Transcriptome Analysis of Gill Tissues from Neptunea cumingii in Different Seasons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Methods
2.2.1. RNA Extraction, Library Preparation, and Illumina Sequencing
2.2.2. Data Filtering and De Novo Assembly
2.2.3. DEGs Screening and Enrichment Analysis
2.2.4. Quantitative Real-Time-PCR Validation
3. Results
3.1. Transcriptome Sequencing Analysis
3.2. Transcription Splicing, Comparison, and Annotation
3.3. Statistics for DEGs
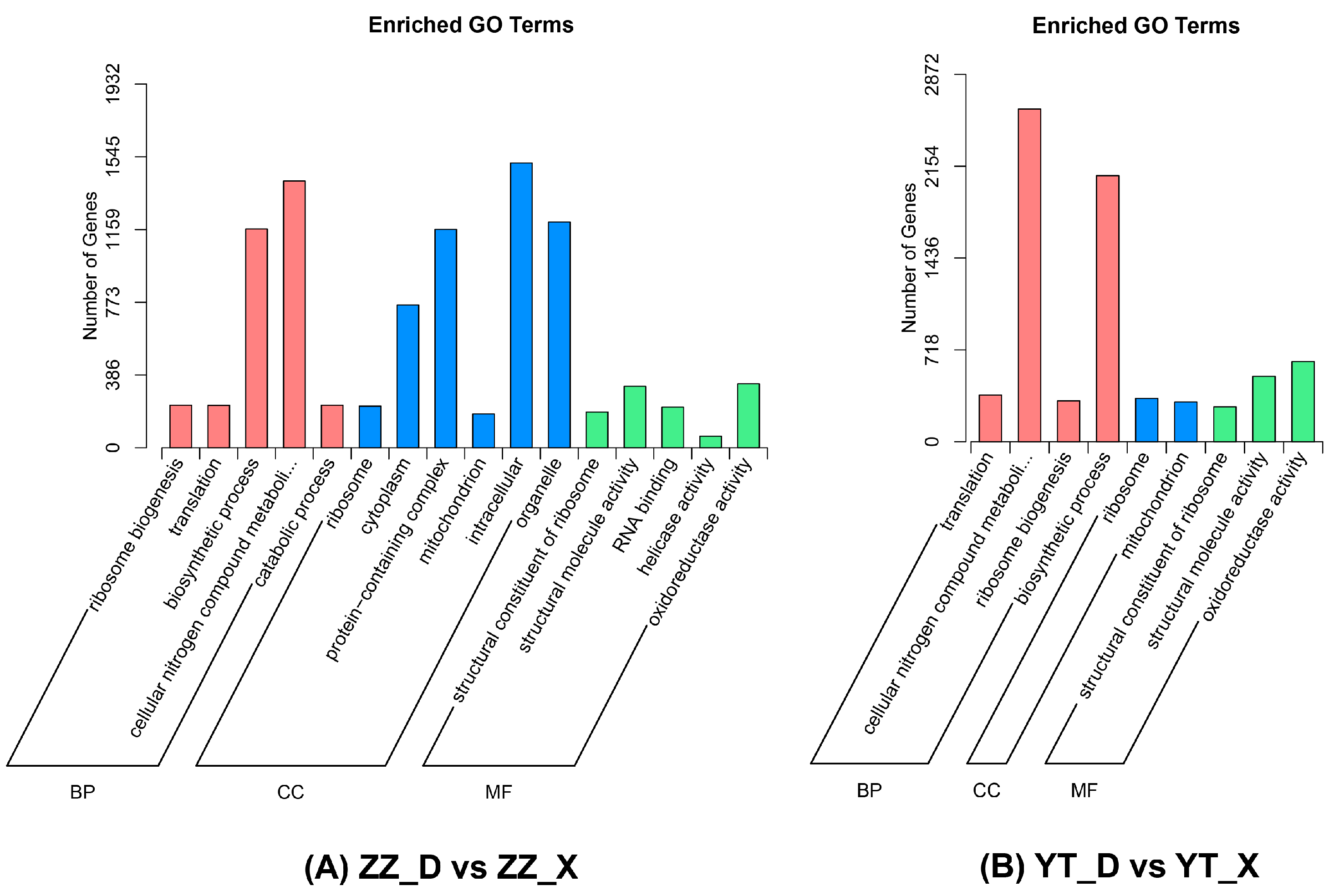
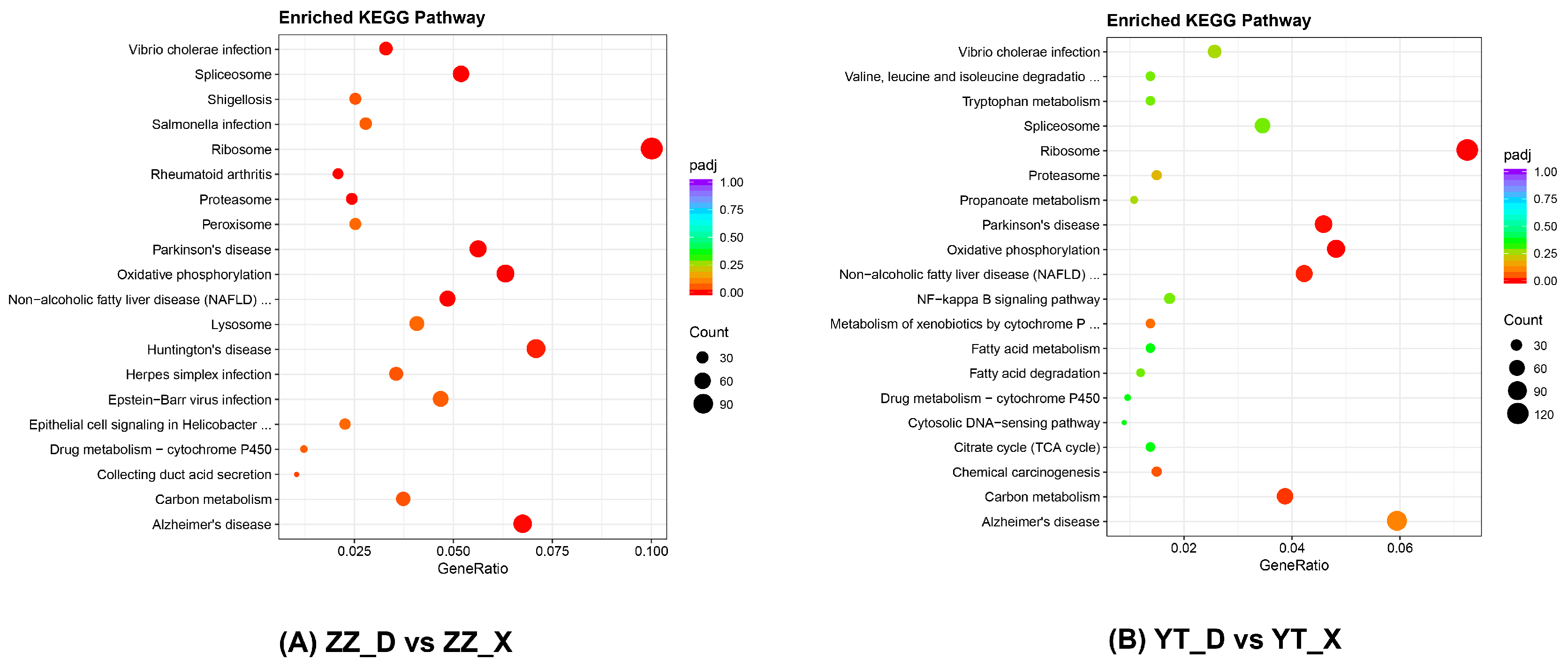
3.4. Enrichment Analysis of DEGs
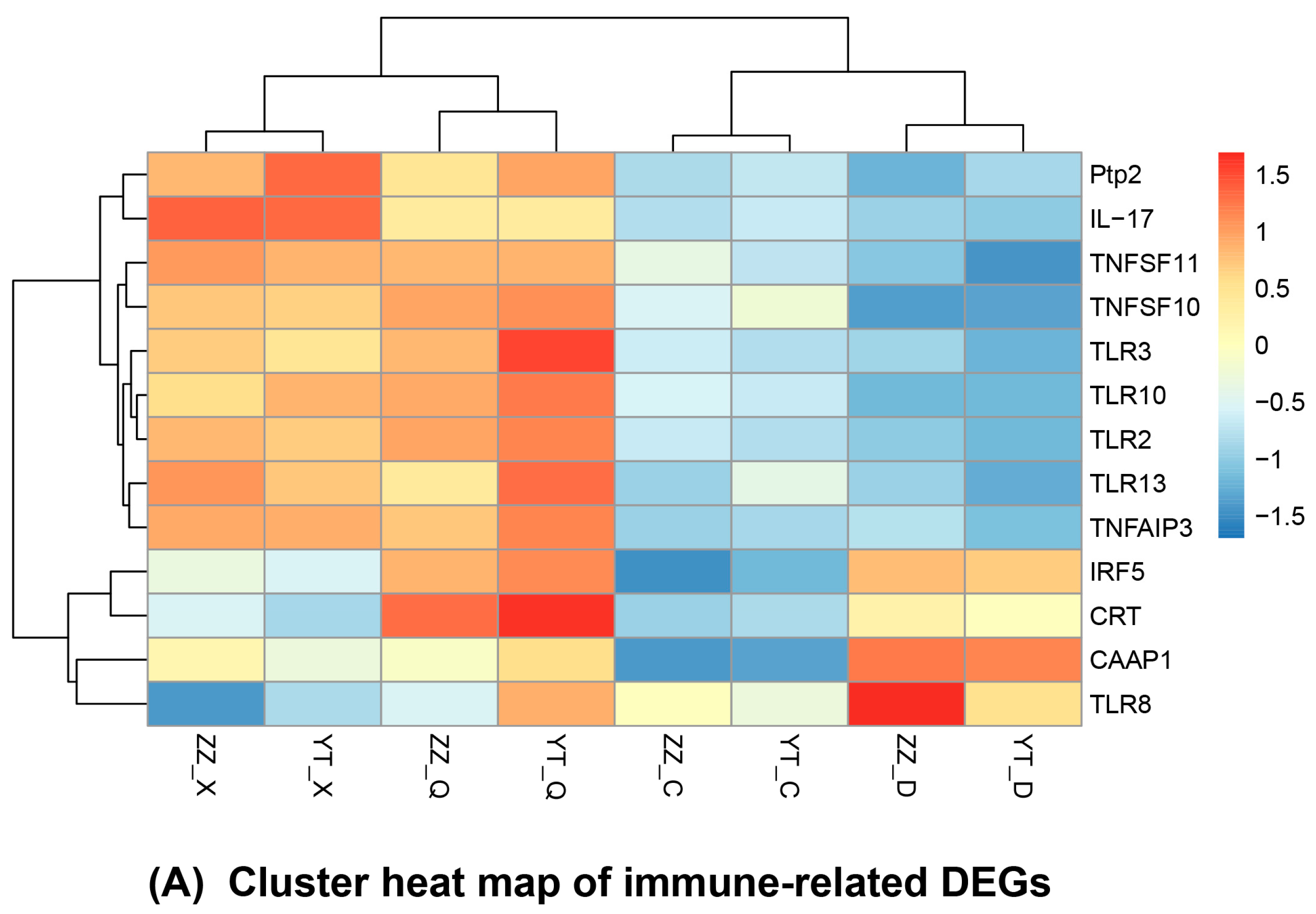
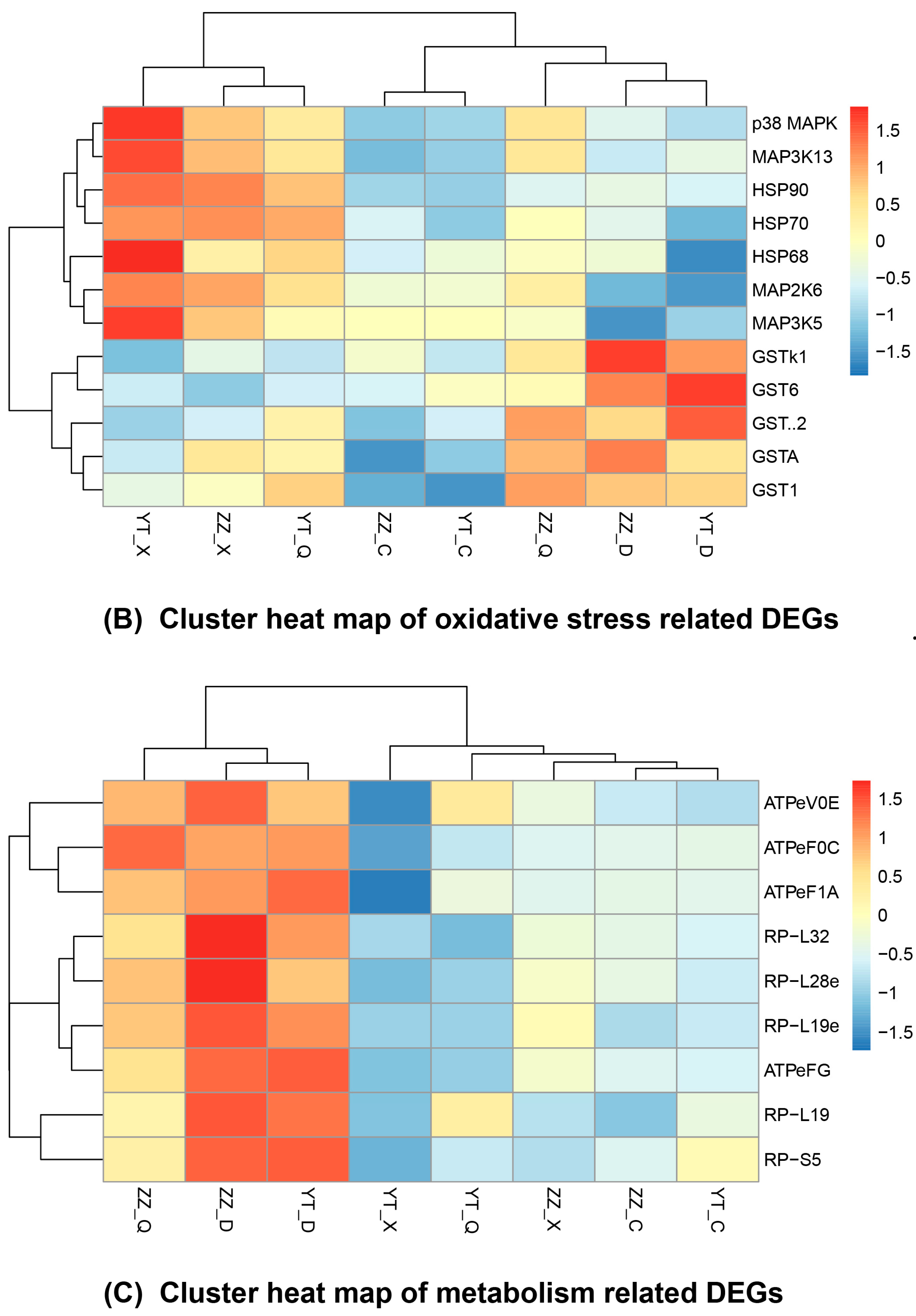
3.5. Seasonal Changes in DEGs
3.6. Validation Snalysis Using qRT-PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qi, Z. Study on the taxonomic flora of benthic invertebrates. Mar. Sci. 1979, 1, 66–69. [Google Scholar]
- Hao, Z.; Wang, Y.; Yu, Y.; Zhan, Y.Y.; Tian, Y.; Wang, L. Analysis and evaluation of nutritive composition in the muscle of Neptunea arthritica cumingii Crosse (Gastropoda: Buccinidae). J. Dalian Univ. 2016, 37, 66–70. [Google Scholar]
- Guo, D.; Liu, X.Z.; Wang, A.Y.; Wang, B.; Li, Y.P.; Dong, J. Stock distribution of whelk Neptunea cumingii Crosse in Liaodong Bay. Fish. Sci. 2015, 34, 718–721. [Google Scholar]
- Ge, X.; Zhao, J.; Liang, Z.; Chi, Q.; Mao, J.; Wang, X.; Chang, Y.; Hao, Z. Comparative analysis of Neptunea cumingii growth, related digestive and immune enzyme indicators, and liver transcriptome under different feeding conditions. Front. Mar. Sci. 2022, 9, 1013180. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Li, J.; Wu, W.; Hao, Z.L. Research progress on the reproductive biology and artificial breeding technology of the Neptunea cumingii. Hebei Fish. 2019, 4, 54–56. [Google Scholar]
- Tan, B.; Zhang, D.; Tian, Y.; Mao, J.; Wang, X.; Wang, L.; Chang, Y.; Hao, Z. Genetic structure and local adaptation of Neptunea cumingii crosse populations in China based on GBS technology. Front. Ecol. Evol. 2023, 11, 1154781. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Wang, S.J.; Tian, Y.; Wang, L.; Mao, J.X.; Wang, X.B.; Wang, Q.; Chang, Y.; Hao, Z. Effects of dissolved oxygen concentration on behavior, antioxidant enzyme activities and tissue structure of whelk Neptunea cumingii Crosse. J. Dalian Ocean Univ. 2022, 37, 643–649. [Google Scholar]
