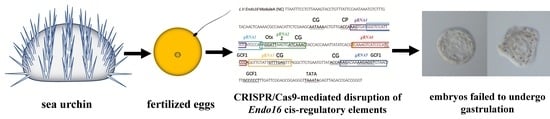
CRISPR/Cas9-Mediated Disruption of Endo16 Cis-Regulatory Elements in Sea Urchin Embryos
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Sea Urchins and Embryos
2.2. gRNAs Preparation
2.3. Microinjection, Drug Treatment and Imaging
2.4. Isolation of Genomic DNA from Single Embryo and Clone Sequencing
2.5. Isolation of RNA from Mixed Embryos and Real-Time PCR Validation
3. Results
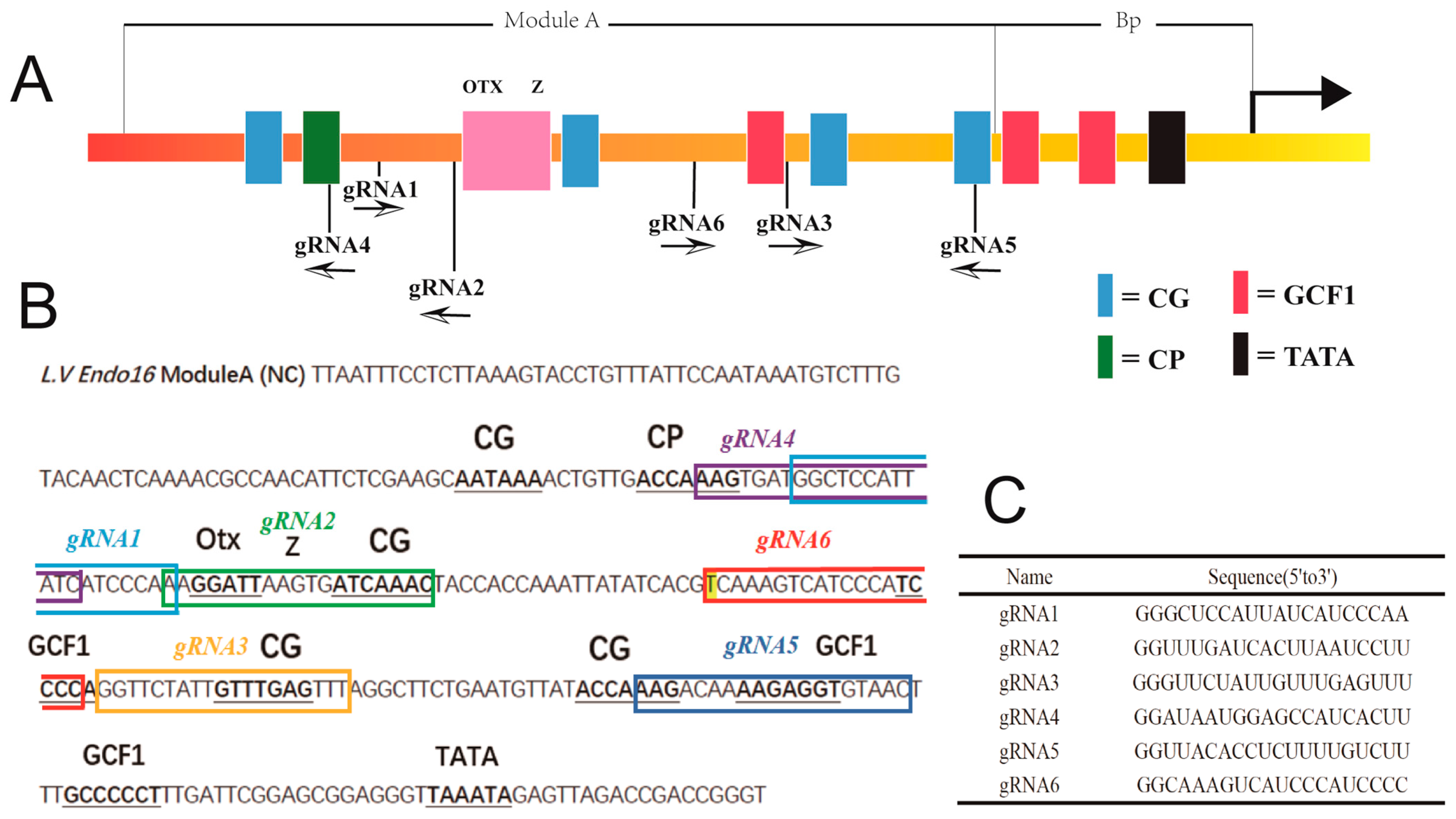
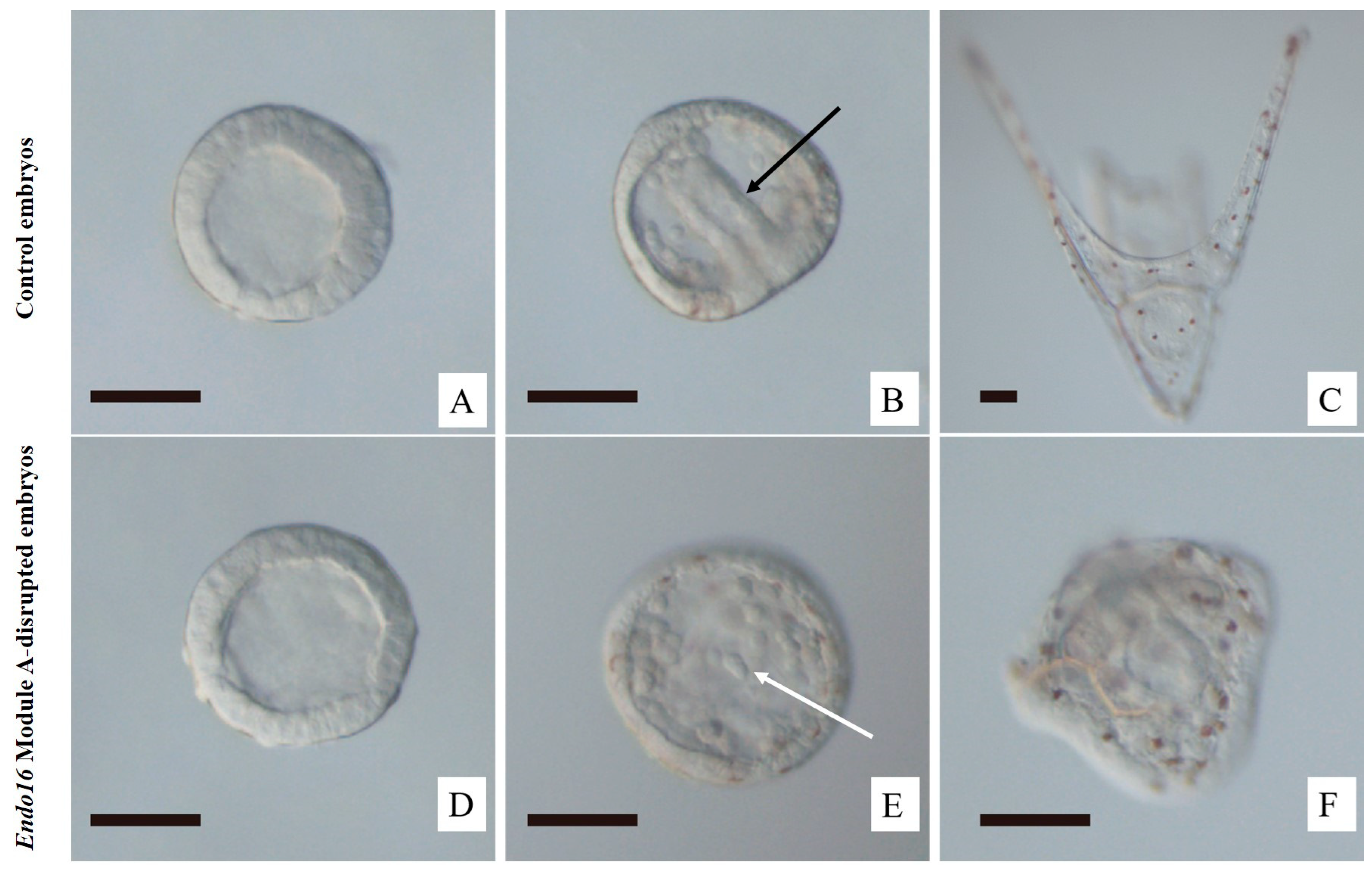
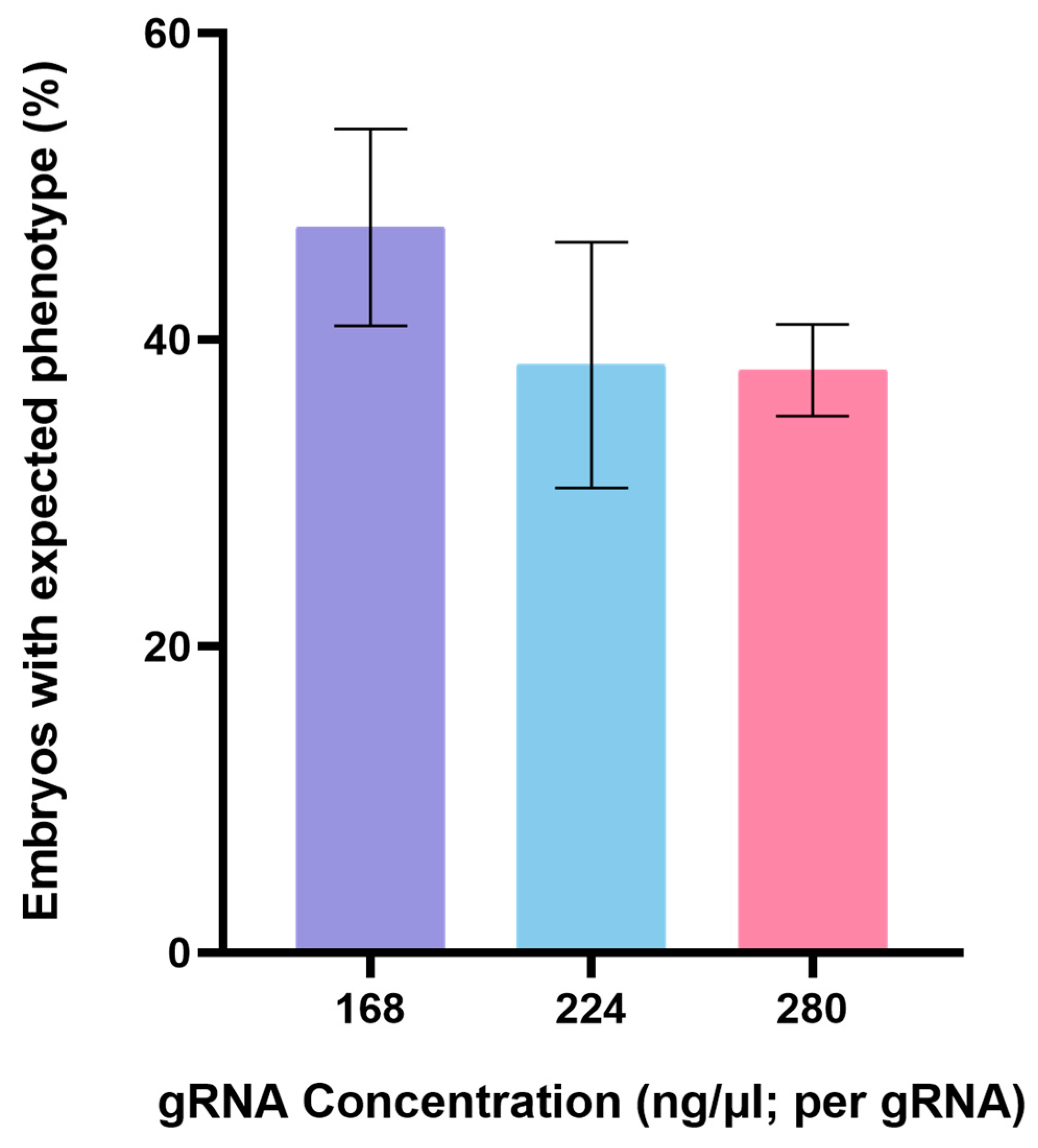
3.1. CRISPR/Cas9-Mediated Genome Editing of Endo16 Module A Produced Mutated Phenotype
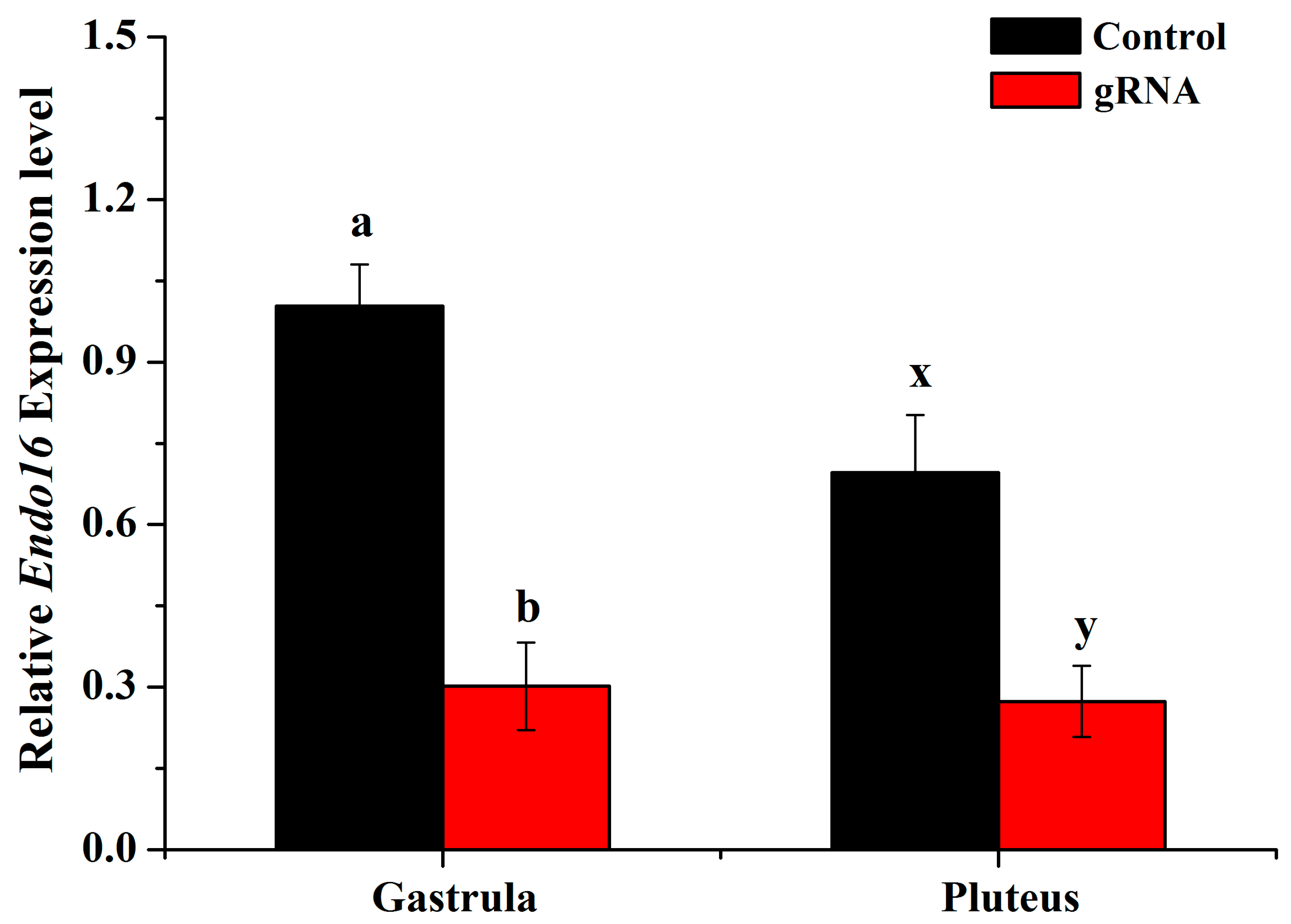
3.2. Disruption of Endo16 Module A Using Cas9 and gRNAs Caused a Downregulation of Endo16 Expression
3.3. Genotype of Embryos Injected with gRNA and Cas9 mRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Golden, C.D.; Koehn, J.Z.; Shepon, A.; Passarelli, S.; Free, C.M.; Viana, D.F.; Matthey, H.; Eurich, J.G.; Gephart, J.A.; Fluet-Chouinard, E.; et al. Aquatic foods to nourish nations. Nature 2021, 598, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Naylor, R.L.; Kishore, A.; Sumaila, U.R.; Issifu, I.; Hunter, B.P.; Belton, B.; Bush, S.R.; Cao, L.; Gelcich, S.; Gephart, J.A.; et al. Blue food demand across geographic and temporal scales. Nat. Commun. 2021, 12, 5413. [Google Scholar] [CrossRef] [PubMed]
