The bZIP Transcription Factor Family Orchestrates the Molecular Response to Nitrite Stress in the Largemouth Bass Spleen
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experimentation
2.2. H&E Staining Procedure
2.3. RNA Extraction and RNA-seq
2.4. Acquisition of Multi-Species Transcriptome Data for Comparison
2.5. Mapping Reads to the Reference Genome
2.6. Gene Expression Quantification
2.7. Differential Expression Analysis
2.8. Functional Enrichment Analysis
2.9. Single-Sample Gene Set Enrichment Analysis (ssGSEA)
2.10. Protein-Protein Interaction Analysis
2.11. Transcription Factor Family Screening
2.12. Identification of msabZIP Family Genes
2.13. Phylogenetic Analysis and Classification of msabZIP Genes
2.14. Predicting Cis-Regulatory Elements and Building Regulatory Networks
3. Results
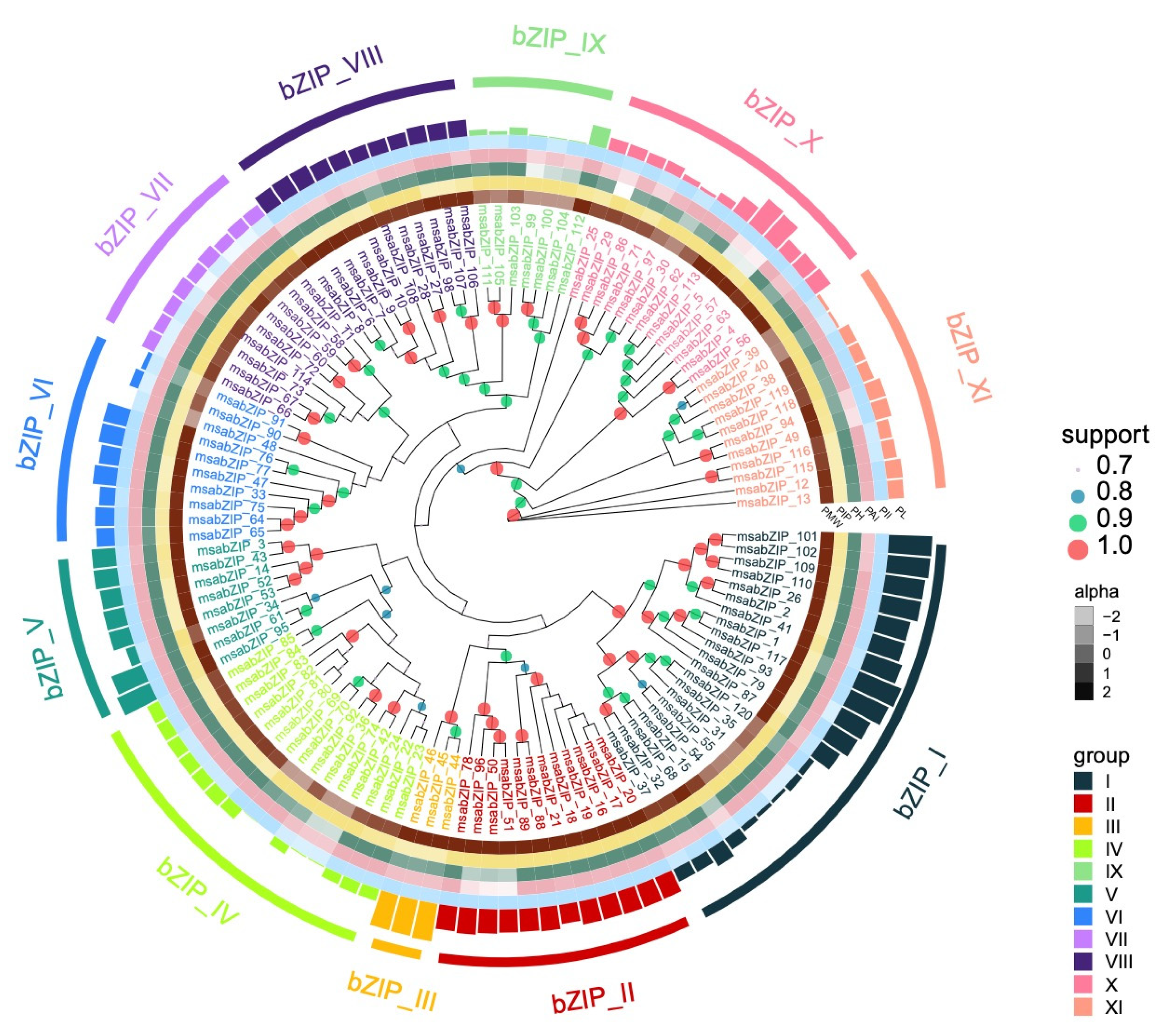
3.1. Identification and Analysis of the LMB bZIP Gene Family
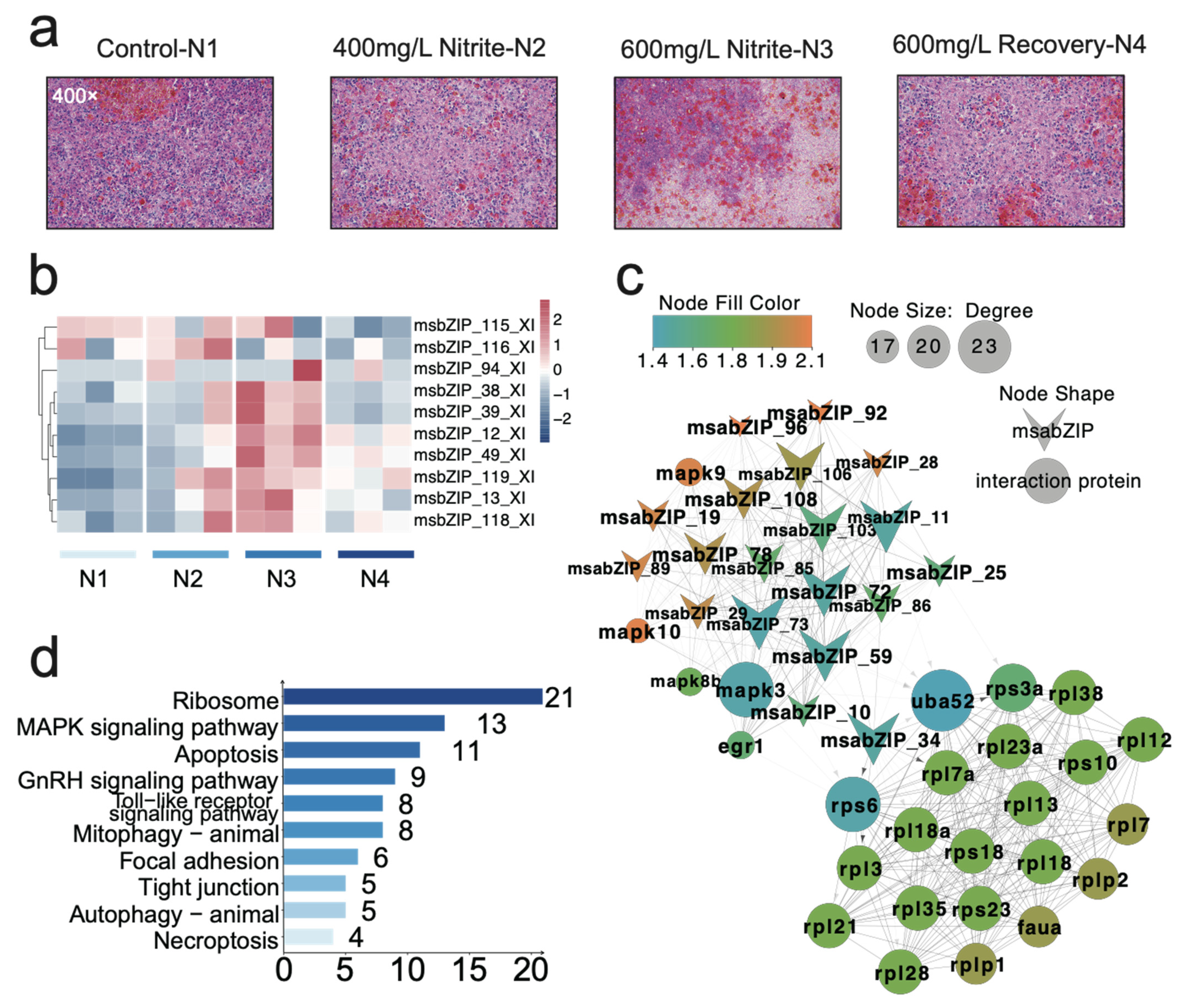
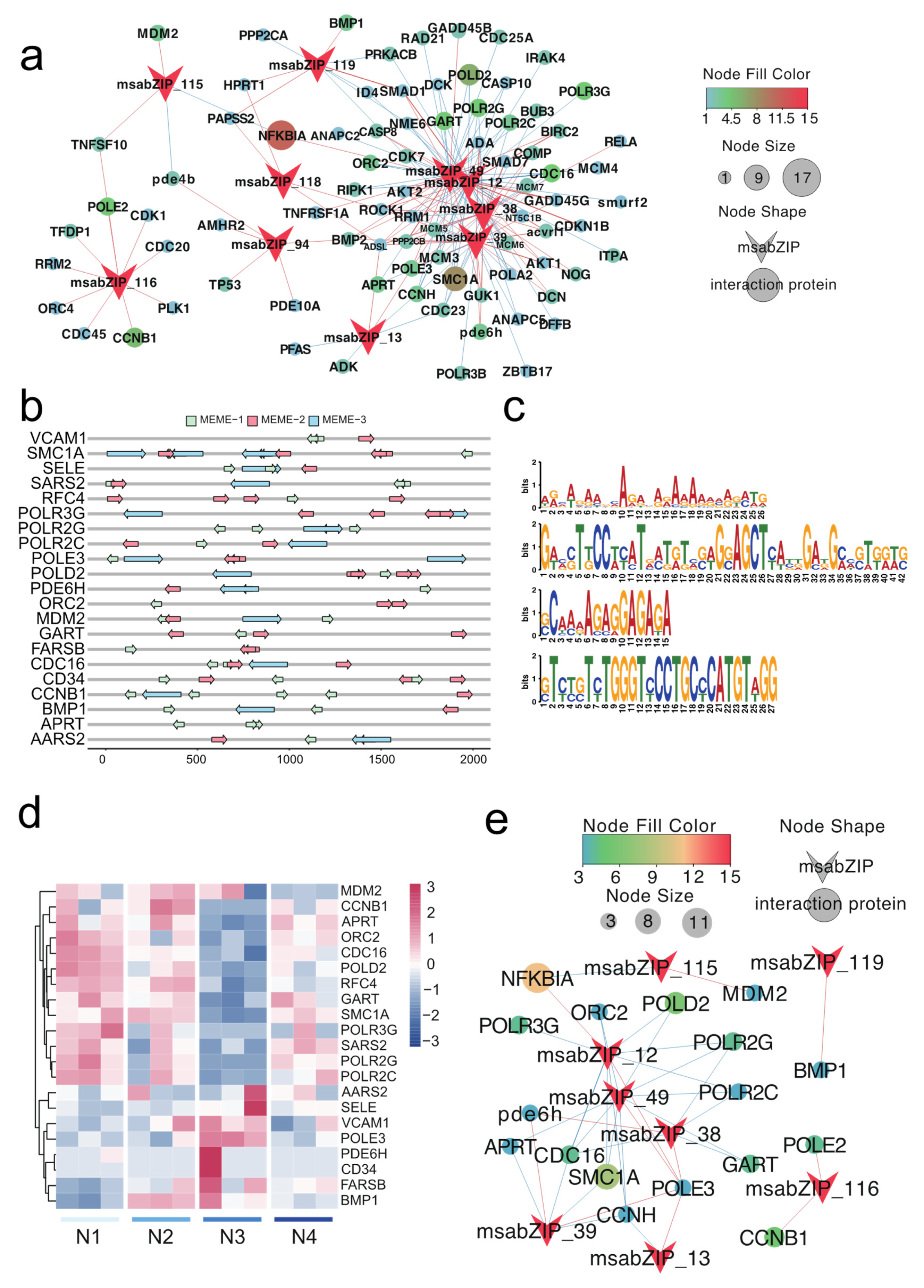
3.2. Analysis of the bZIP Subgroup Associated with Nitrite-Induced Spleen Injury
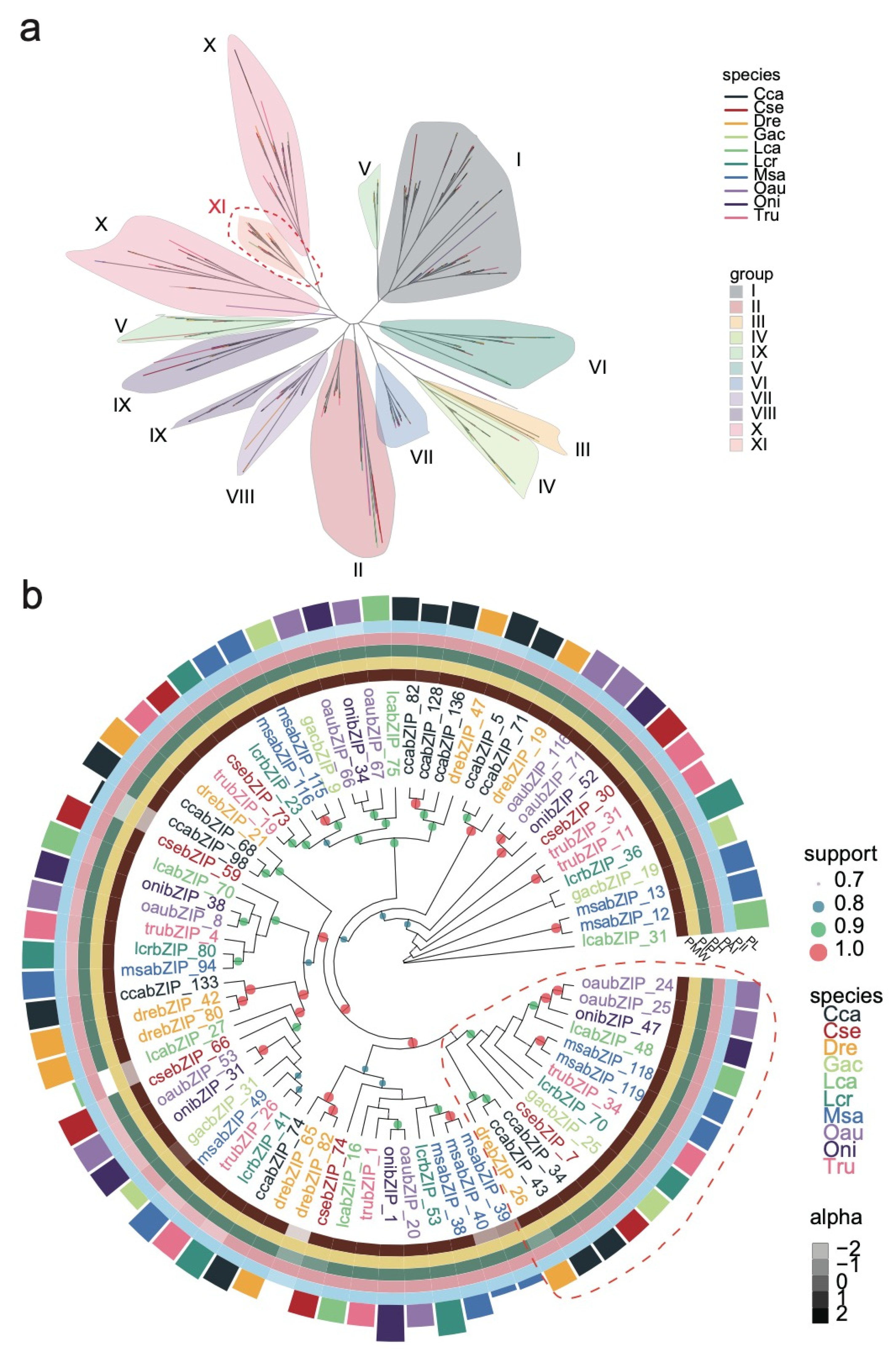
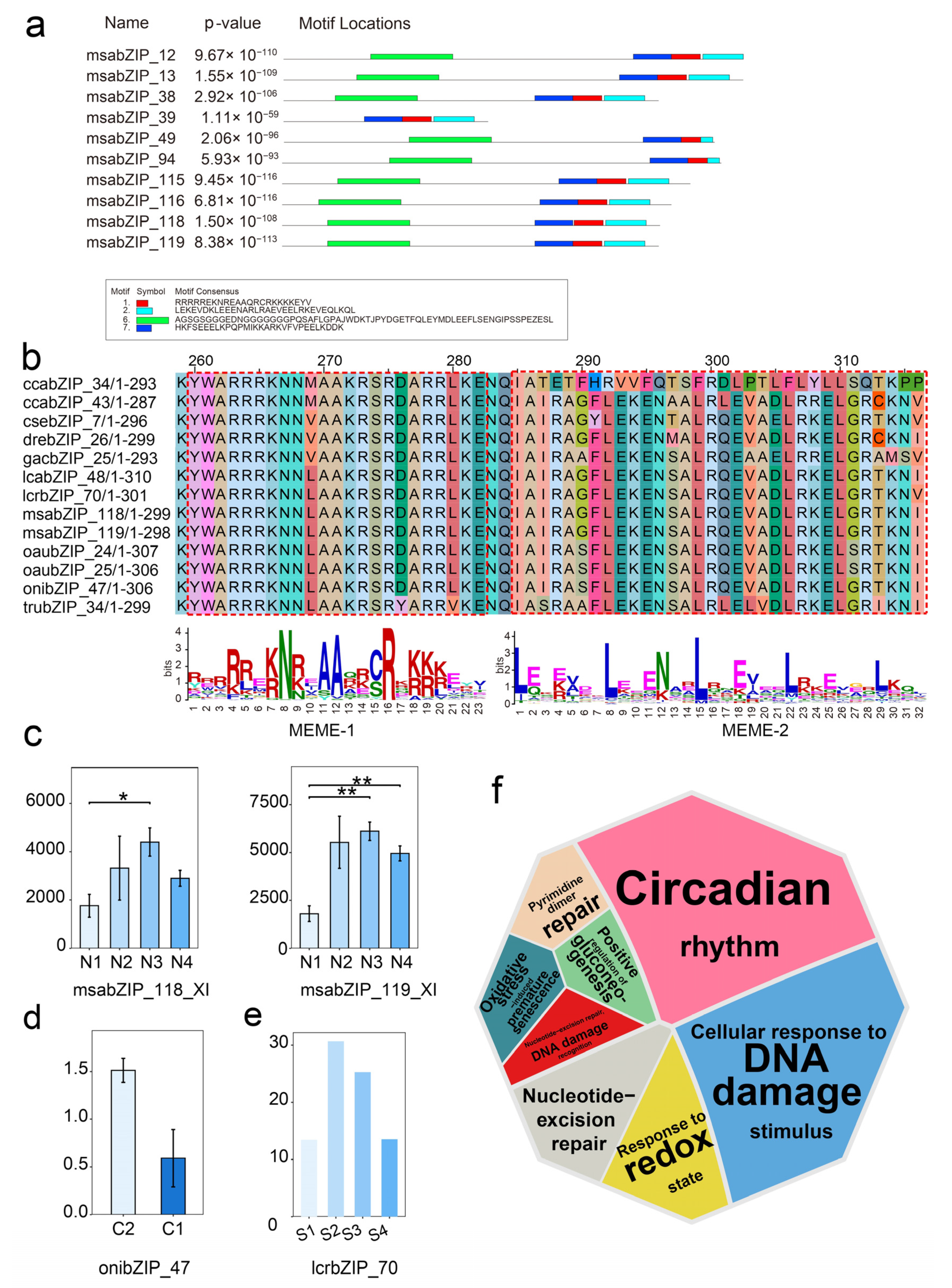
3.3. Phylogenetic and Functional Analysis of the bZIP Subgroup across Species
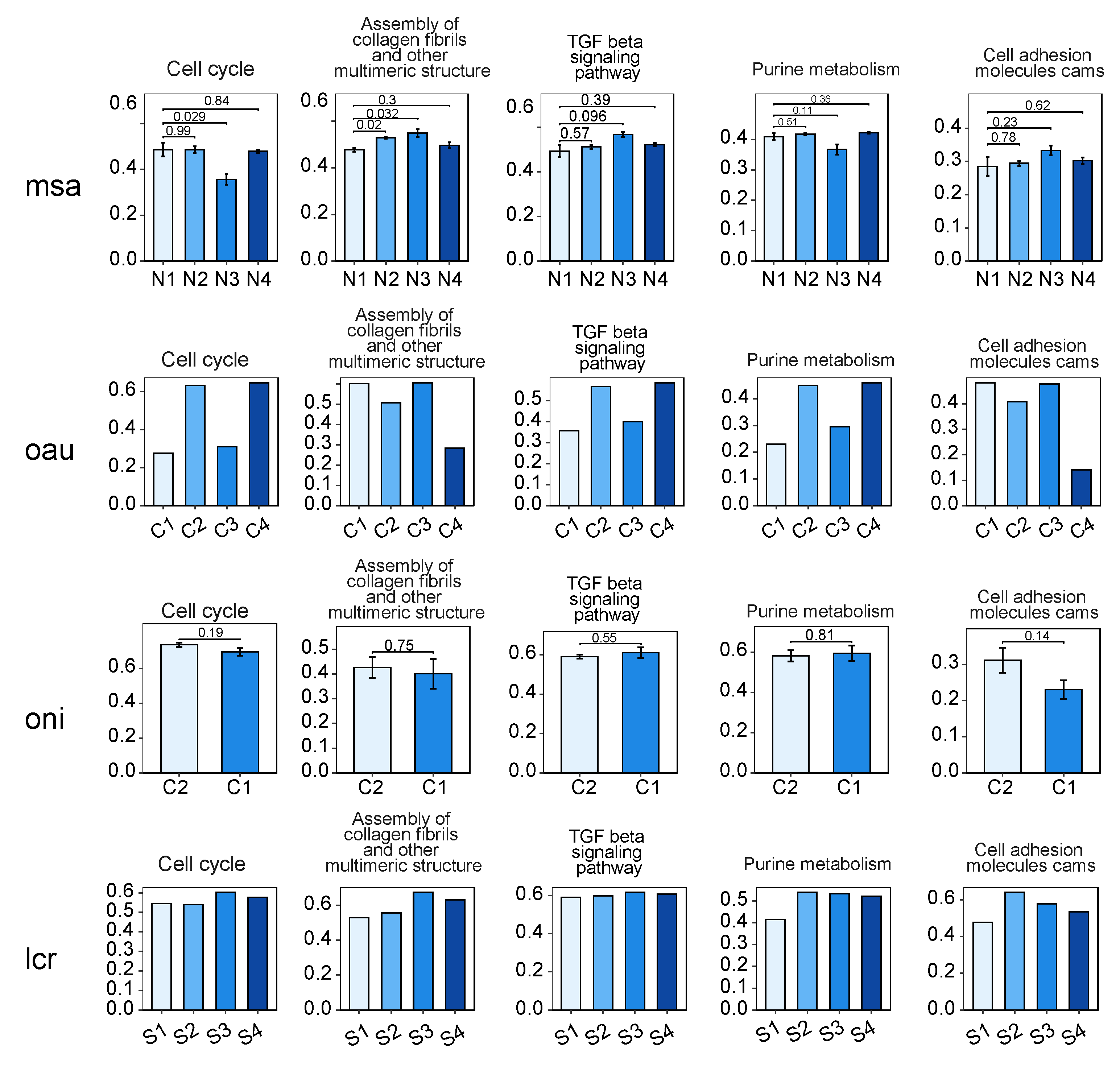
3.4. Key Pathway Screening and Regulatory Network Analysis between the bZIP Subgroup and Nitrite-Induced Splenic Damage
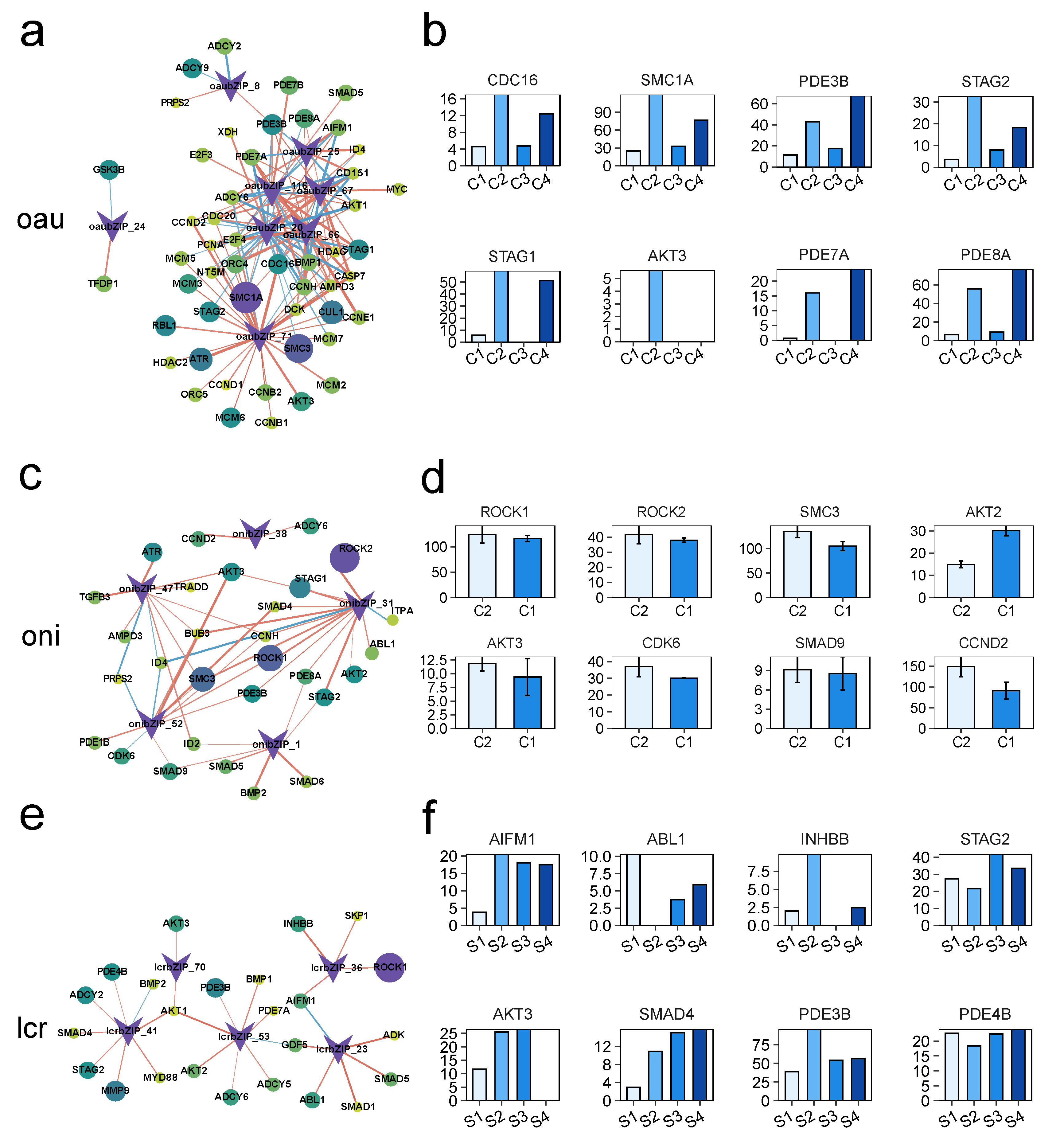