- Zhang, S.Y.; Wang, S.J.; Yang, J.C.; Zhu, J.Y.; Tian, Y.; Wang, L.; Mao, J.; Wang, X.; Chang, Y.; Lu, Z.; et al. Effects of salinity on behavior, activities of antioxidant enzymes and tissue structure of gill and kidney of whelk Neptunea cumingii Crosse. Mar. Sci. 2022, 46, 129–139. [Google Scholar]
- Zhang, D.; Dong, X.; Zhu, J.; Yang, J.; Tian, Y.; Wang, L.; Mao, J.; Wang, X.; Chang, Y.; Hao, Z. Effect of water temperature on the behavior of Neptunea cumingii and the histology, immune enzyme activity, and transcriptome of its gills and kidneys. Invertebr. Surviv. J. 2022, 1–12. [Google Scholar] [CrossRef]
- Jing, H.; Zhou, L.; Gao, Y.; Liu, Z.; Wu, B.; Sun, X.; Tu, K. Transcriptomics and metabolomics reveal the molecular and metabolic adaptation to heat stress in Manila clam Ruditapes philippinarum. Front. Mar. Sci. 2023, 10, 1204598. [Google Scholar] [CrossRef]
- Tan, K.; Zhang, B.; Ma, H.; Li, S.; Zheng, H. Oxidative stress responses of golden and brown noble scallops Chlamys nobilis to acute cold stress. Fish Shellfish Immunol. 2019, 95, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Lin, F.; Fang, J.; Gao, Y.; Du, M.; Fang, J.; Li, W.; Jiang, Z. Transcriptome analysis of the Yesso scallop, Patinopecten yessoensis gills in response to water temperature fluctuations. Fish Shellfish Immunol. 2018, 80, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Huang, X.; Sun, H.; Jin, X.; Guan, W.; Xie, J.; Wang, Y.; Wang, X.; Yin, D.; Hao, Z.; et al. Transcriptome analysis provides insight into adaptive mechanisms of scallops under environmental stress. Front. Mar. Sci. 2022, 9, 971796. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Zhu, B.Q.; Zhang, Y.B.; Wang, H.Y.; Li, C.Y.; Su, Y.H.; Ba, C.F. The research of applying primer premier 5.0 to design PCR primer. J. Jinzhou Med. Univ. 2004, 25, 43–46. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 254, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Sun, Y.; Wang, J.; Xing, Q.; Zou, J.; Li, R.; Wang, Z.; Wang, S.; Hu, X.; Zhang, L.; et al. Sequencing-based gene network analysis provides a core set of gene resource for understanding thermal adaptation in Zhikong scallop Chlamys farreri. Mol. Ecol. Resour. 2014, 14, 184–198. [Google Scholar] [CrossRef]
- Vosloo, D.; Vosloo, A. Response of cold-acclimated, farmed South African abalone (Haliotis midae) to short-term and long-term changes in temperature. J. Therm. Biol. 2010, 35, 317–323. [Google Scholar] [CrossRef]
