Abstract
Heat shock proteins (HSPs) are molecular chaperone proteins that can help maintain cellular protein homeostasis, assist in correcting the folding of cellular proteins, and protect organisms from stress when the body is under stress conditions such as temperature changes or bacterial infections. In this study, the HSP10 and HSP40 genes of Eriocheir hepuensis were cloned and named Eh-HSP10 and Eh-HSP40. The results show that the coding sequence length of the HSP10 and HSP40 genes of E. hepuensis was 309 bp and 1191 bp, encoding 102 and 396 amino acids, respectively. The results of protein domain prediction show that Eh-HSP10 has a Cpn10 domain. The Eh-HSP40 protein contains a DnaJ domain, which is characteristic of the HSP40 gene family. The results of qRT-PCR show that the Eh-HSP10 and Eh-HSP40 genes were expressed in different normal tissues, with the highest expression in the heart. Under Vibrio parahaemolyticus stress, the Eh-HSP10 genes peaked at 6 h, and the Eh-HSP40 peaked at 9 h in the hepatopancreas. In the gill, Eh-HSP10 showed a double peak at 24 and 48 h, and the expression of Eh-HSP40 was time-dependent. In the heart, the expression of Eh-HSP10 increased first and then decreased, whereas Eh-HSP40 peaked at 48 h. The results indicate that the Eh-HSP10 and Eh-HSP40 proteins may play a role in protecting E. hepuensis under V. parahaemolyticus infection and that they may be involved in the innate immune response of E. hepuensis against bacteria.
1. Introduction
Eriocheir hepuensis, commonly known as mitten crab, belongs to Arthropoda, Crustacea, Decapoda, Reptant a, Grapsidae, Varunidae, and Eriocheir [1]. E. hepuensis has its roots in the Hepu subspecies of Japanese mitten crab [2]. This crab comprises large individuals that grow fast and sexually mature in a short period. It is widely distributed in the sea basin of southern Guangxi and the eastern region of Vietnam and is a new breed of mitten crab of high quality [3]. However, in recent years, wild germplasm resources of E. hepuensis have declined rapidly [4]. Vibrio parahaemolyticus infections result in decreased production of E. hepuensis; therefore, fishermen who breed them have faced challenges [1].
Heat shock proteins (HSPs) are a group of highly conserved protein molecules that are part of biological evolution. They can help cells maintain viability under various stress conditions, such as bacteria, temperature, and salinity [5]. They can recognize the hydrophobic regions of denatured proteins, help denatured proteins restore their native conformation, and enhance cell tolerance in the face of stress [6]. HSPs can be divided into groups according to their molecular mass and functions, including HSP110, HSP100, HSP90, HSP70, HSP60, HSP40, and small HSPs (approximately 15–30 kDa) [7]. Under normal circumstances, HSPs can also participate in the process of cell cycle regulation and apoptosis and assist in protein folding and transportation [8,9]. Moreover, HSPs can participate in the innate immune system of the host and have a regulatory effect on the host’s adaptive immune system. Preventing protein aggregation and deformation by inducing the expression of HSPs can help maintain the stability of the intracellular environment of the organism and enhance the antistress and survival abilities of organisms [10].
HSP40 is a subfamily of HSP, and every member has a J domain with 70 amino acids. It is similar to the N-terminal structure of Escherichia coli DnaJ [11,12]. HSP40 is responsible for recognizing and binding unfolded proteins and transferring them to HSP70. Therefore, HSP40 binds with its J domain to HSP70 when HSP70 is in its ATP-bound state [13]. Moreover, HSP40 proteins can be divided into the following three categories based on differences in other conserved regions: the type I proteins contain four domains, including the J domain, the Gly/Phe-rich region (G/F domain), the cysteine-rich domain (CxxCxGxG), and the less-conserved C-terminal domain; the type II proteins lack the cysteine-rich domain; and the type III proteins contain only the J domain [14]. The main functions of HSP40 are manifested in cellular stress protection, protein translation, protein folding, and protein translocation [15]. Moreover, HSP40 can participate in DNA binding and play a role in intracellular signal transduction and apoptosis [16].