3.5. Analysis of the Regulatory Networks of the bZIP Subgroup in Other Species
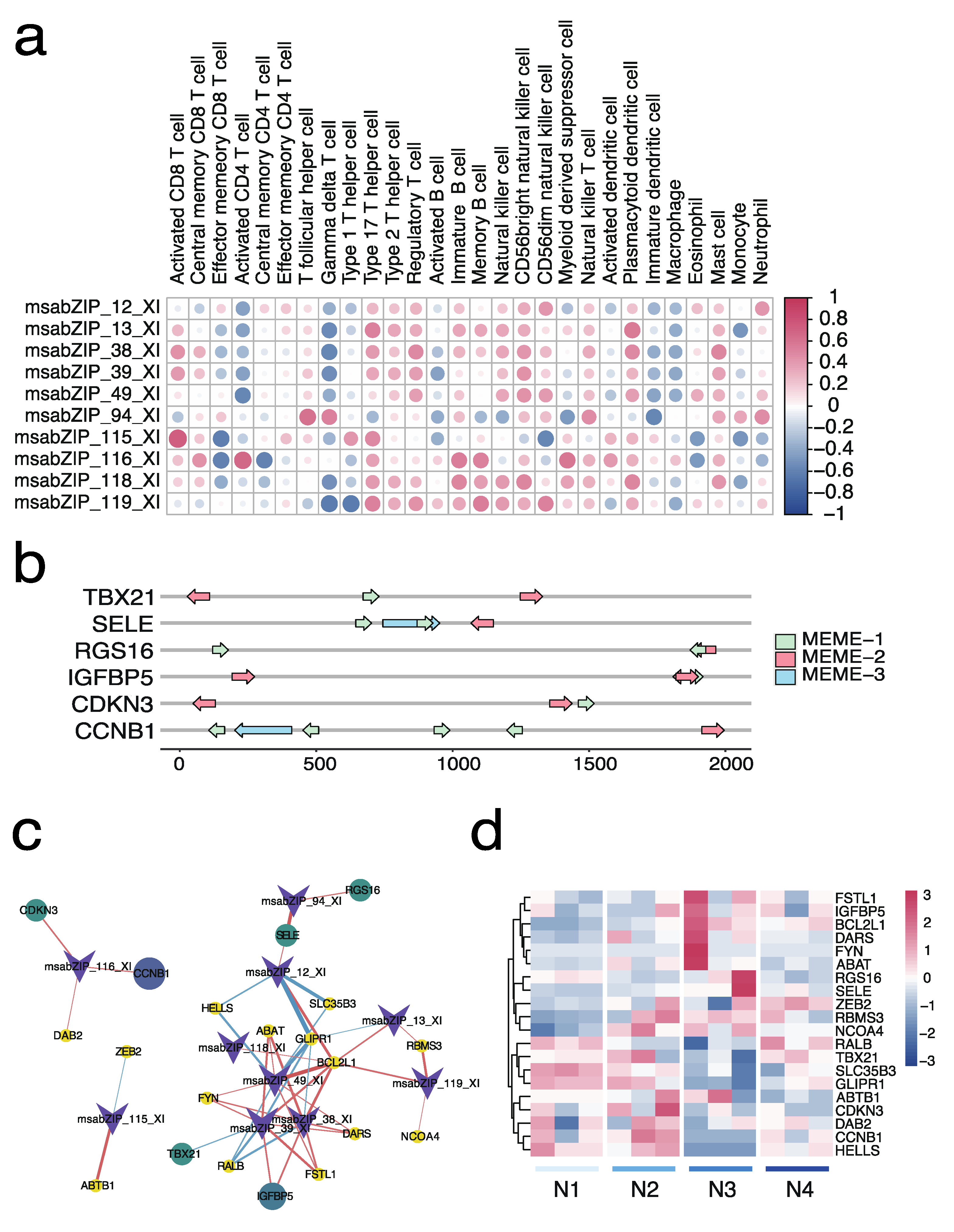
3.6. Relationship and Regulatory Network Analysis between the bZIP Subgroup and Splenic Immune Infiltration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Y.; Wang, Y.; Zhao, H.; Wu, M.; Zhang, J.; Chen, W.; Li, G.; Yang, L. Genome-wide identification and expression analysis of the bZIP transcription factors in the Mycoparasite Coniothyrium minitans. Microorganisms 2020, 8, 1045. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.E.; Timme-Laragy, A.R.; Karchner, S.I.; Stegeman, J.J. Nrf2 and Nrf2-related proteins in development and developmental toxicity: Insights from studies in zebrafish (Danio rerio). Free Radic. Biol. Med. 2015, 88, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Han, K.; Zhang, T.; Qi, G.; Jiang, Z.; Hu, C. Grass carp (Ctenopharyngodon idella) NRF2 alleviates the oxidative stress and enhances cell viability through upregulating the expression of HO-1. Fish Physiol. Biochem. 2020, 46, 417–428. [Google Scholar] [CrossRef] [PubMed]
- van der Hoeven, R.; McCallum, K.C.; Cruz, M.R.; Garsin, D.A. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 2011, 7, e1002453. [Google Scholar]
- McEwan, D.L.; Feinbaum, R.L.; Stroustrup, N.; Haas, W.; Conery, A.L.; Anselmo, A.; Sadreyev, R.; Ausubel, F.M. Tribbles ortholog NIPI-3 and bZIP transcription factor CEBP-1 regulate a Caenorhabditis elegans intestinal immune surveillance pathway. BMC Biol. 2016, 14, 105. [Google Scholar] [CrossRef]