- Golden, C.D.; Allison, E.H.; Cheung, W.W.L.; Dey, M.M.; Halpern, B.S.; McCauley, D.J.; Smith, M.; Vaitla, B.; Zeller, D.; Myers, S.S. Nutrition: Fall in fish catch threatens human health. Nature 2016, 534, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Rimm, E.B.; Appel, L.J.; Chiuve, S.E.; Djoussé, L.; Engler, M.B.; Kris-Etherton, P.M.; Mozaffarian, D.; Siscovick, D.S.; Lichtenstein, A.H. Seafood long-chain n-3 polyunsaturated fatty acids and cardiovascular disease: A science advisory from the American Heart Association. Circulation 2018, 138, e35–e47. [Google Scholar] [CrossRef]
- Gallet, C.A. The demand for fish: A meta-analysis of the own-price elasticity. Aquac. Econ. Manag. 2009, 13, 235–245. [Google Scholar] [CrossRef]
- Vermeulen, S.J.; Park, T.; Khoury, C.K.; Béné, C. Changing diets and the transformation of the global food system. Ann. N. Y. Acad. Sci. 2020, 1478, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, A.S.; Oken, E.; de Ferranti, S.; Lowry, J.A.; Ahdoot, S.; Baum, C.R.; Bole, A.; Byron, L.G.; Landrigan, P.J.; Marcus, S.M.; et al. Fish, shellfish, and children’s health: An assessment of benefits, risks, and sustainability. Pediatrics 2019, 143, e20190999. [Google Scholar] [CrossRef]
- Mcbride, S.C. Sea urchin aquaculture. Am. Fish. Soc. Symp. 2005, 46, 179–208. [Google Scholar]
- Paredes, E. Biobanking of a Marine Invertebrate Model Organism: The Sea Urchin. J. Mar. Sci. Eng. 2016, 4, 7. [Google Scholar] [CrossRef]
- Dvoretsky, A.G.; Dvoretsky, V.G. Aquaculture of green sea urchin in the Barents Sea: A brief review of Russian studies. Rev. Aquac. 2020, 12, 2080–2090. [Google Scholar] [CrossRef]