HSP10 is a protein with a low molecular mass in the HSP family and is classically considered a mitochondrial cochaperonin that interacts with HSP60 [17]. As a common partner of the HSP60 protein, HSP10 is also known as chaperone protein 10 (Cpn10), which can cooperate with HSP60 to help in protein synthesis, folding, and degradation, in addition to correcting misfolded proteins, so as to exert its biological function [18,19]. HSP10 is widely involved in protecting prokaryotic or eukaryotic cells under stress conditions and avoiding infection. As an active component in the apoptosis signal pathway, HSP10 plays a key role in signal transduction. It can upregulate the antiapoptotic gene Bcl2 and downregulate the Caspase3 gene, inhibiting apoptosis and protecting cells [20]. It can also participate in the inflammatory immune response, promote proliferation, etc. [21].
E. hepuensis, an important economical crab found in the Beibu Gulf of Guangxi, is often subjected to environmental stress or Vibrio challenges, resulting in a decreased survival rate and a significant decrease in production [1]. As a crustacean, E. hepuensis lacks an acquired immune system and can only rely on the non-specific immune system to defend against the invasion of external pathogens to protect the body from damage [22]. In recent years, antibiotics and immunopotentiators have been identified as measures for crustaceans to fight against bacteria [23], but strengthening the immunity of E. hepuensis is necessary so that they can resist diseases in order to address the root of the problem. HSPs can effectively activate the innate immune system [24] and can participate in cell signal transduction and hormone response. HSPs also can play an essential role in the survival of cells under conditions of pathogenic attack, stress, or environmental stress [25]. However, it is rarely reported whether HSP genes play a role in the antibacterial infection of E. hepuensis. In this study, we cloned the HSP10 and HSP40 genes of E. hepuensis and analyzed the changes in expression of their tissues after V. parahaemolyticus infections to explore whether they have anti-infection immunity. These results also provide reference materials for further studies on the molecular response mechanism of Eh-HSP10 and Eh-HSP40 after V. parahaemolyticus stress.
2. Materials and Methods
2.1. Experimental Animals
E. hepuensis (15 ± 1 g) were purchased from the Dongfeng market in Qinzhou City, Guangxi Province, China. For tissue expression analysis, the stomach, hepatopancreas, gill, heart, intestine, and muscle from three crabs were collected, quickly frozen in liquid nitrogen, and stored in a −80 °C refrigerator for RNA extraction.
A total of 100 crabs were temporarily raised in a plastic bucket with dimensions of 45 × 30 × 30 cm3 with tap water aerated for 48 h and a water depth of 20 cm for 3 days in the laboratory before processing, with 10 crabs per bucket. During the experimental period, the culture water temperature was 20–22 °C, the dissolved oxygen was 6.8–7.2 mg/L, the pH was 7.5–8.0, and 50% of the water was changed each day [26].
2.2. RNA Extraction and Reverse Transcription
We used the TRIzol method to extract total RNA from the stomach, hepatopancreas, gill, heart, intestine, and muscle. Concentrations were estimated by measuring absorbance at 260 nm with a NanoDrops 2000, and RNA quality and integrity were checked via 1.0% agarose gel electrophoresis [27]. The reverse transcription reaction was performed using a Prime ScriptTM RT reagent kit with gDNA Eraser (TaKaRa, Beijing, China). The reaction process was divided into two steps: (1) gDNA removal at 42 °C for 5 min and (2) reverse transcription of RNA to cDNA at 37 °C for 60 min, at 85 °C for 5 min, and held at 4 °C [28]. The cDNA samples synthesized via reverse transcription were stored at −20 °C for further exploration.