- Dunbar, T.L.; Yan, Z.; Balla, K.M.; Smelkinson, M.G.; Troemel, E.R. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 2012, 11, 375–386. [Google Scholar] [CrossRef]
- Pellegrino, M.W.; Nargund, A.M.; Kirienko, N.V.; Gillis, R.; Fiorese, C.J.; Haynes, C.M. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 2014, 516, 414–417. [Google Scholar] [CrossRef]
- Santos, I.R.; Chen, X.; Lecher, A.L.; Sawyer, A.H.; Moosdorf, N.; Rodellas, V.; Tamborski, J.; Cho, H.-M.; Dimova, N.; Sugimoto, R. Submarine groundwater discharge impacts on coastal nutrient biogeochemistry. Nat. Rev. Earth Environ. 2021, 2, 307–323. [Google Scholar]
- Isaza, D.F.G.; Cramp, R.L.; Franklin, C.E. Living in polluted waters: A meta-analysis of the effects of nitrate and interactions with other environmental stressors on freshwater taxa. Environ. Pollut. 2020, 261, 114091. [Google Scholar] [CrossRef]
- Kim, J.H.; Kang, Y.J.; Kim, K.I.; Kim, S.K.; Kim, J.H. Toxic effects of nitrogenous compounds (ammonia, nitrite, and nitrate) on acute toxicity and antioxidant responses of juvenile olive flounder, Paralichthys olivaceus. Environ. Toxicol. Pharmacol. 2019, 67, 73–78. [Google Scholar] [CrossRef]