- James, P.J. A comparison of roe enhancement of the sea urchin Evechinus chloroticus in sea-based and land-based cages. Aquaculture 2006, 253, 290–300. [Google Scholar] [CrossRef]
- George, S.B.; Lawrence, J.M.; Lawrence, A.L. Complete larval development of the sea urchin Lytechinus variegatus fed an artificial feed. Aquaculture 2004, 242, 217–228. [Google Scholar] [CrossRef]
- Christiansen, J.S.; Silkavuoplo, S.I. The relationship between feed intake and gonad growth. of single and stocked green sea urchin (Strongylocentrotus droebachiensis) in a raceway culture. Aquaculture 2007, 262, 163–167. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fang, W.; Huang, J.; Li, S. The application of genome editing technology in fish. Mar. Life Sci. Technol. 2021, 3, 326–346. [Google Scholar] [CrossRef]
- Square, T.; Romášek, M.; Jandzik, D.; Cattell, M.V.; Klymkowsky, M.; Medeiros, D.M. CRISPR/Cas9-mediated mutagenesis in the sea lamprey, Petromyzon marinus: A powerful tool for understanding ancestral gene functions in vertebrates. Development 2015, 142, 4180–4187. [Google Scholar] [CrossRef]
- Perry, K.J.; Henry, J.Q. CRISPR/Cas9-mediated genome modification in the mollusc, Crepidula fornicata. Genes 2015, 53, 237–244. [Google Scholar] [CrossRef]
- Abe, M.; Kuroda, R. The development of CRISPR for a mollusc establishes the formin Lsdia1 as the long-sought gene for snail dextral/sinistral coiling. Development 2020, 146, dev175976. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Su, Y.-H. Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Dev. Biol. 2016, 409, 420–428. [Google Scholar] [CrossRef]
