Abstract
The melanocortin-4 receptor (MC4R) plays a critical role in homeostasis and the regulation of body weight. Polymorphisms in the mc4r gene have been discovered and linked to growth, carcass composition, and meat quality traits. Therefore, we used the CRISPR-Cas9 system to target the mc4r gene in the most important freshwater aquaculture species in the USA, channel catfish, Ictalurus punctatus. Guide RNAs were designed to direct the Cas9 to the coding sequence of the channel catfish mc4r gene. gRNA(s)-Cas9 mixtures were delivered into one-cell embryos using electroporation and microinjection. For each treatment, the nature and rate of mutations were analyzed. Hatching and survival rates were calculated. The overall mutation rates were 30.6% and 66.7–90.6% for electroporation and microinjection, respectively. Mutated fish generated via electroporation or microinjection exhibited 38% and 20% improvement in body weight, respectively, when compared with the full-sib control. The mean feed conversion ratio of the mutants was 1.18 compared with 1.57 in the control fish. The improved growth and feed conversion indicate that the generation of mc4r-edited fish could economically benefit aquaculture production.
1. Introduction
Melanocortin receptors (MCRs) are transmembrane proteins that belong to the G-protein-coupled receptors superfamily [1]. There are five subtypes of MCRs that are numbered MC1R to MC5R according to the sequence in which they were cloned [2]. These five receptors have been characterized in several vertebrates. Zebrafish, Danio rerio, has six MCRs, including two MC5R orthologous, while pufferfish, Fugu rubripes, possesses only four MCRs, lacking MC3R [3,4,5]. The melanocortin-4 receptor (MC4R) was cloned and characterized first in humans [6]. It is widely expressed in the nervous system [7] and plays a critical role in the regulation of food intake and energy homeostasis in both lower and higher vertebrates [8,9,10,11]. MC4R has been cloned and identified in many fish species including zebrafish [5], goldfish, Carassius auratus [12], spiny dogfish, Squalus acanthias [13], sea bass, Dicentrarchus labrax [14], common carp, Cyprinus carpio [15], grass carp, Ctenopharyngodon idella [16], gibel carp, Carassius gibelio [17], snakehead, Channa argus [18], spotted sea bass, Lateolabrax maculatus [19], and rainbow trout, Oncorhynchus mykiss [20]. In teleost fish, MC4R is expressed in certain tissues other than the central nervous system, including the eyes, gonads, and gastrointestinal tract in zebrafish; the liver and gonads in flounder; the eyes in sea bass; and the ovaries in goldfish, but the respective physiological roles of the MC4Rs in these tissues are not clearly known. The amino acid sequences of MC4R are highly conserved among different species [21].
Polymorphisms in the mc4r gene have been observed and linked to development and growth in medaka, Oryzias latipes, and the cyprinid fish Spinibarbus hollandi [22,23]. In addition, inherited human obesity has been associated with MC4R mutations [24,25]. Knockout of MC4R in animal models increased the size and growth rate in homozygotes, whereas heterozygotes exhibited intermediate weight gain compared with wild-type and mutant homozygotes [26,27,28,29,30,31]. The homozygous knockout mice had maturity-onset obesity, hyperphagia, increased linear growth, hyperinsulinemia, and hyperglycemia. In addition, overexpression of the endogenous melanocortin antagonist agouti-related protein (AgRP) in transgenic zebrafish resulted in obesity and increased linear growth [10]. Thus, MC4R provides a promising target for potential improvement of fish growth.
In channel catfish, Ictalurus punctatus, the growth rate was increased in MC4R-knockout fish [32]. Additional research on MC4R-knockout channel catfish is needed such as the evaluation of additional families to ascertain potential family effects. If there are differences in aggression or competitiveness between mutants and controls, genotype–environment interactions are possible when comparing communal and separate evaluations, potentially affecting the conclusion on the benefits of the mutation. Feed conversion efficiency is a critical trait in aquaculture, and was not measured in a previous study [32] on MC4R-knockout channel catfish.
The use of gene editing methods to generate genetically modified animals has experienced large advances due to the development of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) [33]. The CRISPR-Cas9 system consists of a guide sequence (sgRNA) and an endonuclease (Cas9). Once introduced into the cell, the sgRNA guides the Cas9 to the target site, where a double-strand break is induced. The broken ends are then repaired by non-homologous end joining which results in mutations. The CRISPR-Cas9 system has several advantages including simplicity of design, efficiency of edits, cost effectiveness, and ease of use relative to other gene-editing systems [34]. CRISPR-Cas9 gene editing has been successfully performed in several aquaculture species to enhance performance traits [35,36,37,38,39]. These gene-edited lines have been characterized for growth rate, feed efficiency, disease resistance, and carcass composition.
The channel catfish and its hybrid, channel catfish female × blue catfish (Ictalurus furcatus) male, have been the primary fish grown in the United States aquaculture industry. In the United States, catfish sales reached USD 371 million in 2020; which is 2% lower than 2019 sales, and the total water surface area devoted to catfish production decreased from 24,785 hectares in 2020 to 24,000 hectares in 2021 [40]. Factors contributing to the decline in the catfish industry include increased grain and feed prices, raised fuel costs, and competition from imports [41]. Therefore, genetic improvement of catfish production is a promising approach for the recovery, sustainability, and profitability of catfish production.
In this study, we delivered the CRISPR-Cas9 system into channel catfish zygotes via electroporation or microinjection to edit the mc4r gene of channel catfish and assessed the effects of gene knockout on growth and feed efficiency in an attempt to produce growth-enhanced channel catfish. The potential disadvantage of electroporation with plasmid DNA is the possibility of genomic integration of plasmids, which would result in a transgenic organism not exhibiting one of the advantages of gene editing. Evaluation of potential integration was another objective of the current study.
2. Materials and Methods
2.1. Experiment I: Electroporation
2.1.1. Design of the CRISPR-Cas9 System
Small guided RNAs (sgRNA) were designed (Table 1) and cloned into a pU6dsgRNA (No. 5) plasmid [42] from Addgene (Cambridge, MA, USA). The Cas9 plasmid (pCS2-nCas9n, No. 47929), which was previously tested in zebrafish [43], was obtained from Addgene (Cambridge, MA, USA). The gRNA plasmids were driven by the U6 promoter, while the Cas9 plasmid was driven by the cytomegalovirus (CMV) promoter. All of the plasmids contained the antibiotic selection element for ampicillin.

Table 1.
The sequences of small guided RNAs and the universal (common) primer used to target the channel catfish, Ictalurus punctatus, melanocortin-4 receptor (mc4r) gene.
2.1.2. Preparation of Plasmids for Electroporation
The plasmids were prepared according to Dunham et al. [44] with some modifications. The sgRNA and Cas9 plasmids were transferred to One Shot® Top 10 Chemically Competent E. coli (Invitrogen, Grand Island, NY, USA) following the manufacturer’s guidelines and cultured in Luria–Bertani (LB) broth. One hundred microliters of the transformation mix were spread on the LB agar plate containing 100 μg/mL ampicillin. A single colony was picked from each plate and cultured in 1 mL of LB medium with overnight incubation at 37 °C. Overnight cultures were then transferred to a glass bottle containing 400 mL LB broth with 100 μg/mL ampicillin and incubated overnight. The plasmids were then extracted with an IsoPure Plasmid Maxi II Prep Kit (Denville, Holliston, MA, USA) and the plasmid quantity and quality were inspected with gel electrophoresis and spectrophotometry.
The gRNAs and Cas9 plasmids were prepared for the standard procedure of double electroporation used in our laboratory [44]. Briefly, equal volumes of both gRNAs and Cas9 plasmids were mixed and diluted in 1 mL saline (0.9 % NaCl) solution to the final concentrations of 25 μg/mL for the first electroporation of sperm. The purpose of the saline was to dehydrate the sperm once they were introduced to the solution; when rehydrated, the transformation rates of the embryos can be improved [45,46]. In the meantime, the same concentrations of plasmid solutions were prepared in 3 mL Tris-EDTA (TE) buffer (5 mM Tris–HCl, 0.5 mM EDTA, pH = 8.0) as solvent, which was then used for a second electroporation of embryos.
2.1.3. Channel Catfish Spawning
Channel catfish spawning, egg collection, sperm preparation, and fertilization were performed according to [34]. Sexually mature Kansas random channel catfish females were implanted with 85 μg/kg body weight of luteinizing-hormone-releasing hormone analog (LHRHa) in the dorsal musculature (Reproboost® Implant, Center of Marine Biotechnology, University of Maryland Biotechnology Institute, Rockville, MD, USA). The eggs were hand-stripped into a greased spawning pan by applying gentle pressure to the abdomen. Sexually mature channel catfish males were euthanized, and their testes were collected, crushed, and macerated into saline (0.9 % NaCl) solution to prepare the sperm solution.