2.3. Gene Cloning of Eh-HSP10 and Eh-HSP40
The cDNAs of HSP10 and HSP40 were obtained from E. hepuensis using reverse transcription–polymerase chain reaction (RT-PCR) [29]. According to the transcription of E. hepuensis, specific HSP10 and HSP40 amplification primers (HSP10CDS and HSP40CDS) were designed using Oligo6.0 software, and HSP10 and HSP40 (HSP10DL and HSP40DL), and Ef1 quantitative primers (Ef1DL), were used as control reference genes (Table 1). The amplified cDNA fragments were then combined with specific primers, ligated into a pEASY-Blunt Simple Cloning Vector (Trans GenBiotech, Beijing, China), and transferred into E. coli (Trans-1T1, Beijing, China). The plasmids containing the inserted HSP10 and HSP40 fragments were used as templates and sent to Guangzhou Bioengineering Co., Ltd. (Guangzhou, China) for DNA sequencing [26].

Table 1.
Primer sequence.
2.4. Bioinformatics Analysis
Homologous sequence alignment analysis was conducted using the Blast program at the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 10 October 2022). MEGA11.0 software was used for sequence alignment, and a phylogenetic tree was constructed using the neighbor-joining method. SignalP (https://services.healthtech.dtu.dk/service.php?SignalP-3.0, accessed on 10 October 2022) was used to predict the N-terminal signal peptide. Amino acid composition analysis, molecular mass prediction and isoelectric point prediction were carried out using the Expert Protein Analysis System server (http://web.expasy.org/protparam/, accessed on 10 October 2022) [30]. The SMART program (http://smart.embl—heidelberg.de, accessed on 10 October 2022) was used to predict the possible protein domains of HSP10- and HSP40-encoded protein amino acid sequences. The protein secondary structures of HSP10 and HSP40 were predicted using the Protean program in DNAStar. The SwissModel program (https://swiss-model.expasy.org/interactive, accessed on 10 October 2022) was used to construct the tertiary structures.
2.5. Relative Expression Levels of Eh-HSP10 and Eh-HSP40 in Different Tissues
Six tissues including the stomach, hepatopancreas, gill, heart, intestine, and muscle of E. hepuensis were used as experimental materials. The same methods discussed in Section 2.2 were used to extract the total RNA from the various tissues, with reverse transcription of RNA and cDNA for detection of mRNA expression. Real-time quantitative RT-PCR analysis was performed on a real-time PCR system (BIO-RAD CFX Connect, USA) using SYBR PreMix ExTaqTM (TaKaRa, Beijing, China) [28].
2.6. Relative Expression Levels of Eh-HSP10 and Eh-HSP40 under V. parahaemolyticus Stress
Eighty individuals were randomly divided into two equal experimental and control groups. Approximately 100 μL of phosphate-buffered saline (PBS) solution was mixed with V. parahaemolyticus (2 × 108 CFU/mL), and 100 μL of PBS solution alone were injected into the root of each individual’s fourth walking leg. Crabs were removed from the water as soon as they molted, and three tissues of the hepatopancreas, gills, and heart of three surviving individuals were sampled from the experimental and control groups at 3, 6, 9, 12, 24, and 48 h. The crabs were anesthetized in an ice box for 5–10 min, and the tissues were collected using high-temperature sterilized surgical scissors and tweezers for subsequent RNA extraction and qRT-PCR.
2.7. Statistical Analysis
The 2−ΔΔct method was used to calculate target gene expression levels, and sample data were standardized using Ef1 as the internal reference gene [31]. One-way ANOVA was conducted using SPSS Statistics 26 software for statistical analysis, and the Duncan method was used to compare the significant differences between groups. The significant difference standard was set as p < 0.05, and the highly significant difference was set as p < 0.01.
3. Results
3.1. Cloning and Sequence Analysis of HSP10 and HSP40 CDS Regions of E. hepuensis
The specific bands of Eh-HSP10 and Eh-HSP40 were amplified via PCR amplification (Figure 1), with coding regions of 372 bp and 1316 bp, respectively. The sequences of Eh-HSP10 and Eh-HSP40 were obtained via sequencing.