- Oulhen, N.; Wessel, G.M. Albinism as a visual, in vivo guide for CRISPR/Cas9 functionality in the sea urchin embryo. Mol. Reprod. Dev. 2016, 83, 1046–1047. [Google Scholar] [CrossRef]
- Wessel, G.M.; Kiyomoto, M.; Shen, T.-L.; Yajima, M. Genetic manipulation of the pigment pathway in a sea urchin reveals distinct lineage commitment prior to metamorphosis in the bilateral to radial body plan transition. Sci. Rep. 2020, 10, 1973. [Google Scholar] [CrossRef] [PubMed]
- Wessel, G.M.; Wada, Y.; Yajima, M.; Kiyomoto, M. Sperm lacking Bindin are infertile but are otherwise indistinguishable from wildtype sperm. Sci. Rep. 2021, 11, 21583. [Google Scholar] [CrossRef] [PubMed]
- Pieplow, A.; Dastaw, M.; Sakuma, T.; Sakamoto, N.; Yamamoto, T.; Yajima, M.; Oulhen, N.; Wessel, G.M. CRISPR-Cas9 editing of non-coding genomic loci as a means of controlling gene expression in the sea urchin. Dev. Biol. 2021, 472, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Shevidi, S.; Uchida, A.; Schudrowitz, N.; Wessel, G.M.; Yajima, M. Single nucleotide editing without DNA cleavage using CRISPR/Cas9-deaminase in the sea urchin embryo. Dev. Dyn. 2017, 246, 1036–1046. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef]
- Wray, G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007, 8, 206–216. [Google Scholar] [CrossRef]
- Bauer, D.E.; Kamran, S.C.; Lessard, S.; Xu, J.; Fujiwara, Y.; Lin, C.; Shao, Z.; Canver, M.C.; Smith, E.C.; Pinello, L.; et al. An Erythroid Enhancer of BCL11A Subject to Genetic Variation Determines Fetal Hemoglobin Level. Science 2013, 342, 253–257. [Google Scholar] [CrossRef]