- Tong, C.Y.; Honda, K.; Derek, C.J.C. A review on microalgal-bacterial co-culture: The multifaceted role of beneficial bacteria towards enhancement of microalgal metabolite production. Environ. Res. 2023, 228, 115872. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Cao, J.; Qin, X.; Qiu, W.; Mei, J.; Xie, J. Toxic effects on bioaccumulation, hematological parameters, oxidative stress, immune responses and tissue structure in fish exposed to ammonia nitrogen: A review. Animals 2021, 11, 3304. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cui, C.; Liu, Q.; Sun, J.; He, K.; Adam, A.A.; Luo, J.; Li, Z.; Wang, Y.; Yang, S. Combined exposure to hypoxia and ammonia aggravated biological effects on glucose metabolism, oxidative stress, inflammation and apoptosis in largemouth bass (Micropterus salmoides). Aquat. Toxicol. 2020, 224, 105514. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Q.; Fei, F.; Huo, H.H.; Huang, B.; Meng, X.S.; Zhang, T.; Liu, B.L. Impact of nitrite exposure on plasma biochemical parameters and immune-related responses in Takifugu rubripes. Aquat. Toxicol. 2020, 218, 105362. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Barman, A.S.; Devi, A.L.; Devi, A.G.; Pandey, P.K. Iron mediated hematological, oxidative and histological alterations in freshwater fish Labeo rohita. Ecotoxicol. Environ. Saf. 2019, 170, 87–97. [Google Scholar] [CrossRef]
- Lushchak, V.I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 2011, 101, 13–30. [Google Scholar] [CrossRef]
