The “True Colours” of Golden Loaches (Teleostei: Cobitidae)
Abstract
1. Introduction
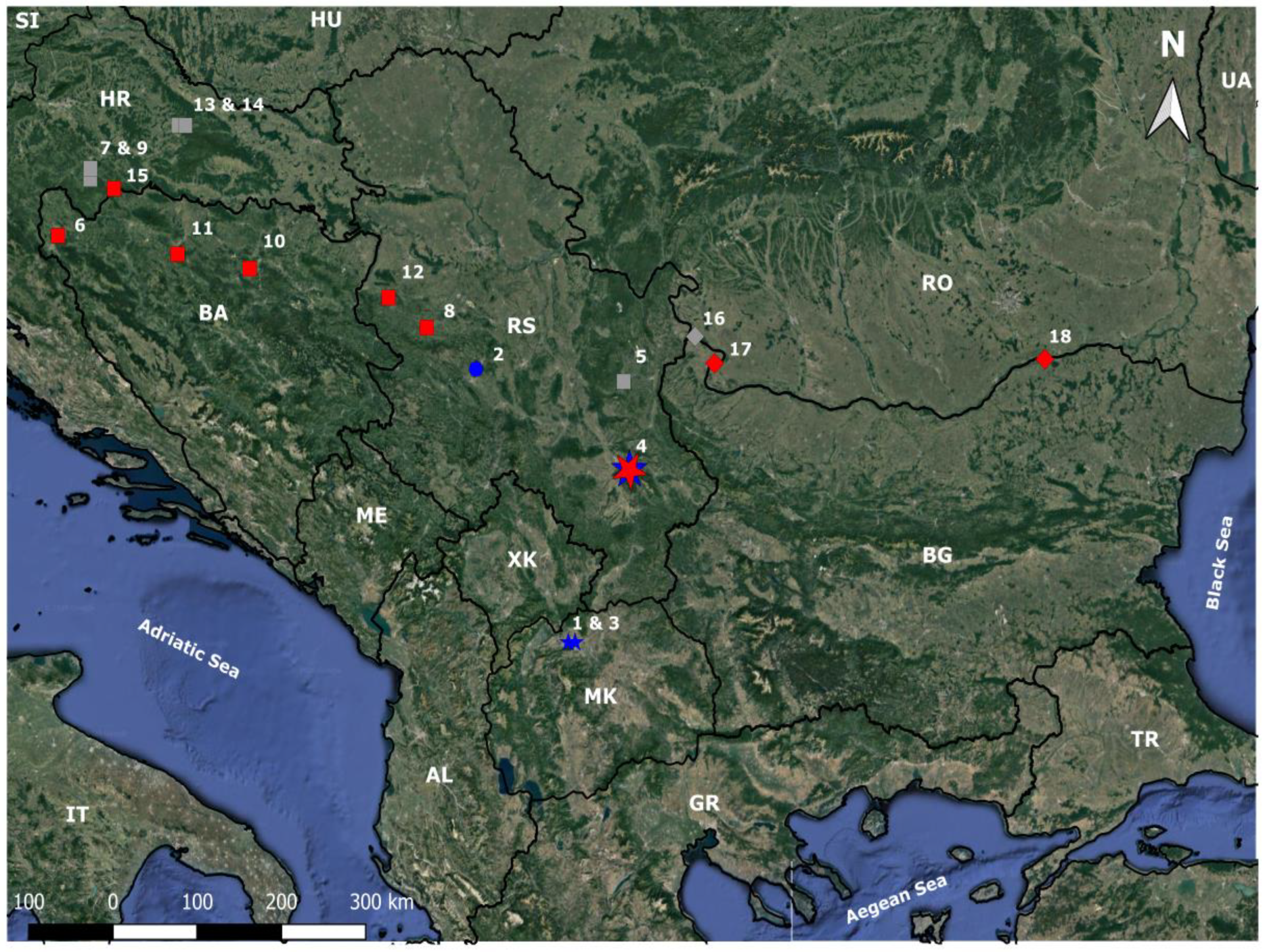
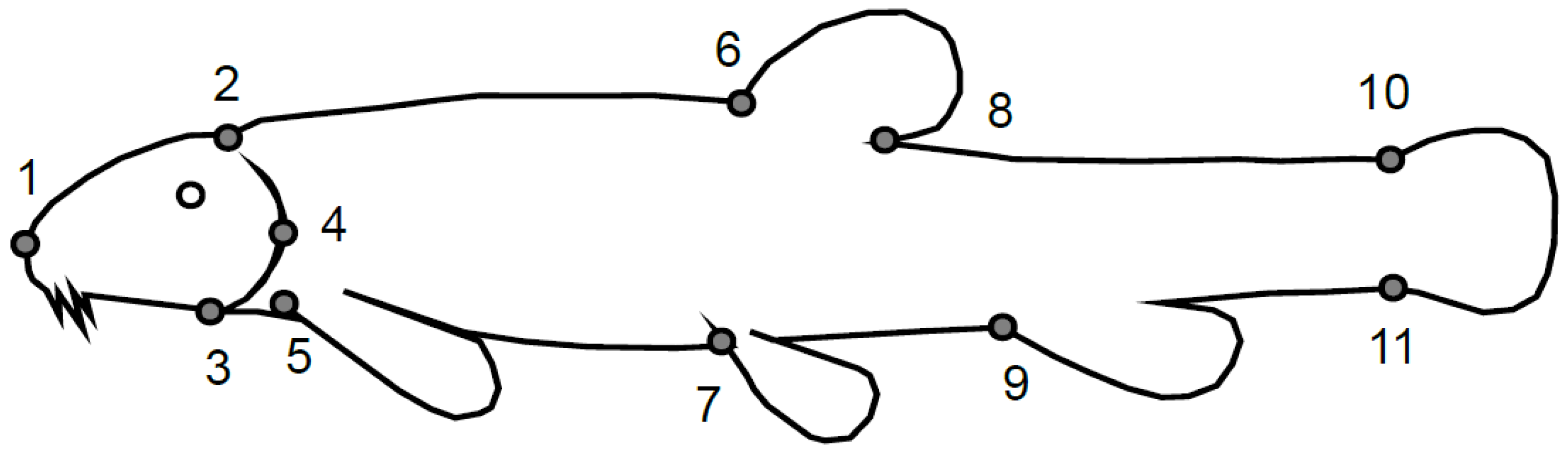
2. Materials and Methods
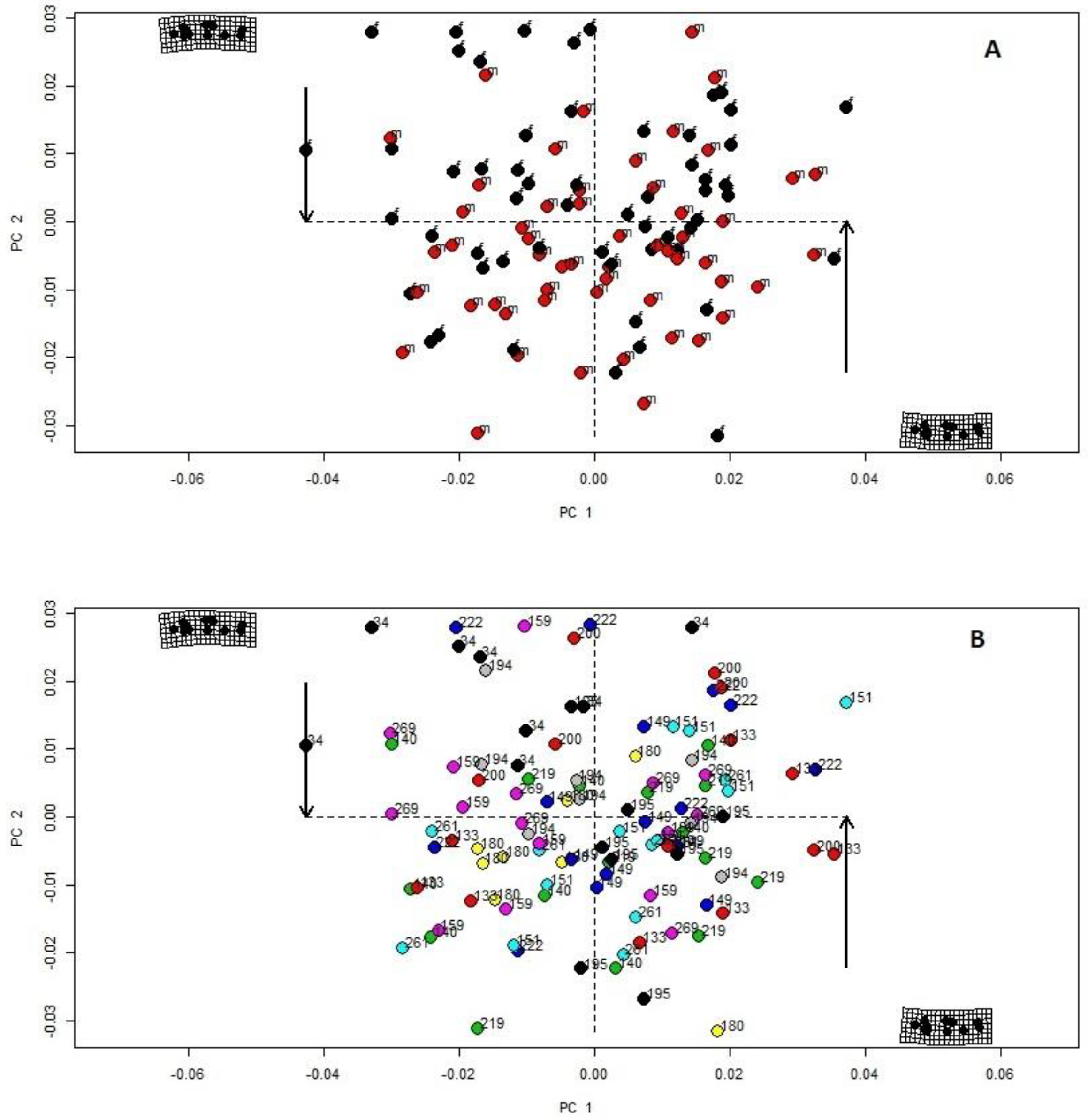
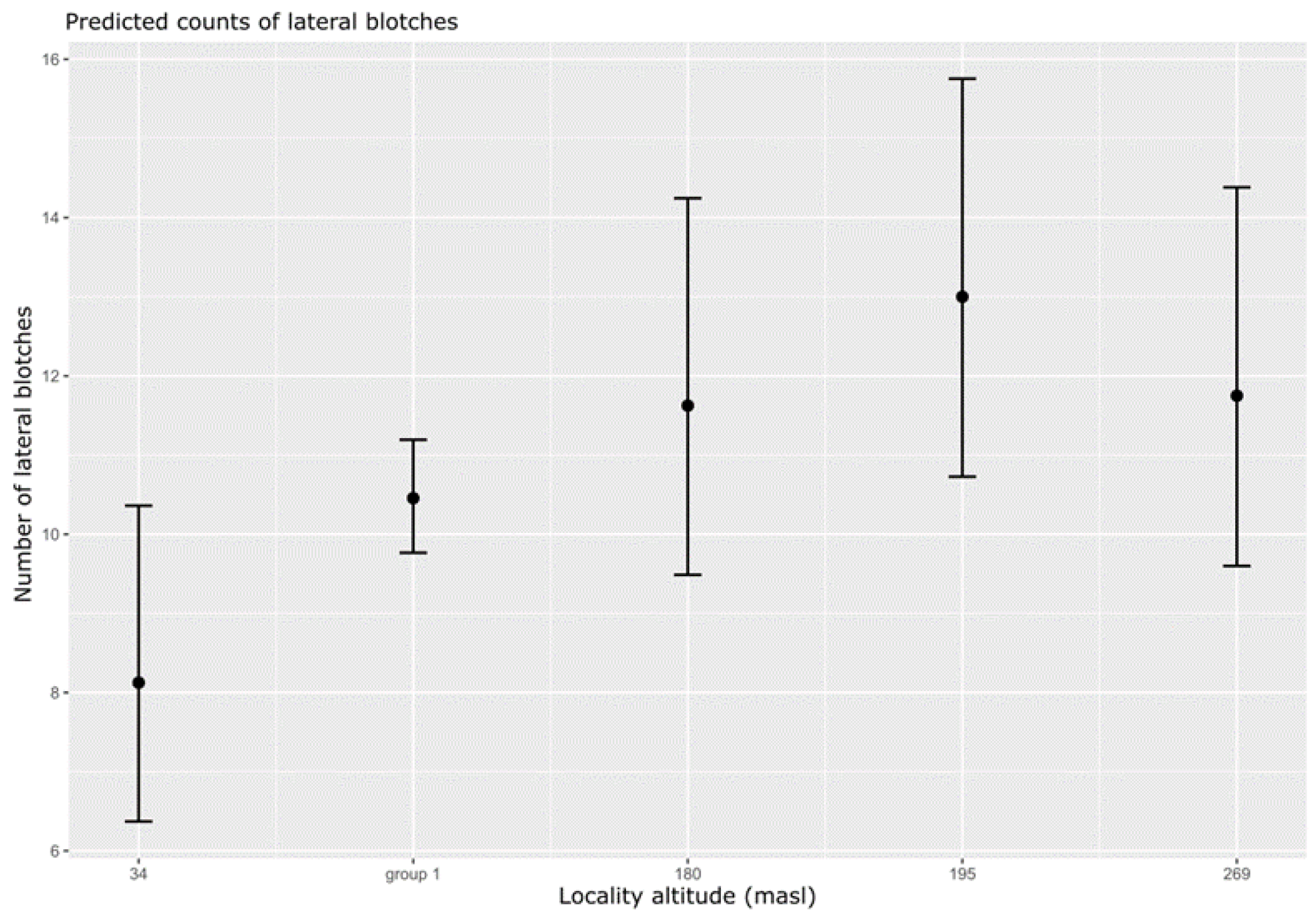
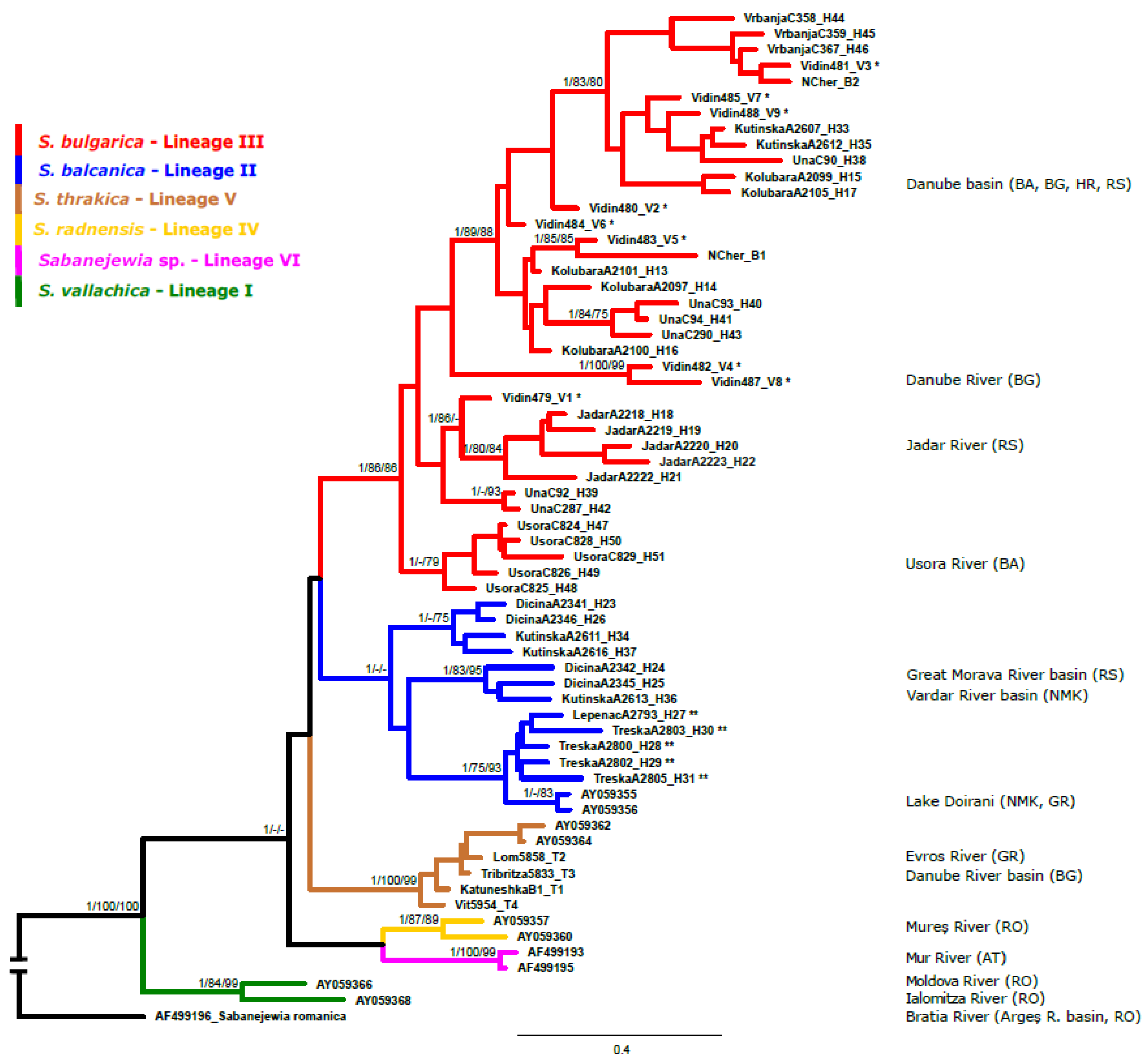
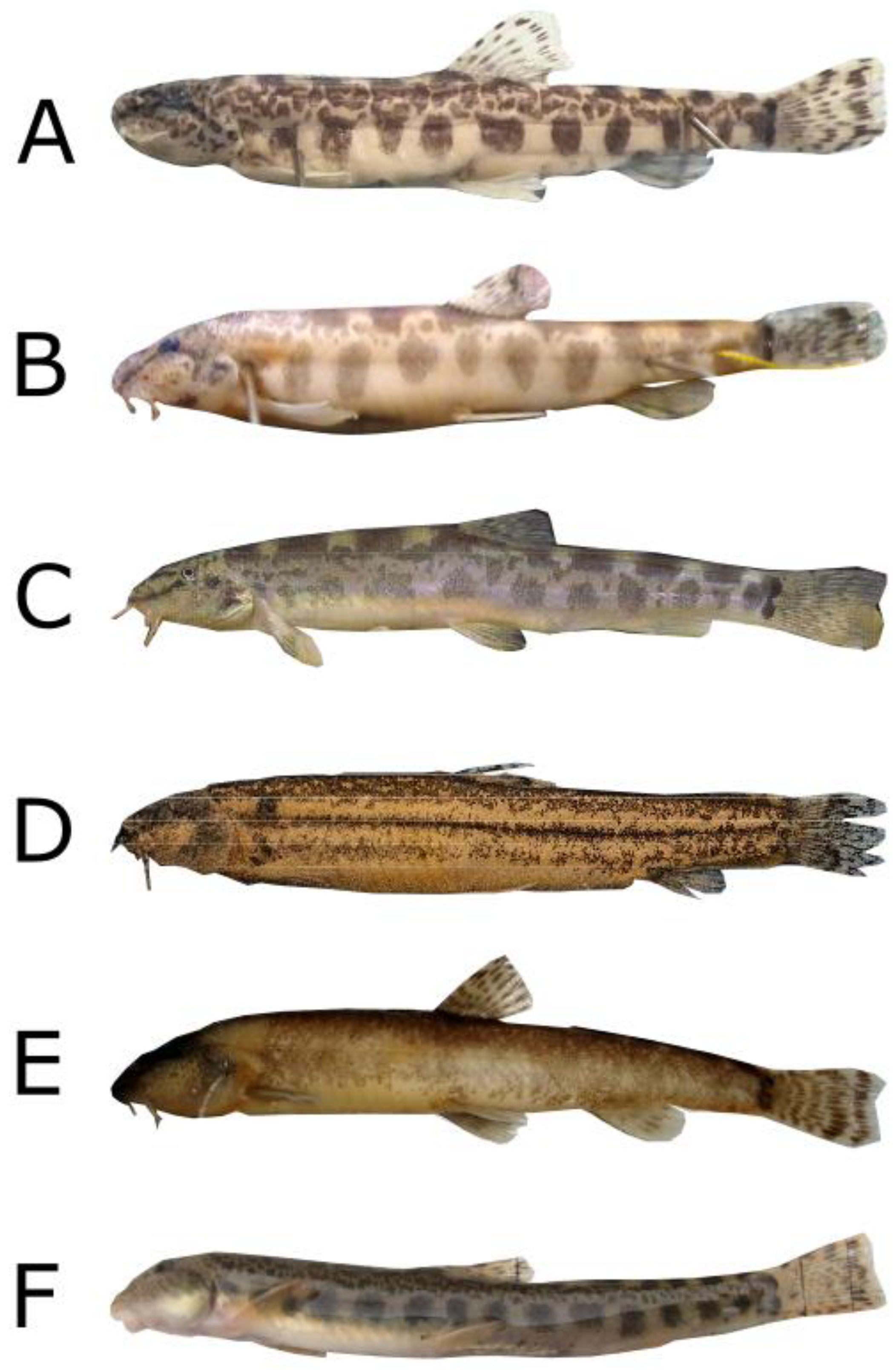
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Vladykov, V. Sur un nouveau genre de Cobitides: Sabanejewia. Bull. Mus. Natl. D’hist. Nat. (Sér. 2) 1929, 2, 85–90. [Google Scholar]
- Banarescu, V.P. Intraspecific variation, subspeciation and speciation in Roumanian fresh-water fishes. J. Zool. Syst. Evol. Res. 1966, 4, 378–396. [Google Scholar] [CrossRef]
- Bănărescu, P.M.; Nalbant, T.T.; Chelmu, S. Revision and geographical variation of Sabanejewia aurata in Romania and the origin of S. bulgarica and S. romanica (Pisces, Cobitidae). Annot. Zool. Bot. Bratisl. 1972, 75, 49. [Google Scholar]