- Harismendy, O.; Notani, D.; Song, X.; Rahim, N.G.; Tanasa, B.; Heintzman, N.; Ren, B.; Fu, X.D.; Topol, E.J.; Rosenfeld, M.G.; et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 2011, 470, 264. [Google Scholar] [CrossRef]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Musunuru, K.; Strong, A.; Frank-Kamenetsky, M.; Lee, N.E.; Ahfeldt, T.; Sachs, K.V.; Li, X.; Li, H.; Kuperwasser, N.; Ruda, V.M.; et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 2010, 466, 714–719. [Google Scholar] [CrossRef]
- Oldridge, D.A.; Wood, A.C.; Weichert-Leahey, N.; Crimmins, I.; Sussman, R.; Winter, C.; McDaniel, L.D.; Diamond, M.; Hart, L.S.; Zhu, S.; et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 2015, 528, 418–421. [Google Scholar] [CrossRef]
- Crisp, P.A.; Bhatnagar-Mathur, P.; Hundleby, P.; Godwin, I.D.; Waterhouse, P.M.; Hickey, L.T. Beyond the gene: Epigenetic and cis-regulatory targets offer new breeding potential for the future. Curr. Opin. Biotechnol. 2021, 73, 88–94. [Google Scholar] [CrossRef]
- Romano, L.A.; Wray, G.A. Endo16 is required for gastrulation in the sea urchin Lytechinus variegatus. Dev. Growth Differ. 2006, 48, 487–497. [Google Scholar] [CrossRef]
- Yuh, C.-H.; Ransick, A.; Martinez, P.; Britten, R.J.; Davidson, E.H. Complexity and organization of DNA-protein interactions in the 5′-regulatory region of an endoderm-specific marker gene in the sea urchin embryo. Mech. Dev. 1994, 47, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Romano, L.A.; Wray, G.A. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development 2003, 130, 4187–4199. [Google Scholar] [CrossRef] [PubMed]