- Yang, L.; Guo, H.; Kuang, Y.; Yang, H.; Zhang, X.; Tang, R.; Li, D.; Li, L. Neurotoxicity induced by combined exposure of microcystin-LR and nitrite in male zebrafish (Danio rerio): Effects of oxidant-antioxidant system and neurotransmitter system. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2022, 253, 109248. [Google Scholar] [CrossRef]
- Garcia-Jaramillo, M.; Beaver, L.M.; Truong, L.; Axton, E.R.; Keller, R.M.; Prater, M.C.; Magnusson, K.R.; Tanguay, R.L.; Stevens, J.F.; Hord, N.G. Nitrate and nitrite exposure leads to mild anxiogenic-like behavior and alters brain metabolomic profile in zebrafish. PLoS ONE 2020, 15, e0240070. [Google Scholar] [CrossRef]
- Parvathy, A.J.; Das, B.C.; Jifiriya, M.J.; Varghese, T.; Pillai, D.; Rejish Kumar, V.J. Ammonia induced toxico-physiological responses in fish and management interventions. Rev. Aquac. 2023, 15, 452–479. [Google Scholar]
- Secombes, C.J.; Wang, T. 1—The innate and adaptive immune system of fish. In Infectious Disease in Aquaculture; Austin, B., Ed.; Woodhead Publishing: Sawston, UK, 2012; pp. 3–68. [Google Scholar]
- Bjørgen, H.; Koppang, E.O. Anatomy of Teleost Fish Immune Structures and Organs. In Principles of Fish Immunology: From Cells and Molecules to Host Protection; Buchmann, K., Secombes, C.J., Eds.; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Zhou, K.; Huang, Y.; Chen, Z.; Du, X.; Qin, J.; Wen, L.; Ma, H.; Pan, X.; Lin, Y. Liver and spleen transcriptome reveals that Oreochromis aureus under long-term salinity stress may cause excessive energy consumption and immune response. Fish Shellfish Immunol. 2020, 107, 469–479. [Google Scholar]
- Xu, C.; Li, E.; Suo, Y.; Su, Y.; Lu, M.; Zhao, Q.; Qin, J.G.; Chen, L. Histological and transcriptomic responses of two immune organs, the spleen and head kidney, in Nile tilapia (Oreochromis niloticus) to long-term hypersaline stress. Fish Shellfish Immunol. 2018, 76, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Li, W.; Wu, B.; Chen, J.; Chen, X. Transcriptome analysis reveals new insights into immune response to hypoxia challenge of large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2020, 98, 738–747. [Google Scholar] [PubMed]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Ahlmann-Eltze, C.; Forbes, K.; Anders, S.; Huber, W. DESeq2: Differential gene expression analysis based on the negative binomial distribution. In Bioconductor Version: Release (312); Love, Heidelberg, Germany, 2021. Available online: https://github.com/mikelove/DESeq2 (accessed on 6 June 2023).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Morgan, M.; Falcon, S.; Gentleman, R. GSEABase: Gene Set Enrichment Data Structures and Methods; R package version 1.58.0; 2023. Available online: https://bioconductor.org/packages/release/bioc/html/GSEABase.html (accessed on 8 June 2023).