2.1.4. Fertilization, Electroporation, and Embryo Incubation
Two drops of sperm solution were added to the plasmid saline solution, mixed, and incubated at room temperature for 5 min. Then, the mixture was poured into a 10-mL petri dish and 2 mL of freshwater were added. This solution was then electroporated with a Baekon 2000 macromolecule transfer system (Baekon, Inc., Saratoga, CA, USA) with the parameters adjusted to 6 kV, 27 pulses, 0.8 s burst, 4 cycles, and 160 µs [47]. The eggs were fertilized with electroporated sperm and incubated at room temperature for one hour. The fertilized eggs were then transferred into a 10 mL petri dish, and enough TE-solution-containing plasmids was introduced to submerge all of the eggs (about 3 mL). After 10 min of incubation at room temperature, the embryos were electroporated again as described above. The same procedure was performed on the control group, using saline and a TE buffer devoid of gRNA/Cas9 plasmids.
The embryos were reared in 10 L tubs filled with Holtfreter’s solution (59 mM NaCl, 0.67 mM KCl, 2.4 mM NaHCO3, 0.76 mM CaCl2, and 1.67 mM MgSO4) [48,49] containing 10 ppm doxycycline and incubated with continuous aeration at 27 °C for 6–7 days until hatching. Dead embryos were removed and recorded, and the solution was changed daily. After hatching, channel catfish fry were moved to 60 L glass aquaria in a recirculating system at a density of 30 fry per aquarium. Water quality parameters were maintained with a dissolved oxygen level above 5 ppm, pH between 6.8 and 7.4, hardness above 30 ppm, less than 2 ppm unionized ammonia, and 0 ppm of nitrite and chlorine. The fry were fed newly hatched brine shrimp four times a day for one week, then gradually switched to Purina® AquaMax® Fry Powder (Purina Mills, LLC, Gray Summit, MO, USA) for one month. The size of the feed increased as the fry grew. They were fed twice a day with Purina® AquaMax® Fry Starter (Purina Mills, LLC, Gray Summit, MO, USA) 100, 200, and 300 according to their age and size. The fry were reared in aquaria for 10 months and sampled for genotyping and individually pit-tagged.
2.2. Experiment II: Microinjection
2.2.1. Design and Preparation of sgRNA and CRISPR/Cas9 System
Based on the results from experiment I where we achieved a single 285 bp deletion in the mc4r gene, we thought changing the gRNA targets might achieve larger deletions and more types of mutation. Therefore, we designed three gRNAs that cover the coding sequence of the channel catfish mc4r gene using the CRISPRscan online tool [50]. The sgRNAs (Table 1 and Figure 1) were selected depending on the scores provided by CRISPRscan. The cloning-free (PCR-based) method to generate sgRNA templates was used. The universal primer containing the sgRNA scaffold as well as ssDNAs templates for gRNAs containing the T7 promoter and the 20 nt gene-specific target sequence without the protospacer-adjacent motif (PAM) were manufactured by Invitrogen (Carlsbad, CA, USA; Table 1). The sgRNAs were generated by T7 run-off as described in [51] with some modifications. The universal primer and ssDNAs template were annealed and filled by Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA). In an RNAse-free environment, the resulting double-stranded DNA served as the template for in vitro transcription to generate sgRNA using a Maxiscript T7 Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s guidelines. Then, the sgRNAs were purified using a Zymo RNA Clean and Concentrator Kit (Zymo Research, Irvine, CA, USA). The Cas9 protein was obtained from PNA BIO, Inc. (Newbury Park, CA, USA) and reconstituted according to the manufacturer’s guidelines. Four sets of injection solutions were prepared: three by mixing each individual gRNA with Cas9 protein separately (MC4R A, B, and C) and the fourth by combining the three gRNAs together with Cas9 protein (MC4R X). Phenol red was added to color the gRNA/Cas9 solutions and mixed up to one third of the final volume [34]. The final concentrations of gRNA and Cas9 protein were 150–200 and 300–350 ng/µL, respectively. The mixtures were then incubated for 10 min on ice before loading into the microinjection needle.
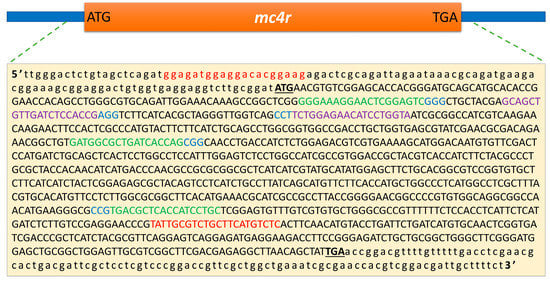
Figure 1.
The genomic target sites of the sgRNAs in the channel catfish, Ictalurus punctatus, melanocortin-4 receptor (mc4r) gene. The coding and non-coding sequences are indicated by uppercase and lowercase letters, respectively. The start and stop codons are represented by bold uppercase letters. The primer sites are indicated in red. The gRNA target sites for electroporation are indicated in purple with a protospacer-adjacent motif (PAM) in blue. The gRNA target sites for microinjection are indicated in green, with PAM in blue. ATG and TGA represent the start and stop codons, respectively.
2.2.2. Fertilization and Microinjection
The sperm and eggs were obtained and prepared as in experiment I. To fertilize the eggs for microinjection, 200–300 eggs were transferred to a greased spawning pan. One to two mL of the normal sperm solution were added to the eggs and mixed gently. Then, freshwater was added to the eggs gently and swirled for 30 s to activate the sperm and eggs. More fresh water was added to the fertilized eggs, and the eggs were allowed to harden for 10–15 min before microinjection. The fertilized eggs were injected according to the procedures of zygote injection developed recently in our laboratory [34]. Fifty nanoliters of the mixture were directly injected into the yolk as close as possible to the blastodisc. One-cell embryos were injected through 15–90 min post fertilization and just before the beginning of the first division [52]. Two controls were used for this experiment: one injected with the solution devoid of Cas9/gRNA (injected control, iCTRL) and the other not injected (non-injected control, nCTRL). All of the injected and control embryos were then incubated and reared as described before [53].
2.3. Mutation Analysis
2.3.1. Genomic DNA Extraction
Fin-clip samples were collected from one-month-old channel catfish fry on ice. The fry and fingerlings were euthanized, and samples from barbel, gills, muscle, and the intestine were collected to study possible mosaicism of the mutations among different tissues. The DNA was extracted according to [54]. Briefly, about 10 mg tissue samples were digested with 0.1-mg/mL proteinase K in an Eppendorf tube containing 600 μL of cell lysis buffer (100 mM NaCl, 10 mM Tris, 25-mM EDTA, and 0.5% sodium dodecyl sulfate) in a shaker incubator 60–90 min. Protein precipitation was achieved by adding 200 μL of a protein precipitation solution (Qiagen, Redwood City, CA, USA) and incubation on ice for 15 min followed by centrifugation at top speed for 10 min at room temperature. The supernatant was transferred to a clean, labeled Eppendorf tube. The DNA was subsequently precipitated from the supernatant with isopropanol, washed with 75% ethanol, air-dried, and dissolved in Tris-EDTA (TE) buffer. The integrity of DNA was examined by using 1% agarose gel electrophoresis. The quantity and purity of DNA were checked with a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.3.2. Polymerase Chain Reaction (PCR) and Surveyor Mutation Detection Assay
The primer sets for the PCR were designed to cover all possible mutation sites (Table 2 and Figure 1). PCR was performed using an Expand High Fidelity PLUS PCR System (Roche, Indianapolis, IN). The PCR amplification procedure was as follows: initial denaturation for 3 min at 94 °C, followed by 34 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 50 s, and a final elongation at 72 °C for 10 min. The resulting PCR product length was verified in 1.5% agarose gel. To detect mutations, the surveyor mutation detection assay was performed on the amplified PCR products without mixing with wild-type DNA since the P1 fish were expected to be mosaic (containing both the mutated and wild-type mc4r gene). We used a Surveyor® mutation detection kit for standard gel electrophoresis (Integrated DNA Technologies, Coralville, IA, USA) according to the manufacturer’s guidelines [55,56]. PCR products were denatured and re-annealed as follows: 95 °C for 10 min; 95 to 85 °C at −2 °C/s; 85 to 35 °C at −0.3 °C/s; cooling to 4 °C. Then, nuclease S, enhancer S, and MgCl2 were mixed, added to the PCR products, and incubated at 42 °C for 1 h. Digested products were separated in 2% agarose gel and compared to those from a full-sib control channel catfish.

Table 2.
Primers used in the polymerase chain reaction (PCR) for molecular cloning of the channel catfish, Ictalurus punctatus, melanocortin-4 receptor (mc4r) gene and detection of pU6dsgRNA and Cas9 plasmids’ integration in gene-edited channel catfish.