- Sivkov, Y. Morphological characteristic of the Danubian loach Sabanejewia bulgarica (Drensky, 1928) (Pisces, Cobitidae). Acta Zool. Bulg. 1991, 42, 34–43. [Google Scholar]
- Iftime, A. Considerations over the taxonomic status of the balkan golden loach (Sabanejewia balcanica) (Pisces: Ostariophysi: Cobitidae) in Romania and Republic of Moldova. Trav. Mus. Natl. D’hist. Nat. Grigore Antipa 2002, 44, 335–355. [Google Scholar]
- Freyhof, J.; Kottelat, M. Handbook of European Freshwater Fishes; Publications Kottelat: Cornol, Switzerland; Berlin, Germany, 2007; pp. 321–327. [Google Scholar]
- Bănărescu, P. Zoogeography of Fresh Waters. Distribution and Dispersal of Freshwater Animals in North America and Eurasia; AULA-Verlag: Wiesbaden, Germany, 1992; Volume 2. [Google Scholar]
- Witkowski, A. Morphological characteristics of Sabanejewia aurata (De Filippi, 1865) from the Odra River Basin, with description of a new subspecies (Teleostei: Cypriniformes: Cobitidae). Zool. Abh. Staatl. Mus. Tierkd. Dresd. 1994, 48, 23–51. [Google Scholar]
- Kottelat, M. Conspectus Cobitidum: An inventory of the loaches of the world (Teleostei: Cypriniformes: Cobitoidei). Raffles Bull. Zool. 2012, 26, 1–199. [Google Scholar]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References; California Academy of Sciences: San Francisco, CA, USA, 2022; Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 17 December 2022).
- Vasil’eva, E.D.; Solovyeva, E.N.; Vasil’ev, V.P. Molecular phylogeny of the Spined Loach Genus Sabanejewia (Osteichthyes: Cobitidae) Revised. J. Ichthyol. 2022, 62, 812–827. [Google Scholar] [CrossRef]