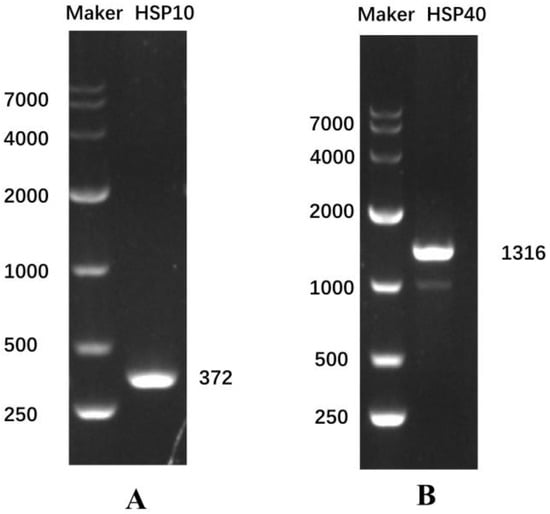
Figure 1.
Electrophoresis map of the Eh-HSP10 gene (A) and Eh-HSP40 gene (B). Marker, DL10000 DNA marker.
The results show that the open reading frame (ORF) of the Eh-HSP10 and Eh-HSP40 genes was 309 bp and 1191 bp, respectively, encoding 102 and 396 amino acids with the molecular weights of 10.99 and 44.63 kDa, respectively. The theoretical isoelectric point of Eh-HSP10 and Eh-HSP40 was 6.28 and 7.15, respectively. HSP10 of E. hepuensis was found to be an acidic protein, whereas Eh-HSP40 was found to be a basic protein. The protein domain prediction data show that Eh-HSP10 contains Cpn10 and GroES domains located at positions 7–99 and 8–100 of the protein sequence, respectively (Figure 2A,E). Eh-HSP40 contained DnaJ (positions 5–60), DnaJ-C (positions 106–322) (Figure 2B), and PTZ00037 domains (positions 60–197) (Figure 2F).
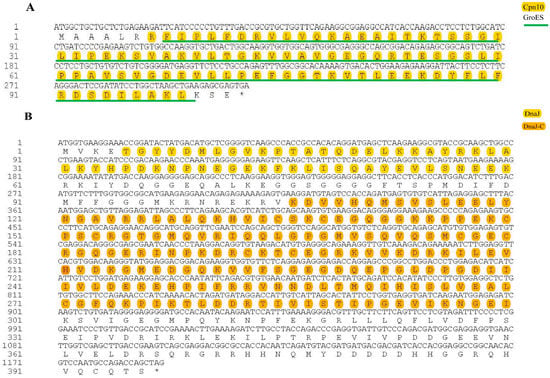
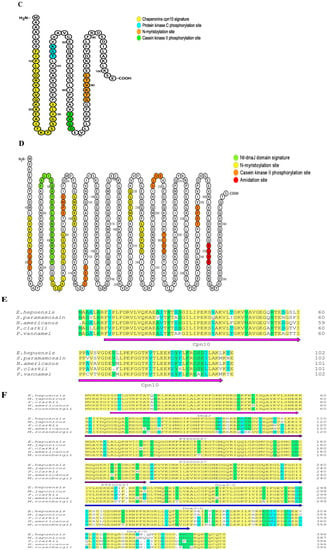
Figure 2.
Nucleotide sequences, amino acid sequences, and functional sites of HSP10 (A,C) and HSP40 (B,D) in E. hepuensis. Amino acid sequence comparison between HSP10 (E), HSP40 (F), and other species.
Functional site prediction analysis of Eh-HSP10 and Eh-HSP40 proteins was performed using the Motif Scan online program. The results show that Eh-HSP10 contains one chaperonin Cpn10 signature (8FIPLFDRVLVQKAEAITKTSSGILI32), one protein kinase C phosphorylation site (42KGT44), one casein kinase II phosphorylation site (65SVGD68), and one N-myristoylation site (77LTVKTG82) (Figure 2C). Eh-HSP40 contains six N-myristoylation sites (12GVKPTA17, 67GGEQAL72, 75GGSGGG80, 96GMKRNR101, 215GMEDGQ220, and 233GLDPGD238), one Nt-DnaJ domain signature (45FKLISQAYEVLSNEEKRKIY64), six casein kinase II phosphorylation sites (18TQDE21, 84SPMD87, 115SLEE118, 265SLVE268, 279TLDD282, and 346EPRT349), and one amidation site (369RGRR372) (Figure 2D).