2.3.3. Cloning and Sequencing
To confirm and identify the mutations in each treatment, genomic DNA was obtained from three positive mutated individuals for each treatment, pooled to save the cost, amplified with PCR, and the resulting amplicons were cloned into a TOPO® TA Cloning® Kit for Sequencing (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions (Invitrogen, Carlsbad, CA). TOPO plasmids were then transformed into One Shot® TOP10 Electrocomp™ E. coli (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA) with some modifications [53]. Fifteen single colonies were selected from each treatment, inoculated into 1.5-mL Eppendorf tubes containing 1 mL LB medium with 100 ppm ampicillin, incubated overnight at 37 °C, and sent to Eurofins Genomics (Louisville, KY, USA) for sequencing. Alignments of nucleotides and amino acid sequences were created and interpreted.
2.3.4. Detection of Plasmid Integration
To determine the integration of plasmids into the channel catfish genome or their persistence in the cytoplasm, two pairs of PCR primers (Table 2) were designed to detect gRNA and Cas9 plasmids in mutated channel catfish by amplifying the vector backbone and/or antibiotic selection gene. In addition, a pair of PCR primers was used to amplify a segment of the channel catfish myostatin gene as an internal control. The PCR procedure was the same as above except that the annealing step was performed at 61 °C. The PCR products were inspected with 1.5 % agarose gel electrophoresis.
2.4. Evaluation of Growth and Feed Efficiency
Ten-month-old mc4r-mutant fish from experiment I were divided into two replicates (eight fish each) and reared in aquaria in a recirculating system. Two control replicates (eight fish each) from normal full-sib channel catfish were reared in the same system under the same feeding regime. The fish were fed ad libitum on a 32% crude protein diet twice a day. The rearing experiment continued for nine months during which feed consumption was recorded daily, and body weight was evaluated at five different time-points. The growth pattern was then determined for the mutated and control channel catfish. The feed conversion ratio (FCR) was calculated for each replicate by dividing the total amount of feed consumed by total weight gain.
2.5. Statistical Analysis
Dead microinjected embryos were collected, recorded, and assigned a value representing the time of death (day post fertilization, dpf). The mortality % was calculated as the number of dead fish in a treatment divided by the total number of embryos in the same treatment and multiplied by 100. The survival curves of embryos and fry and the time to hatch for all treatment and control groups were compared using the Kaplan –Meier test. Multiple comparisons of mean survival and hatch time were performed using the log rank (Mantel–Cox) test [53]. The plots of embryo survival, time to hatch, and fry survival were generated using GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA, USA). Dead embryos were excluded from the calculation of hatch rate and time to hatching. The monthly body weights of mutated and control channel catfish from experiment I were compared using two-way (month and treatment) analysis of variance test with interaction. The statistical significance was set at p < 0.05, and all data are presented as the mean ± standard error (SE). All analyses were performed using IBM SPSS 23.0 software.
3. Results
3.1. Experiment I: Electroporation
3.1.1. Embryo Mortality, Hatchability, and Fry Survival
Two sets of eggs were prepared for electroporation with two hundred eggs in each set. The first set was double electroporated with the CRISPR/Cas9 plasmids targeting the channel catfish mc4r gene. Sixty-two embryos hatched (31% hatch rate), and after five months, 50 fish were still alive (survival rate 83.3%; Table 3). Nineteen of the sixty-two individuals had the modified mc4r gene (30.6% mutation rate). The second set (control) was electroporated in buffer without plasmids, resulting in 87 fry (43.5% hatch rate) with a survival rate of 90.8%.

Table 3.
The mortality, hatchability, and survival of electroporated (experiment I) and microinjected fry (experiment II) from the Kansas random strain of channel catfish, Ictalurus punctatus. In experiment I, one-celled embryos were electroporated with sgRNA and Cas9 plasmids targeting the channel catfish melanocortin-4 receptor (mc4r) gene, while the control group was electroporated with TE buffer without plasmids. In experiment II, one-celled embryos were microinjected with sgRNAs-Cas9 protein targeting the same gene. Three groups were injected with individual sgRNAs (MC4RA, MC4RB, and MC4RC), while MC4RMIX was injected with a mixture of the three sgRNAs and Cas9. Two control groups were used for experiment II, including an injected control (injected with the same injection solution but without sgRNA or Cas9, iCTRL) and a non-injected control (nCTRL).
3.1.2. Identification of Mutations, Mosaicism, and Plasmid Integration
The results of the surveyor mutation detection assay revealed one distinct band in the digested PCR products from control fish, while mutated samples showed three distinct bands (Figure 2A). The PCR product from the mutated P1 fish will likely contain mutated and wild-type sequences. Consequently, the hybridization step in the surveyor assay produces heteroduplex DNA which is composed of wild-type and mutated DNA strands in the same double helix. This results in mismatches in the double helices, which will then be cleaved producing two additional bands besides the original band. PCR products from wild-type fish will not contain mismatches and, therefore, will not be cleaved resulting in a single band. Three of the mutated samples were cloned and sequenced according to [53]. Alignment of both mutated and wild-type sequences verified the deletion of 285 nucleotides at the target sites between sgRNA I and II (Figure 3). The deletion began in the non-coding sequence 14 bp upstream of the start codon with 271 bp deleted from the 5′ end of the coding sequence. Another finding was the insertion of a few nucleotides in different positions and point mutations in 25 positions within the coding sequence. To assess mosaicism, samples from the barbel, muscle, eye, and intestine were analyzed for mc4r mutations. Mutations were detected in all of the tested tissues the from mutated fish (Figure 2B). Inspection of the plasmid integration revealed that the genome of the mc4r-mutated individuals did not possess exogenous DNA fragments from Cas9 or sgRNA plasmids (Figure 2C).
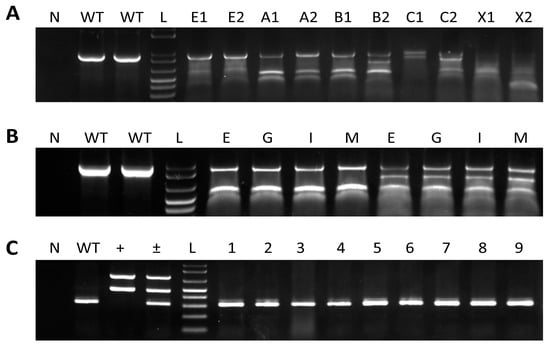
Figure 2.
Identification of mutated melanocortin-4 receptor (mc4r) gene and integration of sgRNA and Cas9 plasmids in channel catfish, Ictalurus punctatus. N indicates the no-template control, while the WT lane represents samples from wild-type channel catfish. L indicates 1 kb plus DNA ladder (Invitrogen). (A) Surveyor-based detection of mc4r gene mutations in pelvic fins from 10 channel catfish. Letters and numbers represent channel catfish tested from different treatments; E1, E2 (electroporated treatment); A1, A2 (MC4RA treatment); B1, B2 (MC4RB treatment); C1, C2 (MC4RC treatment); X1, X2 (MC4RMIX treatment). The presence of two or more additional bands compared to the WT lane indicates the occurrence of mutations. (B) Surveyor-based detection of mc4r gene mutations in different tissues from two mutated channel catfish. E, eye; G, gills; I, intestine; M, muscle. (C) Integration of Cas9 and sgRNA plasmids in electroporated fish. The plus sign (+) indicates the positive controls (Cas9 and sgRNA plasmids). The plus/minus sign (±) indicates the positive controls and wild-type channel catfish. Numbers represent channel catfish individuals carrying mutated mc4r gene.
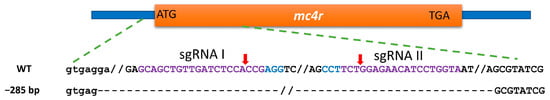
Figure 3.
CRISPR-Cas9-induced mutations in channel catfish, Ictalurus punctatus, mc4r gene via the co-electroporation of the Cas9 plasmid and two sgRNAs plasmids. The wild-type (WT) channel catfish mc4r gene sequence is shown on the top. Sequences in purple and blue colors are the target sites of the sgRNA and protospacer-adjacent motif (PAM), respectively. Uppercase and lowercase letters indicate the coding and non-coding sequences, respectively. Red arrows indicate the expected sites of cleavage by Cas9. Dashes represent the deletion of nucleotides along the mc4r gene.