- Austbø, L.; Aas, I.B.; König, M.; Weli, S.C.; Syed, M.; Falk, K.; Koppang, E.O. Transcriptional response of immune genes in gills and the interbranchial lymphoid tissue of Atlantic salmon challenged with infectious salmon anaemia virus. Dev. Comp. Immunol. 2014, 45, 107–114. [Google Scholar] [CrossRef]
- Havas, M.; Rosseland, B.O. Response of zooplankton, benthos, and fish to acidification: An overview. Water Air Soil Pollut. 1995, 85, 51–62. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Arancibia, S.A.; Beltrán, C.J.; Aguirre, I.M.; Silva, P.; Peralta, A.L.; Malinarich, F.; Hermoso, M.A. Toll-like receptors are key participants in innate immune responses. Biol. Res. 2007, 40, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.W.; Luo, X.C.; Dan, X.M.; Huang, X.Z.; Qiao, W.; Zhong, Z.P.; Li, A.X. Orange-spotted grouper (Epinephelus coioides) TLR2, MyD88 and IL-1β involved in anti-Cryptocaryon irritans response. Fish Shellfish Immunol. 2011, 30, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Li, W.; Wang, X.; Wu, Y.; Liu, F. A house fly TNF ortholog Eiger regulates immune defense via cooperating with Toll and Imd pathways. Dev. Comp. Immunol. 2019, 90, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Tanji, T.; Ip, Y.T. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005, 26, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Meffert, M.K. Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death. Differ. 2006, 13, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Song, L. Research progress of inflammation and its pathogenesis in shellfish. J. Dalian Univ. 2023, 38, 369–379. [Google Scholar]
- Xin, L.; Liu, C.; Zhang, H.; Qiu, L.; Wang, L.; Song, L. The characterization of an interleukin-12 p35 homolog involved in the immune modulation of oyster Crassostrea gigas. Dev. Comp. Immunol. 2021, 123, 104145. [Google Scholar] [CrossRef]
- Huang, X.D.; Zhang, H.; He, M.X. Comparative and evolutionary analysis of the interleukin 17 gene family in invertebrates. PLoS ONE 2015, 10, e0132802. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Zhang, Y.; Xiang, Z.; Tong, Y.; Qu, F.; Yu, Z. Genomic characterization and expression analysis of five novel IL-17 genes in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2014, 40, 455–465. [Google Scholar] [CrossRef]
- Saco, A.; Rey-Campos, M.; Rosani, U.; Novoa, B.; Figueras, A. The evolution and diversity of interleukin-17 highlight an expansion in marine invertebrates and its conserved role in mucosal immunity. Front. Immunol. 2021, 12, 692997. [Google Scholar] [CrossRef]