3.2. Homologous Sequence Analysis of HSP10 and HSP40 in E. hepuensis and Construction of a Phylogenetic Tree
Based on the nucleotide sequence of HSP10 of E. hepuensis, an online alignment was performed using the NCBI. Then, MEGA11.0 was used to construct a phylogenetic tree. The results are shown in Figure 3. HSP10 of E. hepuensis and the homology of Scylla paramamosain were the highest, at 86%, and the homology with Daphnia pulex was the lowest, at 65%.
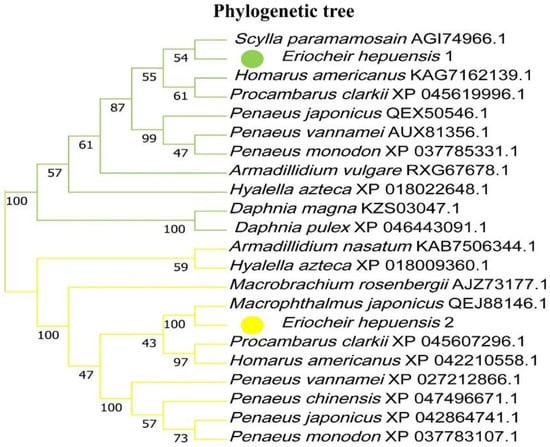
Figure 3.
Neighbor-joining phylogenetic tree based on two HSP gene nucleotide sequences among various species.
The nucleotide sequences of HSP40 of E. hepuensis were aligned on the NCBI database, and multiple sequences of HSP40 of other species with high homology to HSP40 of E. hepuensis were analyzed using DNAMAN software. The protein sequence similarity between HSP40 of E. hepuensis and Macrophthalmus japonicus, Procambarus clarkii, Penaeus japonicus, Penaeus vannamei, and Penaeus chinensis was 97%, 90%, 89%, 89%, and 88%, respectively. The phylogenetic tree in Figure 3 shows that HSP40 of E. hepuensis and M. japonicus was clustered with the highest genetic relationship, and the genetic relationship with Hyalella azteca was more distant.
3.3. Bioinformatics Analysis of Eh-HSP10 and Eh-HSP40 Proteins
Amino acid composition analysis of the Eh-HSP10 protein revealed that the highest content was leucine (11.8%), followed by valine (10.8%), and the lowest was methionine and tyrosine (1.0%). The molecular formula of Eh-HSP10 is C497H820N128O148S1. The prediction results indicate that the HSP10 protein contains no N-terminal signal peptide. The subcellular localization of the HSP10 protein of E. hepuensis was observed in the mitochondrion. The predicted secondary structure of the Eh-HSP10 protein shows that it contains five α helices, five β sheets, two T turns, and three random coils (Figure 4A). The tertiary domain shows that the structure of HSP10 of E. hepuensis (Figure 5A,B) is similar to that of HSP10 of S. paramamosain, with the main conformation via the α helix.
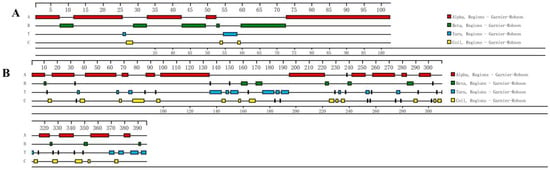
Figure 4.
Secondary structure prediction of HSP10 (A) and HSP40 (B) protein in E. hepuensis.
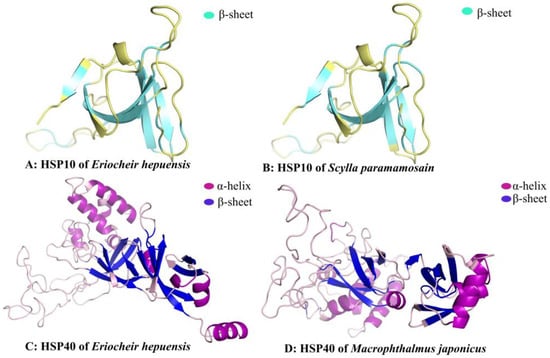
Figure 5.
Prediction of the tertiary structure of the HSP10 (A,B) and HSP40 (C,D) proteins of different species.