3.1.3. Evaluation of Growth and Feed Efficiency in Experiment I
Regardless of the fish genotype, body weights increased over time (F4,120 = 2237.86, p < 0.001). Regardless of the sampling time-points, the mutated fish had higher body weights than the control fish (F1,30 = 8.07, p = 0.008). The body weights of mutated fish were significantly higher than those of the control fish in November and January (Figure 4). At the end of the sampling period, the mutated fish (n = 16) had a 38% increase in their body weights (421.9 ± 35.2 g) compared with the control fish (n = 16; body weight 304.9 ± 15.5 g). The feed conversion ratio was 1.18 in the mc4r-mutated fish compared with 1.57 in the control fish, an observed percent difference of 33% (Table 4). Noting that the sample size for the feed conversion ratio is very small (two replicates), we did not detect a significant difference between the two groups (independent t-test, p = 0.104).
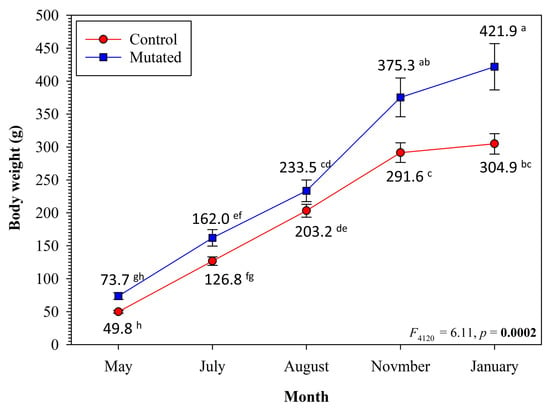
Figure 4.
Growth of melanocortin-4 receptor (mc4r) mutated Kansas random channel catfish, Ictalurus punctatus, compared with the full-sib control fish. The fish were grown in 60 L glass aquaria in a recirculating system at a density of eight fish per aquarium and two replicates per group. All of the fish were fed ad libitum on a 32% crude protein diet. Means are shown for each sampling time-point, where with different superscript letters indicate significant difference (Two-way month and treatment analysis of variance test with interaction).

Table 4.
Feed conversion ratio (FCR) of melanocortin-4 receptor (mc4r) gene-mutated channel catfish, Ictalurus punctatus, compared with the full-sib control fish (experiment I, electroporation). The fish were reared in 60 L glass aquaria in a recirculating system for nine months. Two replicates of eight fish each were used for the control and mc4r-mutated fish. Each of the initial weight, final weight, and weight gain values presented below are the sum of the values of all of the fish per replicate.
3.2. Experiment II: Microinjection
3.2.1. Embryo Mortality, Hatchability, and Fry Survival
Embryo mortality started on the first day post fertilization (dpf) and continued until eight dpf (Figure 5A). Embryo mortality was the lowest in the nCTRL embryos followed by the injected embryos (Table 3, Figure 5A). The higher embryo mortality in the MC4RC group was due to infection with fungus. The mean survival time ranged from 4.8 to 7.2 dpf. Overall comparisons revealed significant differences in the mean survival time among at least two groups (p < 0.0001). With multiple comparisons, the mean survival time for the nCTRL group was significantly longer than those for all other groups (p < 0.005). The MC4RC group had a significantly shorter mean survival time than those for all other groups (p < 0.0001). Survival time was significantly different between the MC4RB and MC4RMIX groups (p = 0.031). All other comparisons were not significant (p > 0.05).
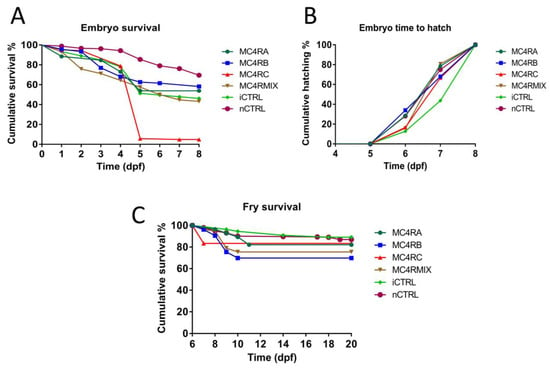
Figure 5.
Embryo survival, hatchability, and fry survival curves of channel catfish, Ictalurus punctatus, embryos from the Kansas random strain microinjected at the one-cell stage with sgRNAs/Cas9 protein targeting the melanocortin-4 receptor (mc4r) gene. Three gRNAs were microinjected individually (MC4RA, MC4RB, and MC4RC) or multiplexed (MC4RMIX). Two control treatments were used. Injected control embryos (iCTRL) were full-sib to the treatment groups and injected with the same solution and volume, but without sgRNA or Cas9 protein. The second control was not injected (nCTRL). (A) Embryo cumulative survival was considered 100% at day 0 then decreased over time as embryos died. (B) The hatch rate was calculated as the fraction of embryos that hatched at a given time point compared to the total live embryos that hatched. (C) Fry survival was calculated for the fry just after hatching and until 20 days post fertilization (dpf).
The embryos started to hatch at 6 dpf and all embryo hatching was completed at 8 dpf (Figure 5B). The embryo mean time to hatch ranged from 6.9 to 7.4 dpf (Table 3). The overall comparison of embryo mean time to hatch revealed significant differences among at least two groups (p = 0.001). Multiple comparisons detected significant differences between the iCTRL group and all of the other groups except MC4RC (p < 0.005). All of the other comparisons were not significant (p > 0.05).
Fry survival curves were plotted (Figure 5C). The fry mean survival time ranged from 16.6 to 19.2 dpf (Table 3). Overall comparison of the fry survival time revealed significant differences between at least two groups (p = 0.01). With multiple comparisons, significant differences were detected between fry mean survival time MC4RB and both iCTRL (p = 0.009) and nCTRL (p = 0.001). Significant differences were also detected between MC4RMIX and both iCTRL (p = 0.048) and nCTRL (p = 0.02).
3.2.2. Identification of Mutations and Mosaicism
Mutated individuals from among both dead embryos and live fry were identified using a surveyor mutation detection assay. More than 87% of dead embryos from all treatments were mutated, while the mutagenesis frequencies in live fry from treatment groups MC4RA, MC4RB, MC4RC, and MC4RMIX were 78.6%, 90.6%, 66.7%, and 84.2%, respectively. The digested PCR products from the control samples produced only one band, while the mutated samples revealed more than one clear band (Figure 2A). The mutated samples of the MC4RMIX group showed the highest variability of banding patterns observed using gel electrophoresis.
The barbel, muscle, eye, and intestine were analyzed for mc4r mutations to assess mosaicism. The mutated fish exhibited the targeted mutation in every tissue examined, with similar banding patterns produced by gel electrophoresis (Figure 2B).
The PCR amplicons from mutated individuals were cloned and sequenced to identify the nature of mutations in each treatment. The nucleic acid alignment of the mutated sequences against the wild-type sequence revealed multiple forms of insertions/deletions caused by the CRISPR/Cas9 system at target sites (Figure 6). The mutated individuals from the MC4RA group showed five types of deletions of 4, 8, 13, 18, and 22 base pairs and only one insertion of 14 base pairs (Figure 6A). In the MC4RB group, only two types of deletions (6 and 10 bp) and one insertion (9 bp) were observed (Figure 6B). The MC4RC group had two types of insertions (4 and 12 bp) and one large deletion (516 bp, Figure 6C). In the 516 bp deletion, approximately half of the exon of the channel catfish mc4r gene was removed. The MC4RMIX group exhibited multiple forms of deletions/insertions, including several large deletions of up to 693 bases (Figure 6D).
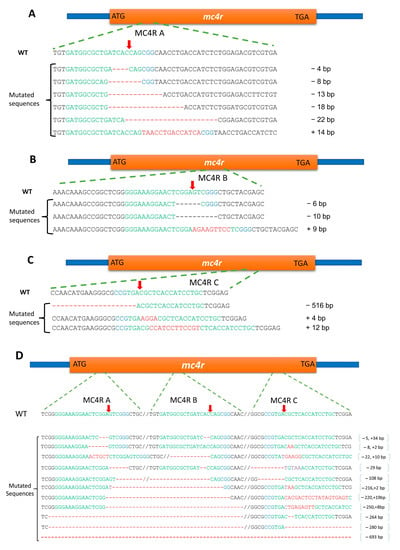
Figure 6.
CRISPR-Cas9-induced mutagenesis in the melanocortin-4 receptor (mc4r) gene of channel catfish, Ictalurus punctatus, Kansas random strain through microinjection of guide RNAs and Cas9 enzyme. The partial sequence of wild-type channel catfish mc4r gene (WT) is the top sequence in each panel. Sequences in green and blue colors are the target sites of the sgRNAs followed by the protospacer-adjacent motif (PAM) sequence, respectively. Red arrows indicate the expected sites of cleavage by Cas9. Dashes indicate the deletion of nucleotides along the mc4r gene. The plus (+) and minus (−) signs indicate insertions and deletions, respectively. (A) MC4RA treatment, (B) MC4RB treatment, (C) MC4RC treatment, and (D) MC4RMIX treatment.