- Wu, S.Z.; Huang, X.D.; Li, Q.; He, M.X. Interleukin-17 in pearl oyster (Pinctada fucata): Molecular cloning and functional characterization. Fish Shellfish Immunol. 2013, 34, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Wang, W.; Fan, S.; Li, J.; Li, Q.; Wu, S.; Wang, L.; Song, L. The receptor CgIL-17R1 expressed in granulocytes mediates the CgIL-17 induced haemocytes proliferation in Crassostrea gigas. Dev. Comp. Immunol. 2022, 131, 104376. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, H.; Li, H.; Wang, A.; Yu, H.Y. Effect of high temperature on immune response of grass carp (Ctenopharyngodon idellus) by transcriptome analysis. Fish Shellfish Immunol. 2016, 58, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, W.; Jiang, W.; Ding, Z. Effects of temperature on immune system of shellfish and its mechanisms. Fish. Sci. 2012, 31, 176–180. [Google Scholar]
- Li, H.; Zhong, R.; Fang, Y.; Yang, L. Research progress of oxidative stress and immunity in animals. Chin. J. Anim. Nutr. 2014, 26, 3217–3221. [Google Scholar]
- Mortensen, S.; Strand, Å.; Bodvin, T.; Alfjorden, A.; Skaar, C.K.; Jelmert, A.; Aspan, A.; Saelemyr, L.; Naustvoll, L.-J.; Albretsen, J. Summer mortalities and detection of ostreid herpesvirus microvariant in Pacific oyster Crassostrea gigas in Sweden and Norway. Dis. Aquat. Organ. 2016, 117, 171–176. [Google Scholar] [CrossRef]
- Lykkesfeldt, J.; Svendsen, O. Oxidants and antioxidants in disease: Oxidative stress in farm animals. Vet. J. 2007, 173, 502–511. [Google Scholar] [CrossRef]
- Jiang, W.; Fang, J.; Li, J.; Jiang, Z.J.; Mao, Y.Z.; Du, M.R.; Gao, Z.K.; Chen, Q.L. Effects of temperature stress on physiological and biochemical activities of Haliotis discus hannai Ino. J. Fish. Sci. China 2017, 24, 220–230. [Google Scholar]
- Hao, S.; Zhang, M. Effects of different cold stress on antioxidant enzyme activity and lipid peroxidation of Sinonovacula constrictus. J. Dalian Univ. 2020, 35, 584–590. [Google Scholar]
- Wang, X.; Cao, S.; Liu, G.; Wang, Y.; Zou, J. Effects of temperature on water-free preservation and antioxidant enzyme activity of Crassadoma gigantea. Mar. Sci. Bull. 2018, 20, 1–20. [Google Scholar]
- Xue, S.; Wang, J.; Li, J.; Ding, J.; Li, Y.; Xu, H.; Mao, Y.; Fang, J. Effects of temperature on energy metabolism and antioxidant enzyme activities of Scapharca broughtonii. J. Fish. China 2019, 43, 573–583. [Google Scholar]