The molecular formula of the Eh-HSP40 protein is C1936H3134N568O595S23. Among its amino acid components, glycine has the highest content (10.6%), followed by lysine (9.1%), and alanine (lowest, 2.0%). The prediction results indicate that the HSP40 protein has no N-terminal signal peptide. Subcellular localization of the HSP40 protein of E. hepuensis was observed in the cytoplasm. The predicted secondary structure of the HSP40 protein of Eriocheir sinensis shows that it contains 16 α helices, 13 β sheets, 31 T turns, and 27 random coils (Figure 4B). The three-dimensional structure of HSP40 shows that it is mainly composed of α helices and β sheets (Figure 5C,D).
3.4. Relative Expression Levels of Eh-HSP10 and Eh-HSP40 in Different Tissues
The expressions of the two HSP genes in different tissues of E. hepuensis are shown in Figure 6, as captured via real-time PCR. The results show that the HSP10 gene is expressed in different tissues of normal E. hepuensis in the following order: heart > gill > hepatopancreas > intestine > stomach > muscle (Figure 6A). The relative expression levels of the HSP40 gene in different tissues of E. hepuensis occur in the following order: heart > intestine > hepatopancreas > gill > stomach > muscle (Figure 6B).
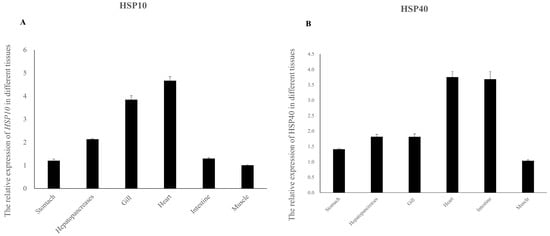
Figure 6.
Relative expression levels of HSP10 (A) and HSP40 (B) genes in different tissues of normal E. hepuensis.
3.5. Relative Expression Levels of Eh-HSP10 and Eh-HSP40 under V. parahaemolyticus Stress
The expression level of the HSP10 gene of E. hepuensis is obviously time-dependent under V. parahaemolyticus stress. In the hepatopancreas, the expression level of the HSP10 gene began to increase gradually at 3 h, and the highest level was recorded at 6 h, which is 1.68 times that of the control group (Figure 7A). Compared with the control group, no significant difference was observed at 9 h (p > 0.05), and there was a significant difference at 24 h (p < 0.05). In the gills, the expression of the Eh-HSP10 gene began to increase at 3–6 h and gradually decreased at 9–12 h; the expression level of the HSP10 gene was the highest at 24 and 48 h, which was 3.33 and 3.26 times that of the control group, respectively. Significant differences were observed between each time period and the control group (p < 0.01) (Figure 7C). In the heart, the expression level of the Eh-HSP10 gene gradually increased within 3–12 h and reached the highest level at 12 h, which was 2.37 times that of the control group. The expression level began to decline at 24 h and returned to the control group level at 48 h. Eh-HSP10 differed significantly from the control group at 3 h (p < 0.05), and no significant difference was observed at 48 h (p > 0.05) (Figure 7E).
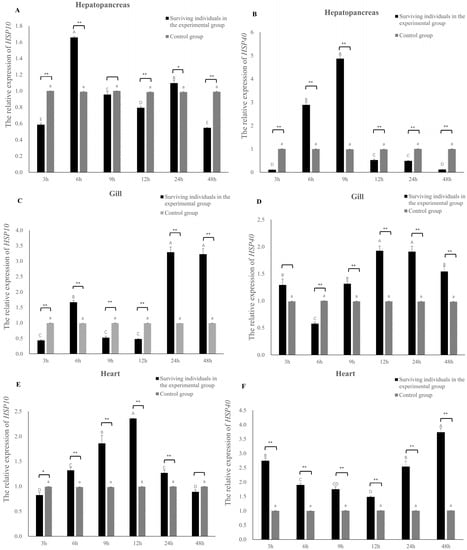
Figure 7.