3.2.3. Evaluation of Growth of Microinjected Fry in Experiment II (Microinjection)
The distribution of body weights of mc4r-mutated fry was shifted towards higher values (Figure 7). The mutated fry exhibited significantly higher body weights (215.8 ± 9.6 mg, n = 72) compared with the control fry (179.4 ± 4.4 mg, n = 117; p = 0.001), which was about a 20% increase in body weight. The comparison of body weight was performed with and without the inclusion of an outlier mutant fry (body weight = 590 mg), but no changes to the significance of the results were detected. No significant differences in body length were detected between the mutated and control fry (p = 0.467).
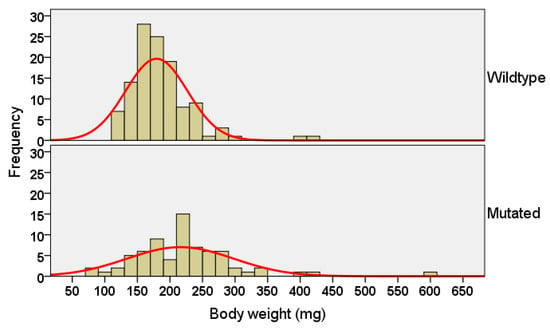
Figure 7.
Distribution of body weights of melanocortin-4 receptor (mc4r)-mutated Kansas random channel catfish, Ictalurus punctatus, at 30 days post fertilization (dpf) compared with full-sib control fish. The fry were reared in fry catchers in a flow-through system and fed Artemia nauplii for one week post fertilization, followed by Purina® AquaMax® Fry Powder. The mean body weight of the mc4r-mutated fry was higher than the control fry (independent t-test, p = 0.001).
4. Discussion
Gene editing of the channel catfish mc4r gene using the CRISPR-Cas9 system was accomplished with both electroporation and microinjection. In the first experiment, the plasmids expressing Cas9 and sgRNAs were delivered into fertilized eggs using electroporation, whereas in the second experiment, the gRNA and Cas9 protein were directly co-injected into the zygote at the one-cell stage. The overall mutation rates were 30.6% and 66.7–90.6% for the electroporation and microinjection, respectively. The mutated fish generated via electroporation or microinjection exhibited 38% and 20% improvement in body weight, respectively, when compared with the full-sib control fish. The feed conversion ratio was also improved from 1.57 in the control fish to 1.18 in the mc4r-edited fish. No potential integration events of plasmid constructs were detected in the gene-edited fish.
CRISPR-Cas9 has been used as a gene editing tool for targeted gene disruption in many living organisms such as the fruit fly [57], zebrafish [43], medaka [58], mice [59], sheep [60], cattle [61], and human cells [62]. The mutation rate differs among studies, ranging from 2–4% [63] to 75–99% [43]. In aquaculture species, CRISPR-Cas9 gene editing was performed in common carp [38], Atlantic salmon, Salmo salar [36], Nile tilapia, Oreochromis niloticus [37], and channel catfish [64]. The mutation rate ranged widely from 2% [35] to 100% [65]. In our study, the mutation rate was 30.6% for electroporation, while the mutation rate varied from 66.7% to 90.6% for microinjection. These results are consistent with these previous studies on channel catfish [34,53,65]. The higher mutation rate for microinjection compared with electroporation was expected. The CRISPR-Cas9 system can be introduced into all microinjected zygotes, whereas in electroporation, the uptake of exogenous nucleic acids by all zygotes is not guaranteed. However, electroporation provides a non-invasive alternative in which gene-editing tools could be applied to large batches of embryos in a short time [34]. In the current study, the overall mutation rates were higher than those obtained in Atlantic salmon [36,66] and common carp [38].
Oligonucleotides are most active when injected directly into the embryonic cells rather than the yolk [67]. However, this procedure needs careful orientation of the embryos and is more time consuming. We co-microinjected Cas9 protein and sgRNA directly into the yolk at the one-cell stage so that it would be transferred to the cytoplasm through cytoplasmic streaming [68,69]. This is usually adequate to edit the genome, requiring less time and effort during microinjection with less disruption of embryos, leading to higher mutation and survival rates [34]. In addition, microinjecting the Cas9 protein instead of its plasmid eliminated the time required for expression of Cas9 plasmids and made genome targeting during the one-cell stage of embryonic development possible.
The CRISPR-Cas9 system showed high gene-targeting specificity, as all the mutations were located within the expected sgRNA binding sites. These mutations should lead to deletion of key amino acids and/or frameshift, generating a premature stop codon and early termination of translation, which should disrupt the molecular functions of the gene. In the design of sgRNA for the current study, our in silico investigation did not discover any possible off-targets. This is supported by the absence of any adverse effects on embryonic development or survival as compared with other gene-editing tools such as the zinc finger nucleases (ZFNs) or transcription activator-like effector nucleases (TALENs) [70]. Compared to the results obtained from TALENs and ZFNs in channel catfish [64,71], the CRISPR-Cas9 system has higher specificity in targeted gene editing, low incidence of off-target effects, and low cell toxicity, which is similar to what was accomplished in zebrafish [43], medaka [58], Nile tilapia [37], Atlantic salmon [36], and channel catfish [53,65].
Electroporated fish showed a 285 bp on-target deletion in addition to substitution mutations that were not at the target sites but were still within the coding sequence. The 285 bp deletion began 14 bp upstream of the start codon and continued for 271 bp in the 5′ end of the coding sequence. The deletion of the 5′ untranslated leader and the start codon from mRNA should reduce ribosome binding strength and translational efficiency [72], and if translated, 90 amino acid residues would be missing from the protein’s N-terminal end in addition to a frameshift mutation. All these consequences of the deletion would likely inhibit MC4R function. Unlike electroporation, which gave only one type of deletion, microinjected fish had several forms of on-target mutations ranging from insertion/deletion of a few base pairs to 693 bp deletion. Deletion or insertion of nucleotides that are not multiples of three is expected to switch the reading frame and change the entire protein. The MC4RA sgRNA resulted in six types of mutations, including 4, 8, 13, 18, and 22 bp deletions and a 14-bp insertion. The 18 bp deletion is expected to eliminate 6 amino acid residues from the protein without changing the frame for translation, while all other mutations should result in a frameshift and a premature stop codon. MC4RB induced two deletions (6 and 10 bp) and 9 bp insertion. A deletion of 516 bp was generated by MC4RC sgRNA in addition to 4 and 12 bp insertions. Eleven mutations were identified in the MC4RMIX group which was simultaneously injected with the three sgRNAs. These mutations included insertions, deletions, or a combination of both. About 82% of these mutations (9 of 11) are expected to shift the reading frame. The other two mutations—the deletions of 264 and 693 bp—would not change the reading frame but are expected to eliminate 88 and 231 amino acid residues from the protein. Compared with the wild type, the patterns and lengths of bands revealed with gel electrophoresis differed depending on the treatment group of the mutated fry. The mutated samples of the MC4RMIX group showed the highest variability in banding patterns revealed by gel electrophoresis. Similarly, the alignment of MC4RMIX group sequences revealed the most variable types of mutation. This may be attributed to the co-injection of the three sgRNAs resulting in several possibilities of double strand break and repair. In several sequencing clones from the MC4RMIX group, the region between two or three sgRNAs was eliminated. This result aligns with those of previous reports of inducing large deletions in channel catfish using multiple sgRNAs simultaneously [53].
The CRISPR/Cas9 system was previously microinjected in channel catfish in our laboratory with variable rates of embryo hatch and fry survival. In the present study, these rates were comparable to those of previous results on channel catfish with considerably high mutation rates [34,53,64,65]. For electroporated fish, hatching, survival, and mutation rates were not greatly changed from what was obtained before [64,71]. In the present study, we assessed the effects of the microinjection technique and introduction of the CRISPR-Cas9 system on embryo survival, hatch rate, and early fry survival. We included an injected control group (without CRISPR-Cas9) in addition to the normal non-injected control group. The injection negatively affected embryo survival while the CRISPR-Cas9 system did not exhibit any noticeable effects on embryonic development or hatch rate, which aligns with the results of Elaswad et al. in channel catfish [53]. In addition, we did not record any abnormalities pre or post hatching, suggesting that off-target mutations did not occur or did not affect the embryonic development or hatching.
Mutations in the mc4r gene have been reported in several species including mammals, birds, and fish [73,74,75], resulting in increased body weight. The mc4r gene was edited in zebrafish [76], channel catfish [32], rats, and mice [77]. In zebrafish, knockout of the mc4r gene improved growth after 2.5 months post fertilization [78]. In a previous study on channel catfish, mc4r mutants exhibited 40% and 30% increase in body weight at the stocker stage (50 g) and the market size, respectively [32]. In the current study, growth was improved by 20% in the one-month-old fry compared with 38% improvement in nineteen-month-old fish. The variations in body weights of the mutated fish were higher than the control fish, which provides the basis for selective breeding to further increase growth and reduce variation. Therefore, future studies are needed to correlate the different types of mutations with body weight. The previous study [32] was conducted with communal evaluation and the current one was conducted with separate evaluation, and the results were similar with no apparent genotype–environment interactions. This is also circumstantial evidence that the aggressiveness of the mc4r mutants and wild type were similar in regard to feeding behavior.