- Kim, M.; Ahn, I.Y.; Cheon, J.; Park, H. Molecular cloning and thermal stress-induced expression of a pi-class glutathione S-transferase (GST) in the Antarctic bivalve Laternula elliptica. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 152, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xiao, Q.; Hao, Q.; Qian, Z.; Li, X.; Li, P.; Li, H.; Chen, L. Genome-wide identification and functional analysis of the glutathione S-transferase (GST) family in Pomacea canaliculata. Int. J. Biol. Macromol. 2021, 193, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ren, R.M.; Yao, C.L. Oxidative stress responses of Mytilus galloprovincialis to acute cold and heat during air exposure. J. Molluscan. Stud. 2018, 84, 285–292. [Google Scholar] [CrossRef]
- Malek, R.L.; Sajadi, H.; Abraham, J.; Grundy, M.A.; Gerhard, G.S. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004, 138, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Canesi, L.; Betti, M.; Ciacci, C.; Lorusso, L.C.; Pruzzo, C.; Gallo, G. Cell signalling in the immune response of mussel hemocytes. Invert. Surviv. J. 2006, 3, 40–49. [Google Scholar]
- Ren, J.; Long, Y.; Liu, R.; Song, G.; Li, Q.; Cui, Z. Characterization of biological pathways regulating acute cold resistance of zebrafish. Int. J. Mol. Sci. 2021, 22, 3028. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.; Wang, Y.; Lv, M.; Li, Y.; Niu, D. Expression of p38MAPK and its regulation of apoptosis under high temperature stress in the razor clam Sinonovacula constricta. Fish Shellfish Immunol. 2022, 122, 288–297. [Google Scholar] [CrossRef]
- Sun, J.; Li, Y.; Li, M.; Liu, Y.; Qu, C.; Wang, L.; Song, L. A novel JNK is involved in immune response by regulating IL expression in oyster Crassostrea gigas. Fish Shellfish Immunol. 2018, 79, 93–101. [Google Scholar] [CrossRef]
- Sun, J.; Wang, L.; Wu, Z.; Han, S.; Wang, L.; Li, M.; Liu, Z.; Song, L. P38 is involved in immune response by regulating inflammatory cytokine expressions in the Pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 2019, 91, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Agius, C.; Saliba, C.; Bossier, P.; Sung, Y.Y. Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: A review. J. Fish. Dis. 2010, 33, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- van Noort, J.M.; Bsibsi, M.; Nacken, P.; Gerritsen, W.H.; Amor, S. The link between small heat shock proteins and the immune system. Int. J. Biochem. Cell. Biol. 2012, 44, 1670–1679. [Google Scholar] [CrossRef] [PubMed]