- Sonja, H.; Castelo, R.; Guinney, J. GSVA: The Gene Set Variation Analysis Package for Microarray and RNA-Seq Data. 2014; pp. 1–20. Available online: Bioconductor.org (accessed on 8 June 2023).
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shen, W.-K.; Chen, S.-Y.; Gan, Z.-Q.; Zhang, Y.-Z.; Yue, T.; Chen, M.-M.; Xue, Y.; Hu, H.; Guo, A.-Y. AnimalTFDB 4.0: A comprehensive animal transcription factor database updated with variation and expression annotations. Nucle Acids Res. 2022, 51, D39–D45. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Evans, T.G.; Kultz, D. The cellular stress response in fish exposed to salinity fluctuations. J. Exp. Zool. A Ecol. Integr. Physiol. 2020, 333, 421–435. [Google Scholar] [CrossRef]
- Shcherbik, N.; Pestov, D.G. The impact of oxidative stress on ribosomes: From injury to regulation. Cells 2019, 8, 1379. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, C.-S.; Lu, C.; Lin, S.-C.; Wu, J.-W.; Wang, Z.-X. A conserved motif in JNK/p38-specific MAPK phosphatases as a determinant for JNK1 recognition and inactivation. Nat. Commun. 2016, 7, 10879. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Lopez, J.M. Understanding MAPK signaling pathways in apoptosis. Int. J. Mol. Sci. 2020, 21, 2346. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.; Whitelaw, M.; Peet, D. The hypoxia-inducible factors: Key transcriptional regulators of hypoxic responses. Cell Mol. Life Sci. 2003, 60, 1376–1393. [Google Scholar] [CrossRef] [PubMed]
- Ossum, C.G.; Wulff, T.; Hoffmann, E.K. Regulation of the mitogen-activated protein kinase p44 ERK activity during anoxia/recovery in rainbow trout hypodermal fibroblasts. J. Exp. Biol. 2006, 209, 1765–1776. [Google Scholar] [CrossRef]
- Liu, F.; Li, X.; Bello, B.K.; Zhang, T.; Yang, H.; Wang, K.; Dong, J. Difenoconazole causes spleen tissue damage and immune dysfunction of carp through oxidative stress and apoptosis. Ecotoxicol. Environ. Saf. 2022, 237, 113563. [Google Scholar] [CrossRef]
- El-Houseiny, W.; Mansour, M.F.; Mohamed, W.A.; Al-Gabri, N.A.; El-Sayed, A.A.; Altohamy, D.E.; Ibrahim, R.E. Silver nanoparticles mitigate Aeromonas hydrophila-induced immune suppression, oxidative stress, and apoptotic and genotoxic effects in Oreochromis niloticus. Aquaculture 2021, 535, 736430. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, W.; Zhang, Y.; Cui, Y.; Xu, S.; Li, S. Bisphenol A regulates cytochrome P450 1B1 through miR-27b-3p and induces carp lymphocyte oxidative stress leading to apoptosis. Fish Shellfish Immunol. 2020, 102, 489–498. [Google Scholar] [CrossRef]
- Green, D.R.; Levine, B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014, 157, 65–75. [Google Scholar] [CrossRef]
- Dikic, I.; Johansen, T.; Kirkin, V. Selective autophagy in cancer development and therapy. Cancer Res. 2010, 70, 3431–3434. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Z.; Yao, H.; Liang, Y.; Xing, H.; Xu, S. Effects of atrazine and chlorpyrifos on oxidative stress-induced autophagy in the immune organs of common carp (Cyprinus carpio L.). Fish Shellfish Immunol. 2015, 44, 12–20. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Shi, Q.; Guo, Y.; Zhang, W.; Zhou, B. A protective role of autophagy in TDCIPP-induced developmental neurotoxicity in zebrafish larvae. Aquat. Toxicol. 2018, 199, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, Z. Nrf2 is involved in osmoregulation, antioxidation and immunopotentiation in Coilia nasus under salinity stress. Biotechnol. Biotechnol. Equip. 2019, 33, 1453–1463. [Google Scholar] [CrossRef][Green Version]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [PubMed]
- Almeer, R.S.; Alarifi, S.; Alkahtani, S.; Ibrahim, S.R.; Ali, D.; Moneim, A. The potential hepatoprotective effect of royal jelly against cadmium chloride-induced hepatotoxicity in mice is mediated by suppression of oxidative stress and upregulation of Nrf2 expression. Biomed. Pharmacother. 2018, 106, 1490–1498. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.-C.; Wang, Z.-Y.; Tang, K.-K.; Zhu, Y.-S.; Fan, R.-F. Trehalose suppresses cadmium-activated Nrf2 signaling pathway to protect against spleen injury. Ecotoxicol. Environ. Saf. 2019, 181, 224–230. [Google Scholar] [CrossRef]
- Dong, J.; Sulik, K.K.; Chen, S.Y. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: Implications for the prevention of fetal alcohol spectrum disorders. Antioxid. Redox Signal. 2008, 10, 2023–2033. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, C.K.; LaVoie, H.A.; DiPette, D.J.; Singh, U.S. Resveratrol restores Nrf2 level and prevents ethanol-induced toxic effects in the cerebellum of a rodent model of fetal alcohol spectrum disorders. Mol. Pharmacol. 2011, 80, 446–457. [Google Scholar] [CrossRef]