Myostatin (MSTN) is another target for growth improvement in fish. MSTN was successfully knocked out in channel catfish using the CRISPR-Cas9 system [39,65]. The improvement in body weight was variable. At one month of age, the mean body weight of the MSTN-mutant fry was 29.7% higher than the full-sib wild-type fry [65]. The P1 MSTN-mutant channel catfish exhibited 88% and 27% higher body weight than the control fish at the stocker stage (100–200 g) and market size, respectively [39]. The F1 MSTN-mutant channel catfish were 218% larger than the control fish at the stocker stage [39]. Considering the improvement of body weight in the mc4r and MSTN mutant fish, it would be interesting to edit these two genes simultaneously in channel catfish and assess the effects in double mutants.
In aquaculture, feeding costs exceed half of the variable operating costs [41]. Therefore, the lower the feeding costs, the higher the profitability of aquaculture production. In our study, the mean FCR was 1.18 in the mc4r-edited fish compared with 1.57 in the control fish. Although the calculated FCR was not statistically different between the mc4r-edited fish and the control fish, it suggests an economically important result. The mc4r-edited fish consumed about 25% less feed to produce the same live weight as the control fish, which can be translated into lower variable operating costs and higher profits. However, production-scale performance trials and in the commercial environment, ponds, are needed to demonstrate this point. Previous studies have reported that certain mutations in the mc4r gene were associated with improved FCR in pigs. In Lithuanian White pigs, a significant association between mc4r mutations and better FCR was reported [79]. Another study indicated that the best FCR was linked with certain polymorphisms in the mc4r gene in Italian Large White and Italian Duroc pigs [80]. The improvement of growth and FCR reported in the current study can be maximized by applying genetic improvement programs, such as selection, to the mc4r-mutant fish. Enhancing growth and improving FCR are desirable traits for aquaculture production and profitability.
Overall, there are fewer concerns regarding commercialization of gene-edited animals than transgenic animals. In the United States, the Food and Drug Administration (FDA) regulates gene-edited animals, and no gene-edited animals have completely navigated the FDA process to date. However, myostatin gene-edited Nile tilapia are now approved in Argentina and Brazil [81]. The Japanese Ministry for Health, Labor and Welfare approved mstn-knockout red sea bream (Pagrus major) and leptin gene-edited tiger puffer (Takifugu rubripes), and they are now sold in Japan.
5. Conclusions
We successfully applied CRISPR-Cas9 technology for targeted gene editing in channel catfish. Both electroporation and microinjection approaches were evaluated, and the channel catfish mc4r gene was mutated. At the fry stage, growth was enhanced by 20% in mc4r-mutated fry. The mutated fish showed 38% improvement in body weight compared with the full-sib control fish. The FCR was 1.18 in the mc4r-edited fish compared with 1.57 in the control fish. Our study goes beyond vertebrate models (zebrafish and medaka) and addresses the utility of CRISPR-Cas9 as an efficient tool in generating gene-edited channel catfish, and potentially other aquaculture species, with high efficiency. Further studies are needed to evaluate the carcass composition and meat-quality traits of mutated fish when they reach food size and to demonstrate results at the production scale. Other physiological parameters and immune status are worthy of being considered and linked to the productivity of these gene-modified strains.
Author Contributions
K.K., A.E., W.C., Z.L., R.D.C. and R.D. conceived and designed the experiment and analyzed the results; K.K., A.E., H.A., M.M., S.L., R.O., Z.Y., D.D., K.V., W.S.B., G.Q., Y.Y. and N.J.C.B. executed the experiments; K.K., A.E., R.O., Z.Y., D.D., K.V., W.S.B., G.Q., Y.Y. and N.J.C.B. collected the samples, gathered the data, and performed the fish care; A.E. and H.A. conducted the statistical analysis; K.K., A.E. and R.D. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the United States Department of Agriculture National Institute of Food and Agriculture award number 2015–67015-23488 to Roger D. Cone and sub-award to Rex Dunham.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, approved by the Auburn University Institutional Animal Care and Use Committee (AU-IACUC; Protocol Review Number: 2016-2871; Approval date: 14 April 2016), and followed the Association for Assessment and Accreditation of Laboratory Animal Care protocols and guidelines.
Data Availability Statement
Not applicable.
Acknowledgments
Karim Khalil was supported by a Ph.D. scholarship funded from the Culture Affairs and Mission Sector, Ministry of Higher Education, Government of Egypt.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Li, Y.; Wang, X.; Lu, L.; Wang, M.; Zhai, Y.; Tai, X.; Dilimulati, D.; Lei, X.; Xu, J.; Zhang, C. Identification of novel GPCR partners of the central melanocortin signaling. Mol. Metab. 2021, 53, 101317. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.-Q.; Hong, Y.; Tao, Y.-X. Melanocortin-5 receptor: Pharmacology and its regulation of energy metabolism. Int. J. Mol. Sci. 2022, 23, 8727. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.W.; Bryson-Richardson, R.J.; Taylor, M.S.; Currie, P.; Jackson, I.J. Sequence characterization of teleost fish melanocortin receptors. Ann. N. Y. Acad. Sci. 2003, 994, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Logan, D.W.; Bryson-Richardson, R.J.; Pagán, K.E.; Taylor, M.S.; Currie, P.D.; Jackson, I.J. The structure and evolution of the melanocortin and MCH receptors in fish and mammals. Genomics 2003, 81, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Ringholm, A.; Fredriksson, R.; Poliakova, N.; Yan, Y.L.; Postlethwait, J.H.; Larhammar, D.; Schiöth, H.B. One melanocortin 4 and two melanocortin 5 receptors from zebrafish show remarkable conservation in structure and pharmacology. J. Neurochem. 2002, 82, 6–18. [Google Scholar] [CrossRef]
- Gantz, I.; Miwa, H.; Konda, Y.; Shimoto, Y.; Tashiro, T.; Watson, S.; DelValle, J.; Yamada, T. Molecular cloning, expression, and gene localization of a fourth melanocortin receptor. J. Biol. Chem. 1993, 268, 15174–15179. [Google Scholar] [CrossRef]
- Mountjoy, K.G.; Mortrud, M.T.; Low, M.J.; Simerly, R.B.; Cone, R.D. Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Mol. Endocrinol. 1994, 8, 1298–1308. [Google Scholar] [CrossRef]
- Schjolden, J.; Schiöth, H.B.; Larhammar, D.; Winberg, S.; Larson, E.T. Melanocortin peptides affect the motivation to feed in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2009, 160, 134–138. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Tsuchiya, K.; Yamanome, T.; Schiöth, H.B.; Kawauchi, H.; Takahashi, A. Food deprivation increases the expression of melanocortin-4 receptor in the liver of barfin flounder, Verasper moseri. Gen. Comp. Endocrinol. 2008, 155, 280–287. [Google Scholar] [CrossRef]