- Frydman, J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu. Rev. Biochem. 2001, 70, 603–647. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Lu, Z.; Liu, L.; Li, R.; Liang, Z.; Shen, M.; Xu, H.; Ren, D.; Ji, M.; Yuan, S.; et al. NOD-like receptor signaling in inflammation-associated cancers: From functions to targeted therapies. Phytomedicine 2019, 64, 152925. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Feng, J.; Song, H.; Zhou, C.; Yu, Z.L.; Yang, M.J.; Shi, P.; Guo, Y.-J.; Li, Y.-R.; Zhang, T. Mechanisms of heat and hypoxia defense in hard clam: Insights from transcriptome analysis. Aquaculture 2022, 549, 737792. [Google Scholar] [CrossRef]
- Menike, U.; Lee, Y.; Oh, C.; Wickramaarachchi, W.D.N.; Premachandra, H.K.A.; Park, S.C.; Lee, J.; De Zoysa, M. Oligo-microarray analysis and identification of stress-immune response genes from manila clam (Ruditapes philippinarum) exposure to heat and cold stresses. Mol. Biol. Rep. 2014, 41, 6457–6473. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, L.; Li, L.; Que, H.; Zhang, G. Expression characterization of stress genes under high and low temperature stresses in the Pacific oyster, Crassostrea gigas. Mar. Biotechnol. 2016, 18, 176–188. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhu, S.G.; Xu, C.F. Biochemistry, 3rd ed.; Higher Education Press: Beijing, China, 2002; pp. 114–146. [Google Scholar]
- Wang, Z.; Ren, X.; Gao, B.; Liu, P.; Li, J.; Wang, L. Changes in oxidative phosphorylation metabolism of Portunus trituberculatus in family inbreeding. J. Fish. Sci. China 2018, 25, 520–535. [Google Scholar]
- Tomanek, L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteomics. 2014, 105, 92–106. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.N.; Liu, Y.; Cai, D.X.; Li, J.Z.; Wang, A.L. Two types of ATPases from the Pacific white shrimp, Litopenaeus vannamei in response to environmental stress. Mol. Biol. Rep. 2012, 39, 6427–6438. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Zheng, Z.; Deng, Y.; Jiao, Y.; Du, X. Adaptive response of pearl oyster Pinctada fucata martensii to low water temperature stress. Fish Shellfish Immunol. 2018, 78, 310–315. [Google Scholar] [CrossRef]
- Johnston, I.A.; Frearson, N.; Goldspink, G. The effects of environmental temperature on the properties of myofibrillar adenosine triphosphatase from various species of fish. Biochem J. 1973, 133, 735–738. [Google Scholar] [CrossRef]