- Cheers, M.S.; Ettensohn, C.A. Rapid Microinjection of Fertilized Eggs. Methods Cell Biol. 2004, 74, 287–310. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.A.; Samanta, M.; Yuan, A.; He, D.; Davidson, E. SpBase: The sea urchin genome database and web site. Nucleic Acids Res. 2009, 37, 750. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Yuh, C.-H.; Bolouri, H.; Davidson, E.H. Genomic Cis-Regulatory Logic: Experimental and Computational Analysis of a Sea Urchin Gene. Science 1998, 279, 1896–1902. [Google Scholar] [CrossRef]
- Cui, M.; Lin, C.-Y.; Su, Y.-H. Recent advances in functional perturbation and genome editing techniques in studying sea urchin development. Briefings Funct. Genom. 2017, 16, 309–318. [Google Scholar] [CrossRef]
- Reinardy, H.C.; Bodnar, A.G. Profiling DNA damage and repair capacity in sea urchin larvae and coelomocytes exposed to genotoxicants. Mutagenesis 2015, 30, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Le Bouffant, R.; Cormier, P.; Cueff, A.; Bellé, R.; Mulner-Lorillon, O. Sea urchin embryo as a model for analysis of the signaling pathways linking DNA damage checkpoint, DNA repair and apoptosis. Cell. Mol. Life Sci. 2007, 64, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yu, Y.; Tay, Y.X.; Yue, G.H. Genome editing and its applications in genetic improvement in aquaculture. Rev. Aquac. 2021, 14, 178–191. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef]