- Song, Y.; Cone, R.D. Creation of a genetic model of obesity in a teleost. FASEB J. 2007, 21, 2042–2049. [Google Scholar] [CrossRef]
- Huszar, D.; Lynch, C.A.; Fairchild-Huntress, V.; Dunmore, J.H.; Fang, Q.; Berkemeier, L.R.; Gu, W.; Kesterson, R.A.; Boston, B.A.; Cone, R.D. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Cerdá-Reverter, J.M.; Ling, M.K.; Schiöth, H.B.; Peter, R.E. Molecular cloning, characterization and brain mapping of the melanocortin 5 receptor in the goldfish. J. Neurochem. 2003, 87, 1354–1367. [Google Scholar] [CrossRef] [PubMed]
- Ringholm, A.; Klovins, J.; Fredriksson, R.; Poliakova, N.; Larson, E.T.; Kukkonen, J.P.; Larhammar, D.; Schiöth, H.B. Presence of melanocortin (MC4) receptor in spiny dogfish suggests an ancient vertebrate origin of central melanocortin system. Eur. J. Biochem. 2003, 270, 213–221. [Google Scholar] [CrossRef]
- Sánchez, E.; Rubio, V.C.; Thompson, D.; Metz, J.; Flik, G.; Millhauser, G.L.; Cerdá-Reverter, J.M. Phosphodiesterase inhibitor-dependent inverse agonism of agouti-related protein on melanocortin 4 receptor in sea bass (Dicentrarchus labrax). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R1293–R1306. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zhang, Y.; Ji, P.; Li, Y.; Xu, P.; Sun, X. Molecular characterization of CART, AgRP, and MC4R genes and their expression with fasting and re-feeding in common carp (Cyprinus carpio). Mol. Biol. Rep. 2012, 39, 2215–2223. [Google Scholar] [CrossRef]
- Li, L.; Yang, Z.; Zhang, Y.-P.; He, S.; Liang, X.-F.; Tao, Y.-X. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in grass carp (Ctenopharyngodon idella). Domest. Anim. Endocrinol. 2017, 59, 140–151. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.; Lei, L.; Deng, X.; Duan, Y.; Fu, S.; Zhang, J.; Yuan, D.; Zhou, C.; He, W. The melanocortin-4 receptor (MC4R) gene in the gibel carp Carassius auratus gibelio: Cloning, tissue distribution, and fasting effects. Aquac. Int. 2022, 30, 2425–2438. [Google Scholar] [CrossRef]
- Wen, Z.-Y.; Liu, T.; Qin, C.-J.; Zou, Y.-C.; Wang, J.; Li, R.; Tao, Y.-X. MRAP2 interaction with melanocortin-4 receptor in snakehead (Channa argus). Biomolecules 2021, 11, 481. [Google Scholar] [CrossRef]
- Zhang, K.-Q.; Hou, Z.-S.; Wen, H.-S.; Li, Y.; Qi, X.; Li, W.-J.; Tao, Y.-X. Melanocortin-4 receptor in spotted sea bass, Lateolabrax maculatus: Cloning, tissue distribution, physiology, and pharmacology. Front. Endocrinol. 2019, 10, 705. [Google Scholar] [CrossRef]
- Haitina, T.; Klovins, J.; Andersson, J.; Fredriksson, R.; Lagerström, M.C.; Larhammar, D.; Larson, E.T.; Schiöth, H.B. Cloning, tissue distribution, pharmacology and three-dimensional modelling of melanocortin receptors 4 and 5 in rainbow trout suggest close evolutionary relationship of these subtypes. Biochem. J. 2004, 380, 475–486. [Google Scholar] [CrossRef]
- Stäubert, C.; Tarnow, P.; Brumm, H.; Pitra, C.; Gudermann, T.; Gruters, A.; Schöneberg, T.; Biebermann, H.; Römpler, H. Evolutionary aspects in evaluating mutations in the melanocortin 4 receptor. Endocrinology 2007, 148, 4642–4648. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Kinoshita, M.; Adolfi, M.C.; Schartl, M. Analysis of the role of the Mc4r system in development, growth, and puberty of medaka. Front. Endocrinol. 2019, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Q.; Shu, H.; Zhou, H.; Li, X.; Hou, L. Characterization of the melanocortin-4 receptor gene from Spinibarbus hollandi and the association between its polymorphisms and S. hollandi growth traits. Fish. Sci. 2017, 83, 967–976. [Google Scholar] [CrossRef]
- Loos, R.J.; Lindgren, C.M.; Li, S.; Wheeler, E.; Zhao, J.H.; Prokopenko, I.; Inouye, M.; Freathy, R.M.; Attwood, A.P.; Beckmann, J.S. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008, 40, 768–775. [Google Scholar] [CrossRef]
- Yeo, G.S.; Farooqi, I.S.; Aminian, S.; Halsall, D.J.; Stanhope, R.G.; O’Rahilly, S. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 1998, 20, 111–112. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.A.; Marks, D.L.; Fan, W.; Kuhn, C.M.; Bartolome, M.; Cone, R.D. Melanocortin-4 receptor is required for acute homeostatic responses to increased dietary fat. Nat. Neurosci. 2001, 4, 605–611. [Google Scholar] [CrossRef]
- Chen, A.S.; Metzger, J.M.; Trumbauer, M.E.; Guan, X.-M.; Yu, H.; Frazier, E.G.; Marsh, D.J.; Forrest, M.J.; Gopal-Truter, S.; Fisher, J. Role of the melanocortin-4 receptor in metabolic rate and food intake in mice. Transgenic Res. 2000, 9, 145–154. [Google Scholar] [CrossRef]
- Garza, J.C.; Kim, C.S.; Liu, J.; Zhang, W.; Lu, X.-Y. Adeno-associated virus-mediated knockdown of melanocortin-4 receptor in the paraventricular nucleus of the hypothalamus promotes high-fat diet-induced hyperphagia and obesity. J. Endocrinol. 2008, 197, 471–482. [Google Scholar] [CrossRef]
- Marie, L.S.; Miura, G.I.; Marsh, D.J.; Yagaloff, K.; Palmiter, R.D. A metabolic defect promotes obesity in mice lacking melanocortin-4 receptors. Proc. Natl. Acad. Sci. USA 2000, 97, 12339–12344. [Google Scholar] [CrossRef]
- Sutton, G.M.; Trevaskis, J.L.; Hulver, M.W.; McMillan, R.P.; Markward, N.J.; Babin, M.J.; Meyer, E.A.; Butler, A.A. Diet-genotype interactions in the development of the obese, insulin-resistant phenotype of C57BL/6J mice lacking melanocortin-3 or-4 receptors. Endocrinology 2006, 147, 2183–2196. [Google Scholar] [CrossRef]
- Weide, K.; Christ, N.; Moar, K.M.; Arens, J.; Hinney, A.; Mercer, J.G.; Eiden, S.; Schmidt, I. Hyperphagia, not hypometabolism, causes early onset obesity in melanocortin-4 receptor knockout mice. Physiol. Genom. 2003, 13, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Coogan, M.; Alston, V.; Su, B.; Khalil, K.; Elaswad, A.; Khan, M.; Johnson, A.; Xing, D.; Li, S.; Wang, J. Improved growth and high inheritance of melanocortin-4 receptor (mc4r) mutation in CRISPR/Cas-9 gene-edited channel catfish, Ictalurus punctatus. Mar. Biotechnol. 2022, 24, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Elaswad, A.; Khalil, K.; Cline, D.; Page-McCaw, P.; Chen, W.; Michel, M.; Cone, R.; Dunham, R. Microinjection of CRISPR/Cas9 protein into channel catfish, Ictalurus punctatus, embryos for gene editing. J. Vis. Exp. 2018, 131, e56275. [Google Scholar] [CrossRef]
- Chakrapani, V.; Patra, S.K.; Panda, R.P.; Rasal, K.D.; Jayasankar, P.; Barman, H.K. Establishing targeted carp TLR22 gene disruption via homologous recombination using CRISPR/Cas9. Dev. Comp. Immunol. 2016, 61, 242–247. [Google Scholar] [CrossRef]
- Edvardsen, R.B.; Leininger, S.; Kleppe, L.; Skaftnesmo, K.O.; Wargelius, A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS ONE 2014, 9, e108622. [Google Scholar] [CrossRef]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci. Rep. 2016, 6, 22953. [Google Scholar] [CrossRef]
- Coogan, M.; Alston, V.; Su, B.; Khalil, K.; Elaswad, A.; Khan, M.; Simora, R.M.; Johnson, A.; Xing, D.; Li, S. CRISPR/Cas-9 induced knockout of myostatin gene improves growth and disease resistance in channel catfish (Ictalurus punctatus). Aquaculture 2022, 557, 738290. [Google Scholar] [CrossRef]
- USDA National Agricultural Statistics Service. Catfish Production. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/cfpd0221.pdf (accessed on 5 February 2023).