- Buckley, B.A.; Gracey, A.Y.; Somero, G.N. The cellular response to heat stress in the goby Gillichthys mirabilis: A cDNA microarray and protein-level analysis. J. Exp. Biol. 2006, 209, 2660–2677. [Google Scholar] [CrossRef]
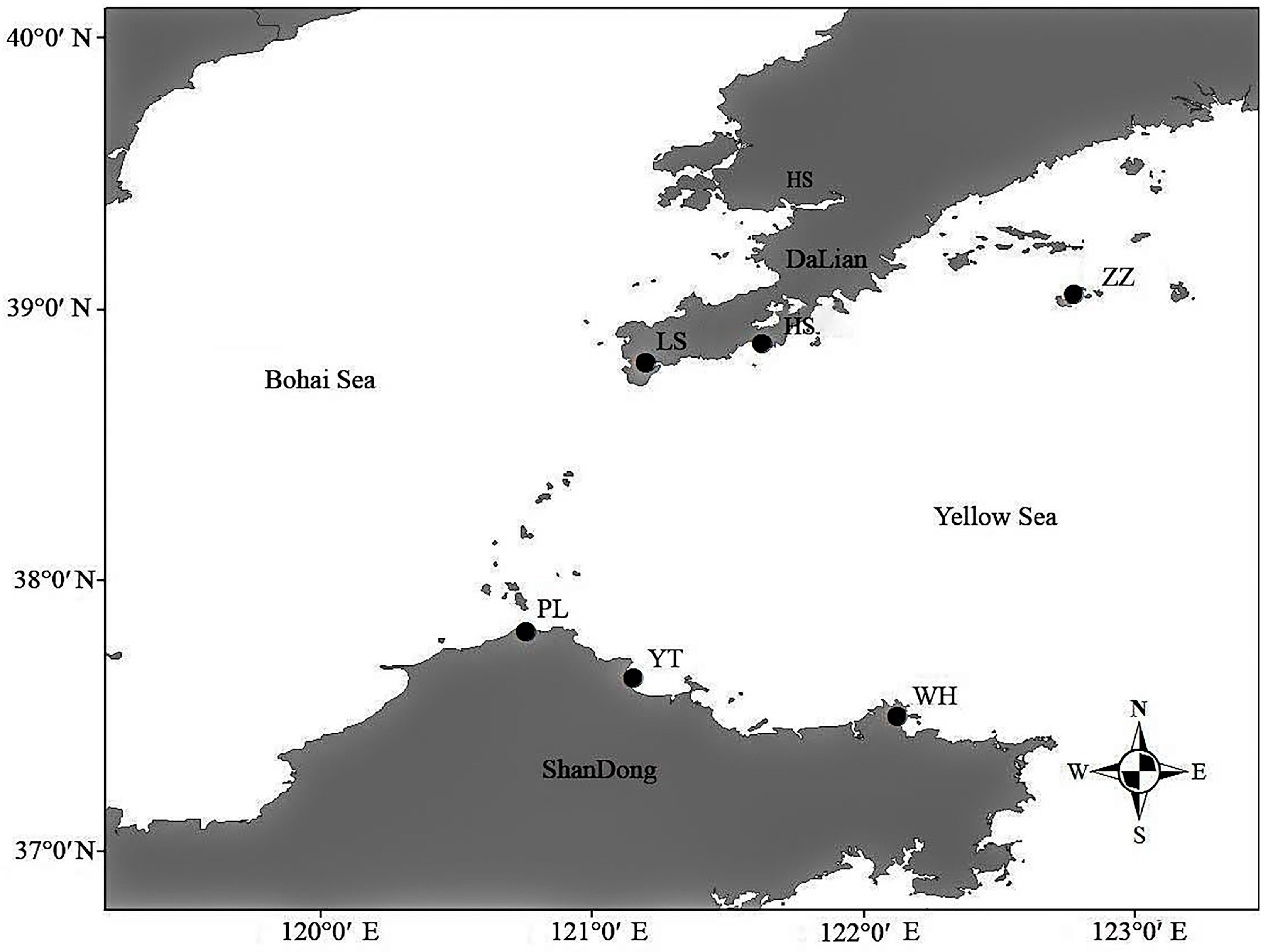

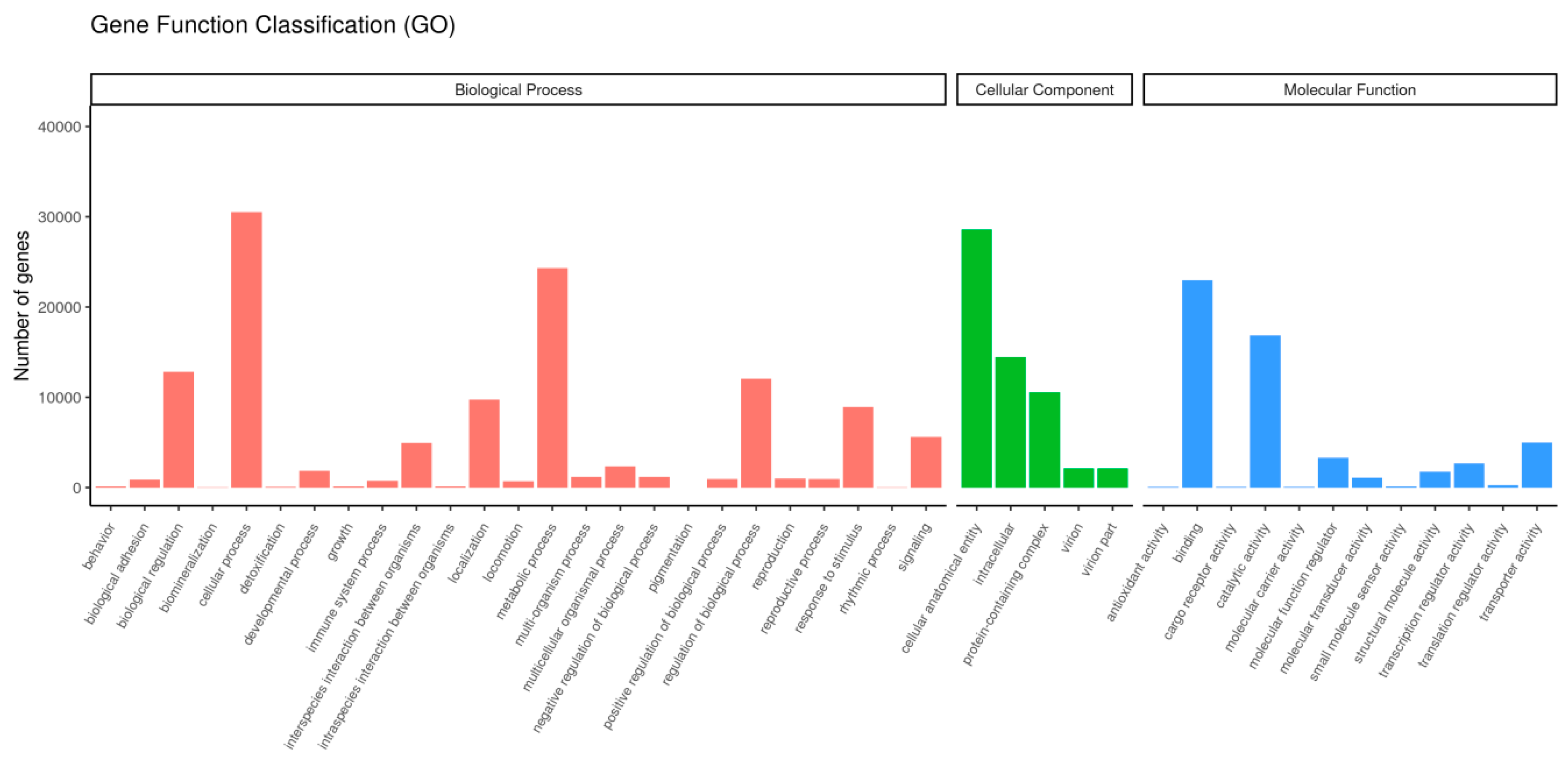
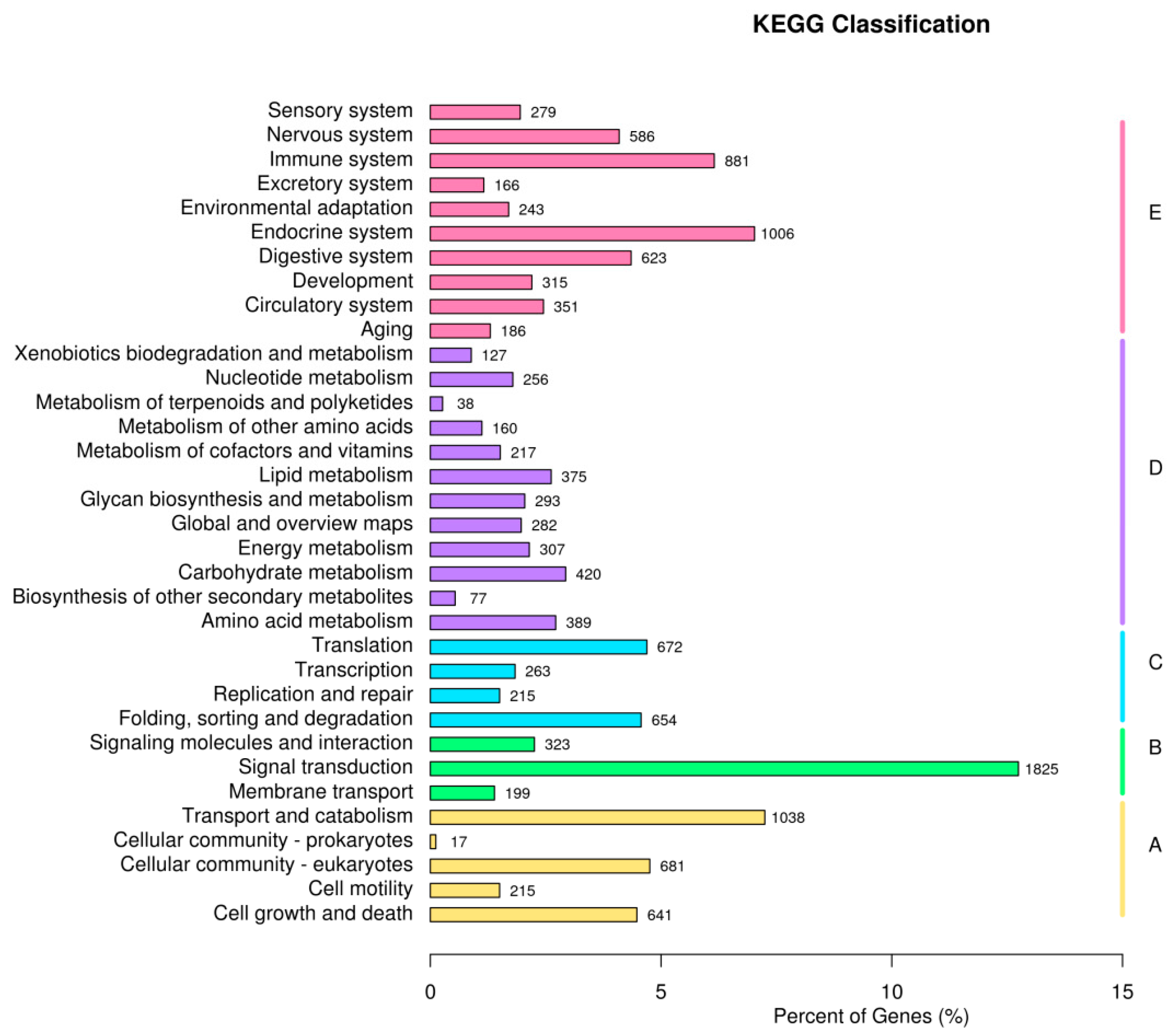
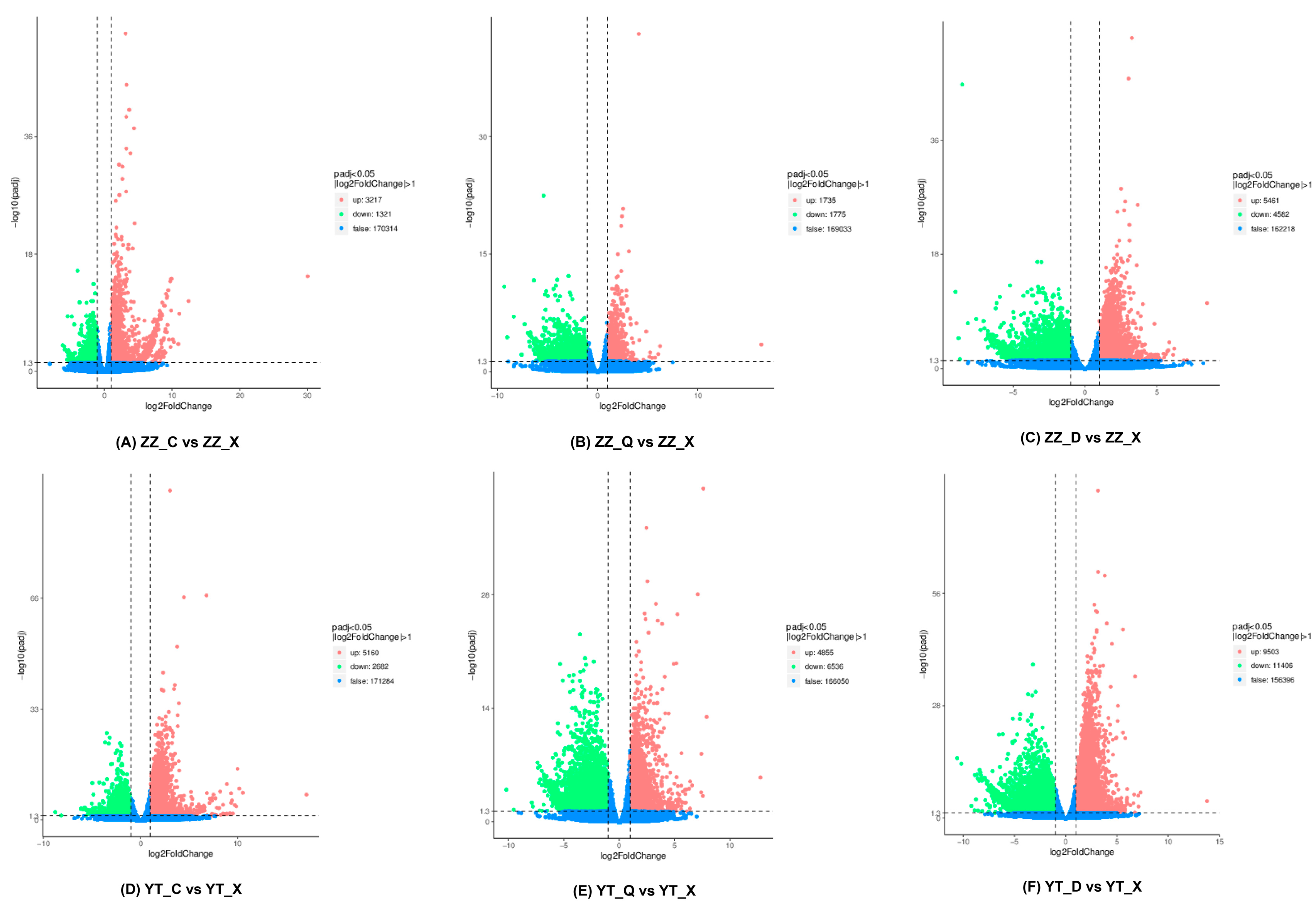

| Locations | Seasons | Number | Date | Temperature |
|---|---|---|---|---|
| Zhangzidao | Spring | 20 | 28 March 2021 | 7 °C |
| Summer | 20 | 25 June 2021 | 21 °C | |
| Autumn | 20 | 23 September 2021 | 18 °C | |
| Winter | 20 | 7 December 2021 | 5 °C | |
| Yantai | Spring | 20 | 28 March 2021 | 8 °C |
| Summer | 20 | 25 June 2021 | 23 °C | |
| Autumn | 20 | 23 September 2021 | 19 °C | |
| Winter | 20 | 7 December 2021 | 6 °C |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| 18S-F | TCTTGATTCGGTGGGTGGTG | CCCGGACATCTAAGGGCATC |
| CAAP1-F | GTATTCGCTGATCCCCAGCA | GTAACTCACGCCGCAGAAAC |
| TLR8-F | CTCATCCGCAGGGAGTTGTT | GACCCGCACACAGTTCTACA |
| TLR13-F | GACCACGGAGGCACACTAAA | CCGGAACCACACAGACAAGA |
| TLR3-F | CAACCTGACCTCGCTGTCTT | TCTTTCAGCCGGTTCAGTCC |
| HSP70-F | AGGAGGGTTCAAGAGGGTGT | AATCTCTGCGGTGTGCTTCA |
| HSP90-F | CCCACCCCGAACTTATTGCT | TGTTCCAGGTTTCCACGCTT |
| CRT-F | GAAAGCCGCGCTGACTAAAG | AAAGCCTCTCTTGTTGCGGA |
| Ptp2-F | GTGCAAAGTTGTGCAGGAGG | AAGATGAGGACGAGGACGGA |
| GSTK1-F | TTGCACACGTCAACGAACAC | ATGGCTGTTCAGGATTGGGG |
| IRF5-F | TTTCTCCGCATCAAGCCACT | GCCATTGCACCAAGGACAAG |
| MAP2K6-F | AGCAGGGAAAAGGCAGACAA | AGCTGCCGTGTTATGTGTGA |
| Sample | Raw_Bases | Clean_Bases | Q20 (%) | Q30 (%) | GC-Content (%) |
|---|---|---|---|---|---|
| ZZ_C_1 | 7.1G | 6.83G | 97.23 | 92.99 | 39.87 |
| ZZ_C_2 | 6.88G | 6.65G | 97.46 | 93.14 | 40.33 |
| ZZ_C_3 | 6.6G | 6.4G | 97.2 | 92.63 | 40.93 |
| ZZ_C_4 | 6.41G | 6.23G | 97.55 | 93.41 | 37.47 |
| YT_C_1 | 6.79G | 6.49G | 96.86 | 92.54 | 43.71 |
| YT_C_2 | 6.96G | 6.58G | 96.51 | 91.48 | 41.71 |
| YT_C_3 | 7.2G | 6.91G | 97.08 | 92.88 | 41.59 |
| YT_C_4 | 6.64G | 6.45G | 97.5 | 93.08 | 38.17 |
| ZZ_X_1 | 7.08G | 6.61G | 96.67 | 92.23 | 44.03 |
| ZZ_X_2 | 7.16G | 6.67G | 96.46 | 91.79 | 43.81 |
| ZZ_X_3 | 6.3G | 5.78G | 96.72 | 92.51 | 44.97 |
| ZZ_X_4 | 6.29G | 5.75G | 96.83 | 92.61 | 45.42 |
| YT_X_1 | 6.88G | 6.32G | 96.37 | 91.7 | 44.52 |
| YT_X_2 | 7.01G | 6.54G | 96.86 | 92.58 | 44.46 |
| YT_X_3 | 6.37G | 5.89G | 96.64 | 92.15 | 44.73 |
| YT_X_4 | 6.96G | 6.29G | 96.61 | 92.11 | 45.11 |