- Marić, S.P.; Bănăduc, D.; Gajić, Đ.D.; Šanda, R.; Veličković, T.Z. Sabanejewia romanica (Băcescu, 1943) (Actinopterygii: Cobitidae), a new species for the ichthyofauna of Serbia. Acta Zool. Bulg. 2022, 74, 369–377. [Google Scholar]
- Perdices, A.; Doadrio, I.; Economidis, P.S.; Bohlen, J.; Bǎnǎrescu, V.P. Pleistocene effects on the European freshwater fish fauna: Double origin of the cobitid genus Sabanejewia in the Danube basin (Osteichthyes: Cobitidae). Mol. Phylogenet. Evol. 2003, 26, 289–299. [Google Scholar] [CrossRef]
- Buj, I.; Podnar, M.; Mrakovčić, M.; Ćaleta, M.; Mustafić, P.; Zanella, D.; Marčić, Z. Morphological and genetic diversity of Sabanejewia balcanica in Croatia. Folia Zool. 2008, 57, 100–110. [Google Scholar]
- Križek, P.; Mendel, J.; Fedorčák, J.; Koščo, J. In the foothill zone—Sabanejewia balcanica (Karaman 1922), in the lowland zone—Sabanejewia bulgarica (Drensky, 1928): Myth or reality? Ecol. Evol. 2021, 10, 7929–7947. [Google Scholar] [CrossRef] [PubMed]
- Kottelat, M. European freshwater fishes. Biologia 1997, 52, 1–271. [Google Scholar] [CrossRef]
- Marešová, E.; Delić, A.; Kostov, V.; Marić, S.; Mendel, J.; Šanda, R. Genetic diversity of Sabanejewia balcanica (Actinopterygii: Cobitidae) in the western Balkans and comparison with other regions. Folia Zool. 2011, 60, 335–342. [Google Scholar] [CrossRef]
- Mousavi-Sabet, H.; Anvarifar, H. Landmark-based morphometric variation between Cobitis keyvani and Cobitis faridpaki (Pisces: Cobitidae), with new habitat for C. faridpaki in the southern Caspian Sea basin. Folia Zool. 2013, 62, 167–175. [Google Scholar] [CrossRef]
- Rohlf, F.J. TpsDig; Version 1.4; Stony Brook University: Stony Brook, NY, USA, 2004; Available online: https://life.bio.sunysb.edu/morph/ (accessed on 1 January 2020).