- Dunham, R.A.; Elaswad, A. Catfish biology and farming. Annu. Rev. Anim. Biosci. 2018, 6, 305–325. [Google Scholar] [CrossRef]
- Yin, L.; Maddison, L.A.; Li, M.; Kara, N.; LaFave, M.C.; Varshney, G.K.; Burgess, S.M.; Patton, J.G.; Chen, W. Multiplex conditional mutagenesis using transgenic expression of Cas9 and sgRNAs. Genetics 2015, 200, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Jao, L.-E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [PubMed]
- Dunham, R.A.; Elaswad, A.; Qin, Z. Gene Editing in Channel Catfish via Double Electroporation of Zinc-Finger Nucleases. In Zinc Finger Proteins: Methods and Protocols; Liu, J., Ed.; Springer: New York, NY, USA, 2018; pp. 201–214. [Google Scholar]
- Kang, J.-H.; Yoshizaki, G.; Homma, O.; Strüssmann, C.A.; Takashima, F. Effect of an osmotic differential on the efficiency of gene transfer by electroporation of fish spermatozoa. Aquaculture 1999, 173, 297–307. [Google Scholar] [CrossRef]
- Dunham, R.A.; Winn, R.N. Production of Transgenic Fish. In Transgenic Animal Technology: A Laboratory Handbook; Elsevier BV: Amsterdam, The Netherlands, 2014; pp. 308–336. [Google Scholar]
- Powers, D.A.; Hereford, L.; Cole, T.; Chen, T.T.; Lin, C.; Kight, K.; Creech, K.; Dunham, R. Electroporation: A method for transferring genes into the gametes of zebrafish (Brachydanio rerio), channel catfish (Ictalurus punctatus), and common carp (Cyprinus carpio). Mol. Mar. Biol. Biotechnol. 1991, 1, 301–308. [Google Scholar]
- Bart, A.; Dunham, R. Effects of sperm concentration and egg number on fertilization efficiency with channel catfish (Ictalurus punctatus) eggs and blue catfish (I. furcatus) spermatozoa. Theriogenology 1996, 45, 673–682. [Google Scholar] [CrossRef]
- Armstrong, J.; Duhon, S.; Malacinski, G. Raising the Axolotl in Captivity. In Developmental Biology of The Axolotl; Oxford University Press: New York, NY, USA, 1989; pp. 220–227. [Google Scholar]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.-D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.C.; Robinson, E.H. Channel Catfish Farming Handbook; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1990. [Google Scholar]
- Elaswad, A.; Khalil, K.; Ye, Z.; Liu, Z.; Liu, S.; Peatman, E.; Odin, R.; Vo, K.; Drescher, D.; Gosh, K. Effects of CRISPR/Cas9 dosage on TICAM1 and RBL gene mutation rate, embryonic development, hatchability and fry survival in channel catfish. Sci. Rep. 2018, 8, 16499. [Google Scholar] [CrossRef]
- Elaswad, A.; Khalil, K.; Ye, Z.; Alsaqufi, A.; Abdelrahman, H.; Su, B.; Perera, D.A.; Dong, S.; Abass, N.; Dunham, R. Effects of cecropin transgenesis and interspecific hybridization on the resistance to Ichthyophthirius multifiliis in channel catfish and female channel catfish × male blue catfish hybrids. N. Am. J. Aquac. 2019, 81, 242–252. [Google Scholar] [CrossRef]
- Gerard, G.; Shandilya, H.; Qiu, P.; Shi, Y.; Lo, J. Genetic Variance Detection Using Surveyor Nuclease. In Genetic Variance Detection-Technologies for Pharmacogenomics; Hecker, K.H., Ed.; DNA Press: Eagleville, PA, USA, 2006; pp. 95–129. [Google Scholar]
- Qiu, P.; Shandilya, H.; D’Alessio, J.M.; O’Connor, K.; Durocher, J.; Gerard, G.F. Mutation detection using Surveyor™ nuclease. Biotechniques 2004, 36, 702–707. [Google Scholar] [CrossRef]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013, 194, 1029–1035. [Google Scholar] [CrossRef]
- Ansai, S.; Kinoshita, M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 2014, 3, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhang, J.; Wu, H.; Wang, J.; Ma, K.; Li, Z.; Zhang, X.; Zhang, P.; Huang, X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013, 23, 720. [Google Scholar] [CrossRef] [PubMed]
- Hongbing, H.; Yonghe, M.; Tao, W.; Ling, L.; Xiuzhi, T.; Rui, H.; Shoulong, D.; Kongpan, L.; Feng, W.; Ning, L. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agric. Sci. Eng. 2014, 1, 2–5. [Google Scholar] [CrossRef]
- Ikeda, M.; Matsuyama, S.; Akagi, S.; Ohkoshi, K.; Nakamura, S.; Minabe, S.; Kimura, K.; Hosoe, M. Correction of a disease mutation using CRISPR/Cas9-assisted genome editing in Japanese black cattle. Sci. Rep. 2017, 7, 17827. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Qin, G.; Qin, Z.; Lu, C.; Ye, Z.; Elaswad, A.; Bangs, M.; Li, H.; Zhang, Y.; Huang, Y.; Shi, H. Gene editing of the catfish gonadotropin-releasing hormone gene and hormone therapy to control the reproduction in channel catfish, Ictalurus punctatus. Biology 2022, 11, 649. [Google Scholar] [CrossRef]
- Khalil, K.; Elayat, M.; Khalifa, E.; Daghash, S.; Elaswad, A.; Miller, M.; Abdelrahman, H.; Ye, Z.; Odin, R.; Drescher, D. Generation of myostatin gene-edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 system. Sci. Rep. 2017, 7, 7301. [Google Scholar] [CrossRef]
- Wargelius, A.; Leininger, S.; Skaftnesmo, K.O.; Kleppe, L.; Andersson, E.; Taranger, G.L.; Schulz, R.W.; Edvardsen, R.B. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 2016, 6, 21284. [Google Scholar] [CrossRef]
- Westerfield, M.; Zon, L.I. Methods in Cell Biology: Genetics, Genomics, and Informatics. The Zebrafish Genetics, Genomics, and Informatics; Elsevier Academic Press: New York, NY, USA, 2004. [Google Scholar]
- Abraham, V.C.; Gupta, S.; Fluck, R.A. Ooplasmic segregation in the medaka (Oryzias latipes) egg. Biol. Bull. 1993, 184, 115–124. [Google Scholar] [CrossRef]
- Leung, C.F.; Webb, S.E.; Miller, A.L. On the mechanism of ooplasmic segregation in single-cell zebrafish embryos. Dev. Growth Differ. 2000, 42, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, H.; Yamamoto, T. Genome Editing Using Zinc-Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs). In Targeted Genome Editing Using Site-Specific Nucleases; Springer: Tokyo, Japan, 2015; pp. 3–24. [Google Scholar]
- Qin, Z.; Li, Y.; Su, B.; Cheng, Q.; Ye, Z.; Perera, D.A.; Fobes, M.; Shang, M.; Dunham, R.A. Editing of the luteinizing hormone gene to sterilize channel catfish, Ictalurus punctatus, using a modified zinc finger nuclease technology with electroporation. Mar. Biotechnol. 2016, 18, 255–263. [Google Scholar] [CrossRef]
- O’Donnell, S.M.; Janssen, G.R. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of c I mRNA with or without the 5′ untranslated leader. J. Bacteriol. 2001, 183, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Brouwers, B.; de Oliveira, E.M.; Marti-Solano, M.; Monteiro, F.B.; Laurin, S.-A.; Keogh, J.M.; Henning, E.; Bounds, R.; Daly, C.A.; Houston, S. Human MC4R variants affect endocytosis, trafficking and dimerization revealing multiple cellular mechanisms involved in weight regulation. Cell Rep. 2021, 34, 108862. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.J.; Fang, X.T.; Liu, Y.; Zhang, C.L.; Liu, X.X.; Zhao, J.; Li, J.J.; Chen, H. Effects of single and combined genotypes of MC4R and POU1F1 genes on two production traits in Langshan chicken. Mol. Biol. Rep. 2013, 40, 4645–4650. [Google Scholar] [CrossRef]
- Yang, Z.; Liang, X.-F.; Li, G.-L.; Tao, Y.-X. Biased signaling in fish melanocortin-4 receptors (MC4Rs): Divergent pharmacology of four ligands on spotted scat (Scatophagus argus) and grass carp (Ctenopharyngodon idella) MC4Rs. Mol. Cell. Endocrinol. 2020, 515, 110929. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.-L.; Bian, W.-P.; Wang, C.; Junaid, M.; Zou, J.-X.; Pei, D.-S. A novel technique based on in vitro oocyte injection to improve CRISPR/Cas9 gene editing in zebrafish. Sci. Rep. 2016, 6, 34555. [Google Scholar] [CrossRef]
- Li, D.; Qiu, Z.; Shao, Y.; Chen, Y.; Guan, Y.; Liu, M.; Li, Y.; Gao, N.; Wang, L.; Lu, X. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 681–683. [Google Scholar] [CrossRef]
- Fei, F.; Sun, S.-Y.; Yao, Y.-X.; Wang, X. Generation and phenotype analysis of zebrafish mutations of obesity-related genes lepr and mc4r. Acta Physiol. Sin. 2017, 69, 61–69. [Google Scholar]
- Jokubka, R.; Maak, S.; Kerziene, S.; Swalve, H.H. Association of a melanocortin 4 receptor (MC4R) polymorphism with performance traits in Lithuanian White pigs. J. Anim. Breed. Genet. 2006, 123, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Davoli, R.; Braglia, S.; Valastro, V.; Annarratone, C.; Comella, M.; Zambonelli, P.; Nisi, I.; Gallo, M.; Buttazzoni, L.; Russo, V. Analysis of MC4R polymorphism in Italian Large White and Italian Duroc pigs: Association with carcass traits. Meat Sci. 2012, 90, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Hallerman, E.M.; Dunham, R.; Houston, R.D.; Walton, M.; Wargelius, A.; Wray-Cahen, D. Towards production of genome-edited aquaculture species. Rev. Aquac. 2022, 1–5. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).