| ZZ_Q_1 | 6.95G | 6.55G | 96.91 | 92.61 | 44.64 |
| ZZ_Q_2 | 6.99G | 6.6G | 96.98 | 92.59 | 45.15 |
| ZZ_Q_3 | 6.83G | 6.42G | 97.02 | 92.68 | 44.86 |
| ZZ_Q_4 | 6.92G | 6.44G | 96.75 | 92.29 | 45.8 |
| YT_Q_1 | 6.83G | 6.35G | 96.69 | 92.31 | 45.61 |
| YT_Q_2 | 6.95G | 6.41G | 96.59 | 92.09 | 45.95 |
| YT_Q_3 | 7.02G | 6.56G | 96.77 | 92.37 | 44.19 |
| YT_Q_4 | 6.73G | 6.15G | 96.51 | 91.96 | 45.43 |
| ZZ_D_1 | 7.13G | 6.74G | 96.04 | 90.74 | 44.98 |
| ZZ_D_2 | 6.79G | 6.39G | 95.64 | 90.12 | 44.21 |
| ZZ_D_3 | 6.97G | 6.55G | 95.94 | 90.6 | 44.69 |
| ZZ_D_4 | 6.81G | 6.36G | 95.92 | 90.52 | 44.42 |
| YT_D_1 | 6.47G | 6.08G | 95.64 | 90.11 | 44.56 |
| YT_D_2 | 7.43G | 6.95G | 95.99 | 90.51 | 44.89 |
| YT_D_3 | 7.1G | 6.67G | 96.15 | 90.97 | 44.29 |
| YT_D_4 | 7.16G | 6.67G | 96.01 | 90.62 | 42.31 |
| Total | 300–500 bp | 500–1k bp | 1k–2k bp | >2k bp | Total Nucleotides | Max Length | Min Length | N50 | N90 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Transcript | 398,505 | 156,348 | 130,839 | 71,522 | 39,796 | 383,685,494 | 30,252 | 301 | 1335 | 420 |
| Unigene | 187,640 | 80,055 | 62,261 | 30,404 | 14,920 | 165,201,498 | 30,252 | 301 | 1147 | 399 |
| Number of Unigenes | Percentage (%) | |
|---|---|---|
| NR | 38,761 | 20.65 |
| NT | 14,670 | 7.81 |
| KO | 12,238 | 6.52 |
| SwissProt | 19,358 | 10.31 |
| PFAM | 51,868 | 27.64 |
| GO | 51,864 | 27.64 |
| KOG | 9929 | 5.29 |
| Annotated in all databases | 3842 | 2.04 |
| Annotated in at least one database | 73,085 | 38.94 |
| Total unigenes | 187,640 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, D.; Tian, Y.; Mao, J.; Liu, Y.; Hao, Z. Transcriptome Analysis of Gill Tissues from Neptunea cumingii in Different Seasons. Fishes 2023, 8, 549. https://doi.org/10.3390/fishes8110549
Zhang Y, Zhang D, Tian Y, Mao J, Liu Y, Hao Z. Transcriptome Analysis of Gill Tissues from Neptunea cumingii in Different Seasons. Fishes. 2023; 8(11):549. https://doi.org/10.3390/fishes8110549
Chicago/Turabian StyleZhang, Yifan, Dandan Zhang, Ying Tian, Junxia Mao, Yang Liu, and Zhenlin Hao. 2023. "Transcriptome Analysis of Gill Tissues from Neptunea cumingii in Different Seasons" Fishes 8, no. 11: 549. https://doi.org/10.3390/fishes8110549
APA StyleZhang, Y., Zhang, D., Tian, Y., Mao, J., Liu, Y., & Hao, Z. (2023). Transcriptome Analysis of Gill Tissues from Neptunea cumingii in Different Seasons. Fishes, 8(11), 549. https://doi.org/10.3390/fishes8110549