- Baken, E.; Collyer, M.; Kaliontzopoulou, A.; Adams, D. geomorph v4.0 and gmShiny: Enhanced analytics and a new graphical interface for a comprehensive morphometric experience. Methods Ecol. Evol. 2021, 12, 2355–2363. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 16 February 2023).
- Park, P.J.; Aguirre, W.E.; Spikes, D.A.; Miyazaki, J.M. Landmark-Based Geometric Morphometrics: What fish shapes can tell us about fish evolution. Proc. Assoc. Biol. Lab. Educ. 2013, 34, 361–371. [Google Scholar]
- Adams, D.; Collyer, M.; Kaliontzopoulou, A. Geomorph: Software for Geometric Morphometric Analyses. Version 3.2.1. 2020. Available online: https://cran.r-project.org/package=geomorph (accessed on 1 February 2023).
- Chessel, D.; Dufour, A.; Thioulouse, J. The ade4 package—I: One-table methods. R News 2004, 4, 5–10. Available online: https://cran.r-project.org/doc/Rnews/ (accessed on 1 February 2023).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019; Available online: https://socialsciences.mcmaster.ca/jfox/Books/Companion/ (accessed on 1 February 2023).
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Palumbi, S.R. Nucleic acids II: The polymerase chain reaction. Mol. Syst. 1996, 205–247. [Google Scholar]
- Perdices, A.; Doadrio, I. The Molecular systematics and biogeography of the European Cobitids Based on mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2001, 19, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Leford, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods); Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Posada, D. jModeltest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Yang, J.; He, S.; Freyhof, J.; Witte, K.; Liu, H. The phylogenetic relationships of the Gobioninae (Teleostei: Cyprinidae) inferred from mitochondrial cytochrome b gene sequences. Hydrobiologia 2006, 553, 255–266. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA sequence polymorphism analyses of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Marić, D.; Milošević, D. First record and morphological characteristics of the Balkan golden loach Sabanejewia balcanica (Cobitidae) in Montenegro. Period. Biol. 2010, 112, 149–152. [Google Scholar]
- Ratschan, C.; Gumpinger, C.; Schauer, M.; Wanzenböck, J.; Zauner, G. Arten-schutzprojekt Kleinfische und Neunaugen in Oberösterreich. Teil 2: Balkan-Goldsteinbeißer (Sabanejewia balcanica Karaman, 1922). Osterr. Fisch. 2011, 64, 174–188. [Google Scholar]
- Pekárik, L.; Koščo, J.; Košuthová, L.; Košuth, P. Coenological and habitat affinities of Cobitis elongatoides, Sabanejewia balcanica and Misgurnus fossilis in Slovakia. Folia Zool. 2008, 57, 172. [Google Scholar]
- Năstase, A.; Oţel, V.; Năvodaru, I. Ecological status of fish fauna in arms of the Danube Delta (Danube Delta Biosphere Reserve, Romania) at the beginning of the third Millennium. Acta Zool. Bulg. 2017, 69, 349–360. [Google Scholar]
- Sayyadzadeh, G.; Abbasi, K.; Esmaeili, H.R. Review and re-description of Sabanejewia species in Iran (Teleostei: Cobitidae). Iran. J. Ichthyol. 2019, 5, 277–292. [Google Scholar]
- Kaya, C.; Bayçelebi, E.; Turan, D. Taxonomic assessment and distribution of fishes in upper Kura and Aras river drainages. Zoosyst. Evol. 2020, 96, 325–344. [Google Scholar] [CrossRef]
- Erős, T.; Botta-Dukat, Z.; Grossman, G.D. Assemblage structure and habitat use of fishes in a Central European submontane stream: A patch-based approach. Ecol. Freshw. Fish 2003, 12, 141–150. [Google Scholar] [CrossRef]
- Kutsokon, Y. Northern golden loach (Sabanejewia baltica Witkowski, 1994) in Ubort River. Stud. Biol. 2014, 8, 255–258. [Google Scholar] [CrossRef]
- Bajrić, A.; Buj, I.; Adrović, A.; Hajdarević, E. Morphological and genetic diversity of Sabanejewia balcanica (Cobitidae, Actinopterygii) in Bosnia and Herzegovina. J. Appl. Ichthyol. 2020, 37, 89–98. [Google Scholar] [CrossRef]
- Drensky, P. Die Fische der Familie Cobitidae in Bulgarien. Izv. Tsarskite Prir. Inst. Sofia 1928, 8, 156–181. [Google Scholar]
- Shuai, F.; Yu, S.; Lek, S.; Li, X. Habitat effects on intra-species variation in functional morphology: Evidence from freshwater fish. Ecol. Evol. 2018, 8, 10902–10913. [Google Scholar] [CrossRef]
- Bohlen, J. First report of the spawning behaviour of a golden spined loach, Sabanejewia vallachica (Teleostei: Cobitidae). Folia Zool. 2008, 57, 139–146. [Google Scholar]
- Economidis, P.S.; Nalbant, T.T. A study of the loaches of the genera Cobitis and Sabanejewia (Pisces, Cobitidae) of Greece, with description of six new taxa. Trav. Mus. Natl. D’hist. Nat. Grigore Antipa 1996, 36, 297–347. [Google Scholar]
- Fedorčák, J.; Šanda, R.; Stefanov, T.; Mendel, J.; Koščo, J. Influence of habitat on the external morphology of Sabanejewia (Cypriniformes: Cobitidae) specimens. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Nowak, M.; Klaczak, A.; Szczerbik, P.; Popek, W. Diel differences in the exploitation of shallow inshore habitats by three small bottom-dwelling fishes with implications for their monitoring in a large lowland river. Arch. Pol. Fish. 2018, 26, 159–164. [Google Scholar] [CrossRef]
- Nowak, M.; Klaczak, A.; Koščo, J.; Szczerbik, P.; Fedorčák, J.; Hajdú, J.; Popek, W. Diel changeover of fish assemblages in shallow sandy habitats of lowland rivers of different sizes. Knowl. Manag. Aquat. Ecosyst. 2019, 420, 41. [Google Scholar] [CrossRef]
- Ittiofauna. Available online: https://tinyurl.com/45jyrwhc (accessed on 7 February 2023).