- Lopes, R.; Korkmaz, G.; Agami, R. Applying CRISPR–Cas9 tools to identify and characterize transcriptional enhancers. Nat. Rev. Mol. Cell Biol. 2016, 17, 597–604. [Google Scholar] [CrossRef]

| Primer | Sequence (5’ to 3’) | Annealing Temperature | Product Size | Aim |
|---|---|---|---|---|
| 1-F | GACAGAGACCGTATCGAATTAACATGCG | 69 °C | 406 bp | Endo16 Module A cloning |
| 1-R | TTCGACCACGCCACGGCCAGCACGG | |||
| 2-F | GACCTGTAGCGAACACACAAAGCCG | 60 °C | 441 bp | Endo16 mRNA expression level analysis |
| 2-R | TCACGGCAGTGCAGATGGCCTCG | |||
| 3-F | CACAGGCAAGACCATCACA | 147 bp | Housekeeping gene—Ubiquitin | |
| 3-R | GAGAGAGTGCGACCATCCTC |
| Combination | gRNA Concentration (ng/μL; per gRNA) | Embryos with Microinjection | Dead Embryos % (n) | Alive Normal Embryos % (n) | Embryos with Expected Phenotype % (n) |
|---|---|---|---|---|---|
| gRNA 1 and 2 and 3 | 112 | 187 | 15 (28) | 75 (119) | 25 (40) |
| gRNA 4 and 5 and 6 | 112 | 33 | 40 (13) | 60 (12) | 40 (8) |
| gRNA 1 and 2 and 3 | 168 | 100 | 37 (37) | 52 (33) | 48 (30) |
| gRNA 4 and 5 and 6 | 168 | 66 | 32 (21) | 56 (25) | 44 (20) |
| gRNA 2 and 4 and 5 | 168 | 195 | 15 (30) | 65 (108) | 35 (57) |
| gRNA 1 and 3 and 6 | 168 | 179 | 26 (46) | 70 (93) | 30 (40) |
| gRNA 4 and 5 | 168 | 190 | 19 (37) | 48 (73) | 52 (80) |
| gRNA 1 and 3 | 168 | 175 | 22 (38) | 50 (68) | 50 (69) |
| gRNA 2 and 6 | 168 | 161 | 13 (21) | 60 (84) | 40 (56) |