- Singh, V.; Kaur, R.; Kumari, P.; Pasricha, C.; Singh, R. ICAM-1 and VCAM-1: Gatekeepers in various inflammatory and cardiovascular disorders. Clin. Chim. Acta 2023, 548, 117487. [Google Scholar] [CrossRef]
- Sans, M.; Panés, J.; Ardite, E.; Elizalde, J.I.; Arce, Y.; Elena, M.; Palacín, A.; Fernández–Checa, J.C.; Anderson, D.C.; Lobb, R. VCAM-1 and ICAM-1 mediate leukocyte-endothelial cell adhesion in rat experimental colitis. Gastroenterology 1999, 116, 874–883. [Google Scholar] [CrossRef]
- Abonia, J.P.; Hallgren, J.; Jones, T.; Shi, T.; Xu, Y.; Koni, P.; Flavell, R.A.; Boyce, J.A.; Austen, K.F.; Gurish, M.F. Alpha-4 integrins and VCAM-1, but not MAdCAM-1, are essential for recruitment of mast cell progenitors to the inflamed lung. Blood 2006, 108, 1588–1594. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Cotterill, S. Association of mutations in replicative DNA polymerase genes with human disease: Possible application of Drosophila models for studies. Int. J. Mol. Sci. 2023, 24, 8078. [Google Scholar] [CrossRef]
- Mao, X.; Wu, J.; Zhang, Q.; Zhang, S.; Chen, X.; Liu, X.; Wei, M.; Wan, X.; Qiu, L.; Zeng, M. Requirement of WDR70 for POLE3-mediated DNA double-strand breaks repair. Sci. Adv. 2023, 9, eadh2358. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Qiu, L.; Ru, S.; Yang, Y.; Wang, J.; Zhang, X. Bisphenol S disrupts opsins gene expression and impairs the light-sensing function via antagonizing TH-TRbeta signaling pathway in zebrafish larvae. Food Chem. Toxicol. 2023, 172, 113588. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.R.; Moga, S.; Singh, B.; Aulakh, G.K. CD34 Protein: Its expression and function in inflammation. Cell Tissue Res. 2023, 393, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Krenke, K.; Szczaluba, K.; Bielecka, T.; Rydzanicz, M.; Lange, J.; Koppolu, A.; Ploski, R. FARSA mutations mimic phenylalanyl-tRNA synthetase deficiency caused by FARSB defects. Clin. Genet. 2019, 96, 468–472. [Google Scholar] [CrossRef]
- Anastasi, C.; Rousselle, P.; Talantikite, M.; Tessier, A.; Cluzel, C.; Bachmann, A.; Mariano, N.; Dussoyer, M.; Alcaraz, L.B.; Fortin, L. BMP-1 disrupts cell adhesion and enhances TGF-β activation through cleavage of the matricellular protein thrombospondin-1. Sci. Signal 2020, 13, eaba3880. [Google Scholar] [CrossRef]
- Zhuang, M.; Deng, Y.; Zhang, W.; Zhu, B.; Yan, H.; Lou, J.; Zhang, P.; Cui, Q.; Tang, H.; Sun, H. LncRNA Bmp1 promotes the healing of intestinal mucosal lesions via the miR-128-3p/PHF6/PI3K/AKT pathway. Cell Death Dis. 2021, 12, 595. [Google Scholar] [CrossRef]
- Gao, X.-Q.; Fei, F.; Huo, H.H.; Huang, B.; Meng, X.S.; Zhang, T.; Liu, B.-L. Effect of acute exposure to nitrite on physiological parameters, oxidative stress, and apoptosis in Takifugu rubripes. Ecotoxicol. Environ. Saf. 2020, 188, 109878. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Yu, H.; Dong, A. The oncogenic role of γ-aminobutyrate aminotransferase in human tumor: A pan-cancer analysis. Chemotherapy 2022, 11, 1–11. [Google Scholar] [CrossRef]
- Dominguez, C.X.; Amezquita, R.A.; Guan, T.; Marshall, H.D.; Joshi, N.S.; Kleinstein, S.H.; Kaech, S.M. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 2015, 212, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, C.S.; Bronner, M.E. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-beta signaling. J. Cell Biol. 2012, 199, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Lee, H.Y.; Li, W.; Platt, R.J.; Barrasa, M.I.; Ma, Q.; Elmes, R.R.; Rosenfeld, M.G.; Lodish, H.F. Thyroid hormone receptor beta and NCOA4 regulate terminal erythrocyte differentiation. Proc. Natl. Acad. Sci. USA 2017, 114, 10107–10112. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Huang, Y.; Wang, Y.; Wang, Y.; Hao, G.; Jiang, C.; Huang, Z. The bZIP Transcription Factor Family Orchestrates the Molecular Response to Nitrite Stress in the Largemouth Bass Spleen. Fishes 2023, 8, 540. https://doi.org/10.3390/fishes8110540
Sun Y, Huang Y, Wang Y, Wang Y, Hao G, Jiang C, Huang Z. The bZIP Transcription Factor Family Orchestrates the Molecular Response to Nitrite Stress in the Largemouth Bass Spleen. Fishes. 2023; 8(11):540. https://doi.org/10.3390/fishes8110540
Chicago/Turabian StyleSun, Yan, Yi Huang, Ying Wang, Yanqun Wang, Guiying Hao, Changwei Jiang, and Zhiqiu Huang. 2023. "The bZIP Transcription Factor Family Orchestrates the Molecular Response to Nitrite Stress in the Largemouth Bass Spleen" Fishes 8, no. 11: 540. https://doi.org/10.3390/fishes8110540
APA StyleSun, Y., Huang, Y., Wang, Y., Wang, Y., Hao, G., Jiang, C., & Huang, Z. (2023). The bZIP Transcription Factor Family Orchestrates the Molecular Response to Nitrite Stress in the Largemouth Bass Spleen. Fishes, 8(11), 540. https://doi.org/10.3390/fishes8110540