- Siti. Biosphere Reserve Nomination Form Collina Po. Ente di Gestione Aree Protette Po e Collina Torinese. 2015. Available online: www.parchipocollina.to.it/PDF/mab_collinapo.pdf (accessed on 7 February 2023).
- Buj, I.; Mustafić, P.; Ćaleta, M.; Marčić, Z.; Ivić, L.; Žalac, S.; Raguž, L. Peculiar occurrence of Cobitis bilineata Canestrini, 1865 and Sabanejewia larvata (De Filippi, 1859) (Cobitidae, Actinopteri) in the Danube River basin in Croatia. Fundam. Appl. Limnol. 2021, 194, 201–213. [Google Scholar] [CrossRef]
- Mezhzherin, S.V.; Pukhtayevych, P.P.; Tsyba, A.A. Absorbing hybridization of Cobitis taenia and Sabanejewia aurata (Cypriniformes, Cobitidae) in water reservoirs of northern Ukraine connected with diploid-polyploid complex formation. Vestn. Zool. 2014, 48, 503–510. [Google Scholar] [CrossRef]
- Vasil’eva, E.D.; Vasil’ev, V.P. Natural hybridization in spined loaches of the genera Cobitis and Sabanejewia (Cobitidae). J. Ichthyol. 2019, 59, 776–785. [Google Scholar] [CrossRef]
- Saitoh, K.; Aizawa, H. Local differentiation within the striated spined loach (the striata type of Cobitis taenia complex). Jpn. J. Ichthyol. 1987, 34, 334–345. [Google Scholar] [CrossRef]
- Poisot, T.; Verneau, O.; Desdevises, Y. Morphological and molecular evolution are not linked in Lamellodiscus (Plathyhelminthes, Monogenea). PLoS ONE 2011, 6, e26252. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, K.; Franklin, C.E.; Moritz, C.; Blows, M.W. Adaptation of rainbow fish to lake and stream habitats. Evolution 2003, 57, 104–118. [Google Scholar] [CrossRef]
| Main Effects | d.f. | SS | MS | Pseudo F | R2 |
|---|---|---|---|---|---|
| Elevation | 13 | 0.032 | 0.003 | 2.468 | <0.001 |
| Sex | 1 | 0.005 | 0.005 | 4.828 | <0.001 |
| Slope | 1 | 0.0008 | 0.0008 | 0.821 | >0.076 |
| Residuals | 95 | 0.096 | 0.001 | ||
| Total | 110 | 0.134 | |||
| Interaction terms | d.f. | SS | MS | pseudo F | R2 |
| Elevation | 13 | 0.032 | 0.003 | 2.543 | <0.05 |
| Sex | 1 | 0.004 | 0.005 | 4.975 | <0.001 |
| Slope | 1 | 0.0008 | 0.0008 | 0.847 | 0.033 |
| Elevation:sex | 13 | 0.016 | 0.001 | 1.239 | >0.06 |
| Sex:slope | 1 | 0.0008 | 0.0008 | 0.783 | >0.33 |
| Residuals | 81 | 0.079 | 0.001 | ||
| Total | 110 | 0.134 |
| Observed Variances by Group | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.00053 | 0.00086 | 0.00068 | 0.00056 | 0.00072 | 0.00076 | 0.00077 | 0.00053 | 0.00044 | 0.00087 | 0.00086 | 0.00085 | 0.00066 | 0.00101 | ||
| Elevation | 34 | 133 | 140 | 149 | 151 | 159 | 180 | 194 | 195 | 200 | 219 | 222 | 261 | 269 | |
| Pairwise P-values between variances | 34 | 1 | |||||||||||||
| 133 | 0.1069 | 1 | |||||||||||||
| 140 | 0.4537 | 0.3722 | 1 | ||||||||||||
| 149 | 0.8656 | 0.1542 | 0.5719 | 1 | |||||||||||
| 151 | 0.333 | 0.5124 | 0.8185 | 0.4335 | 1 | ||||||||||
| 159 | 0.2417 | 0.647 | 0.6647 | 0.3221 | 0.8451 | 1 | |||||||||
| 180 | 0.2402 | 0.6701 | 0.6458 | 0.3146 | 0.8215 | 0.9811 | 1 | ||||||||
| 194 | 1 | 0.1108 | 0.4723 | 0.8704 | 0.3411 | 0.2494 | 0.2355 | 1 | |||||||
| 195 | 0.6642 | 0.0401 | 0.2455 | 0.5447 | 0.1653 | 0.1127 | 0.1062 | 0.6664 | 1 | ||||||