| gRNA 4 and 5 | 224 | 89 | 19 (17) | 54 (39) | 46 (33) |
| gRNA 1 and 3 | 224 | 70 | 20 (14) | 61 (34) | 39 (22) |
| gRNA 2 and 6 | 224 | 63 | 10 (6) | 70 (40) | 30 (17) |
| gRNA 4 and 5 | 280 | 202 | 16 (32) | 59 (100) | 41 (70) |
| gRNA 1 and 3 | 280 | 198 | 12 (24) | 62 (107) | 38 (67) |
| gRNA 2 and 6 | 280 | 187 | 20 (38) | 65 (97) | 35 (52) |
| gRNA1 | 168 | 113 | 23 (27) | 35 (30) | 65 (56) |
| gRNA2 | 168 | 145 | 20 (29) | 39 (45) | 61 (71) |
| gRNA3 | 168 | 124 | 24 (30) | 74 (70) | 26 (24) |
| gRNA4 | 168 | 43 | 53 (23) | 70 (14) | 30 (6) |
| gRNA5 | 168 | 41 | 61 (25) | 38 (6) | 62 (10) |
| gRNA6 | 168 | 40 | 50 (20) | 100 (20) | 0 (0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, L.; Wang, L.; Roos, F.; Lee, M.; Wray, G.A. CRISPR/Cas9-Mediated Disruption of Endo16 Cis-Regulatory Elements in Sea Urchin Embryos. Fishes 2023, 8, 118. https://doi.org/10.3390/fishes8020118
Xing L, Wang L, Roos F, Lee M, Wray GA. CRISPR/Cas9-Mediated Disruption of Endo16 Cis-Regulatory Elements in Sea Urchin Embryos. Fishes. 2023; 8(2):118. https://doi.org/10.3390/fishes8020118
Chicago/Turabian StyleXing, Lili, Lingyu Wang, Femke Roos, Michelle Lee, and Gregory A. Wray. 2023. "CRISPR/Cas9-Mediated Disruption of Endo16 Cis-Regulatory Elements in Sea Urchin Embryos" Fishes 8, no. 2: 118. https://doi.org/10.3390/fishes8020118
APA StyleXing, L., Wang, L., Roos, F., Lee, M., & Wray, G. A. (2023). CRISPR/Cas9-Mediated Disruption of Endo16 Cis-Regulatory Elements in Sea Urchin Embryos. Fishes, 8(2), 118. https://doi.org/10.3390/fishes8020118