [CrossRef]
- Yasruddin, M.L.; Hakim Ismail, M.A.; Husin, Z.; Tan, W.K. Feasibility Study of Fish Disease Detection Using Computer Vision and Deep Convolutional Neural Network (DCNN) Algorithm. In Proceedings of the 2022 IEEE 18th International Colloquium on Signal Processing and Applications (CSPA), 12 May 2022; IEEE: Selangor, Malaysia, 2022; pp. 272–276. [Google Scholar] [CrossRef]


| Models | Feature Extraction Network | Activation Functions | Use of Depthwise Separable Convolution | Number of Parameters |
|---|---|---|---|---|
| YOLOv4 | CSPDarknet | ReLU | False | 64,363,101 |
| MobileNetv1 | 40,952,893 | |||
| MobileNetv2 | 39,062,013 | |||
| MobileNetv3 | 39,989,933 | |||
| MobileNetv1 | True | 12,692,029 | ||
| MobileNetv2 | 10,801,149 | |||
| MobileNetv3 | 11,729,069 | |||
| MobileNetv3 | GELU | 11,428,545 |
| Models | Feature Extraction Network | Activation Functions | Precision/% | Recall/% | mAP/% | FPS |
|---|---|---|---|---|---|---|
| YOLOv4 | CSPDarknet | ReLU | 95.83 | 85.39 | 87.25 | 20.31 |
| MobileNetv1 | 98.42 | 86.45 | 94.28 | 54.14 | ||
| MobileNetv2 | 98.79 | 94.92 | 95.47 | 47.36 | ||
| MobileNetv3 | 98.90 | 94.83 | 95.7 | 38.31 | ||
| MobileNetv3 | GELU | 98.98 | 98.65 | 99.64 | 39.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.; Zhang, J.; Chen, A.; Wan, R. Detection and Identification of Fish Skin Health Status Referring to Four Common Diseases Based on Improved YOLOv4 Model. Fishes 2023, 8, 186. https://doi.org/10.3390/fishes8040186
Yu G, Zhang J, Chen A, Wan R. Detection and Identification of Fish Skin Health Status Referring to Four Common Diseases Based on Improved YOLOv4 Model. Fishes. 2023; 8(4):186. https://doi.org/10.3390/fishes8040186
Chicago/Turabian StyleYu, Gangyi, Junbo Zhang, Ao Chen, and Rong Wan. 2023. "Detection and Identification of Fish Skin Health Status Referring to Four Common Diseases Based on Improved YOLOv4 Model" Fishes 8, no. 4: 186. https://doi.org/10.3390/fishes8040186
APA StyleYu, G., Zhang, J., Chen, A., & Wan, R. (2023). Detection and Identification of Fish Skin Health Status Referring to Four Common Diseases Based on Improved YOLOv4 Model. Fishes, 8(4), 186. https://doi.org/10.3390/fishes8040186