| 200 | 0.1059 | 0.9546 | 0.3666 | 0.1473 | 0.4917 | 0.6234 | 0.6309 | 0.1118 | 0.0436 | 1 | |||||
| 219 | 0.11 | 0.9903 | 0.374 | 0.1518 | 0.5194 | 0.6557 | 0.6691 | 0.1147 | 0.0456 | 0.9473 | 1 | ||||
| 222 | 0.1197 | 0.9542 | 0.4058 | 0.1678 | 0.5491 | 0.6929 | 0.7053 | 0.1229 | 0.0491 | 0.914 | 0.9611 | 1 | |||
| 261 | 0.4992 | 0.3404 | 0.9529 | 0.6062 | 0.7781 | 0.6153 | 0.6109 | 0.5072 | 0.264 | 0.3318 | 0.3453 | 0.3737 | 1 | ||
| 269 | 0.0211 | 0.4488 | 0.0964 | 0.031 | 0.1597 | 0.2322 | 0.2305 | 0.0215 | 0.0055 | 0.4998 | 0.4412 | 0.4147 | 0.0897 | 1 | |
| GLMM Family | Response | Model Fit | Explanatory | df | Chi2 | p | ANOVA | AIC | Df |
|---|---|---|---|---|---|---|---|---|---|
| Poisson | Lateral blotches | Intercept (full model) | 1 | 3.9 | <0.05 | Type 3 | 500 | 6 | |
| 3 (ANOVA summary) | Elevation2 (groups) | 4 | 10.9 | <0.05 | |||||
| Model summary | Est | SE | Pr (>|z|) | ||||||
| Intercept (Elevation 34) | 2.09 | 0.12 | <0.01 | ||||||
| (Elevation group1) | 0.25 | 0.13 | <0.05 | ||||||
| (Elevation 180) | 0.36 | 0.16 | <0.05 | ||||||
| (Elevation 195) | 0.47 | 0.16 | <0.01 | ||||||
| (Elevation 269) | 0.37 | 0.16 | <0.05 | ||||||
| 2 | Intercept | 1 | 3.9 | <0.05 | Type 3 | 517 | 18 | ||
| Elevation | 13 | 13.97 | >0.37 | ||||||
| Sex | 1 | 0.79 | >0.77 | ||||||
| Slope | 1 | 0.58 | >0.44 | ||||||
| SL | 1 | 0.68 | >0.40 | ||||||
| 1 | Elevation | 13 | 13.97 | >0.37 | Type 2 | 569 | 46 | ||
| Sex | 1 | 0.03 | >0.86 | ||||||
| Slope | 1 | 0.16 | >0.68 | ||||||
| SL | 1 | 0.72 | >0.39 | ||||||
| Double int. (all exp.) | no sig. effect of interaction | ||||||||
| S. bulgarica | S. balcanica | “S. radnensis” | “S. thrakica” | Lin. VI. | S. vallachica | |
| S. bulgarica | 0.003 | 0.003 | 0.003 | 0.004 | 0.003 | 0.004 |
| S. balcanica | 0.018 | 0.003 | 0.002 | 0.003 | 0.003 | 0.004 |
| “S. radnensis” | 0.014 | 0.013 | 0.003 | 0.003 | 0.002 | 0.004 |
| “S. thrakica” | 0.019 | 0.018 | 0.013 | 0.002 | 0.003 | 0.005 |
| Lineage VI. | 0.015 | 0.015 | 0.009 | 0.014 | 0.001 | 0.004 |
| S. vallachica | 0.027 | 0.026 | 0.019 | 0.026 | 0.02 | 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedorčák, J.; Šanda, R.; Stefanov, T.; Mendel, J.; Nowak, M.; Križek, P.; Perdices, A.; Vukić, J.; Koščo, J. The “True Colours” of Golden Loaches (Teleostei: Cobitidae). Fishes 2023, 8, 119. https://doi.org/10.3390/fishes8020119
Fedorčák J, Šanda R, Stefanov T, Mendel J, Nowak M, Križek P, Perdices A, Vukić J, Koščo J. The “True Colours” of Golden Loaches (Teleostei: Cobitidae). Fishes. 2023; 8(2):119. https://doi.org/10.3390/fishes8020119
Chicago/Turabian StyleFedorčák, Jakub, Radek Šanda, Tihomir Stefanov, Jan Mendel, Michal Nowak, Peter Križek, Anabel Perdices, Jasna Vukić, and Ján Koščo. 2023. "The “True Colours” of Golden Loaches (Teleostei: Cobitidae)" Fishes 8, no. 2: 119. https://doi.org/10.3390/fishes8020119
APA StyleFedorčák, J., Šanda, R., Stefanov, T., Mendel, J., Nowak, M., Križek, P., Perdices, A., Vukić, J., & Koščo, J. (2023). The “True Colours” of Golden Loaches (Teleostei: Cobitidae). Fishes, 8(2), 119. https://doi.org/10.3390/fishes8020119