Expression of the HSP gene in the hepatopancreas, heart, and gill tissues of E. hepuensis under 2 × 108 CFU/mL V. parahaemolyticus stress. * (A) The relative expression of HSP10 gene in the hepatopancreas; (B) The relative expression of HSP40 gene in the hepatopancreas; (C) The relative expression of HSP10 gene in the hepatopancreas; (D) The relative expression of HSP40 gene in the hepatopancreas; (E) The relative expression of HSP10 gene in the hepatopancreas; (F) The relative expression of HSP40 gene in the hepatopancreas. Different lower and upper case letters above columns indicate statistical differences. Different asterisks above the columns show significant differences between control group and exprimental group at each time (* p < 0.05, ** p < 0.01).
The expression level of the HSP40 gene of E. hepuensis changed after V. parahaemolyticus stress. The expression level of the hepatopancreas began to increase significantly at 6 h and reached the highest level at 9 h, which was 2.88 times and 4.92 times that of the control group, respectively (Figure 7B). Compared with the control group, extremely significant differences were observed at 6, 9, 12, 24, and 48 h (p < 0.01). In the gills, the Eh-HSP40 gene began to be induced at 3 h, gradually increased from 6 to 24 h, and peaked at 12 h, which was 1.95 times that of the control group (Figure 7D). No significant difference was observed at 3 h compared with the control group (p > 0.05), and the rest of the time periods were significantly different from the control group (p < 0.01). In the heart, the expression level of HSP40 increased significantly at 3 h and, then gradually decreased at 6–12 h. At 12–48 h, its expression began to gradually increase, reaching the highest value at 48 h, which was 3.7 times that of the control group. Significant differences were observed at 3, 6, 9, 12, 24, and 48 h compared with the control group (p < 0.01) (Figure 7F).
4. Discussion
HSP plays an important role in biological functions and can be rapidly induced to express and participate in immune defense under the influence of various stresses and pathogens [5]. Our findings indicate that ORF of the Eh-HSP10 and Eh-HSP40 genes is 309 bp and 1191 bp, respectively, encoding 102 and 396 amino acids with molecular weights of 10.99 and 44.63 kDa, respectively. The theoretical isoelectric point of the Eh-HSP10 and Eh-HSP40 genes is 6.28 and 7.15, respectively. HSP10 of E. hepuensis is an acidic protein, whereas the Eh-HSP40 gene is a basic protein, as reported in a study were also by Nitnavare et al. (2016) [32]. Bioinformatic analysis showed that the Eh-HSP10 protein sequence contains a conserved domain of Cpn10, has no signal peptide, and forms an α helix, which enables the protein to pass through the mitochondrial membrane to perform its functions, as reported in humans by David [33]. The Eh-HSP10 protein has the classic domain of the HSP10 family, but the HSP10 protein of E. hepuensis and S. paramamosain has five different bases in the classic domains of the Cpn10 [22], indicating that the difference of HSP10 gene between freshwater crab and marine crab. A comparison of tertiary structures shows that the α-helix and β-sheet structures of HSP10 of E. hepuensis and S. paramamosain are similar [22], indicating that the structure of HSP10 was highly conserved during the process of evolution and might be vital for Eh-HSP10 to perform its biological functions.
The Eh-HSP40 protein sequence contains a DnaJ domain, an HPD motif, a cysteine-rich domain (CxxCxGxG), and a C-terminal domain, with typical structural features of the HSP40 family [34,35]. Moreover, the Eh-HSP40 domain, in combination with other protein structures, performs various functions in different species. For example, the ERdj5 protein in mammals contains a thioredoxin structure and a J domain, which could promote ERdj5 to form disulfide bonds during the folding of endoplasmic reticulum protein [36], playing a role in stabilizing the spatial structure of the peptide chain. The Eh-HSP40 protein has a typical family domain and a similar tertiary structure, but the composition and protein structures of HSP40 protein differ depending on the species, which might lead to varying biological functional significance. It was found that the secondary structure of the Eh-HSP40 protein and the HSP40 protein of M. japonicus differ by one α helix [37], possibly owing to the structural changes in the HSP40 among different crab species caused by the geographic environment of freshwater vs. seawater.
The results of this study show that the expression levels of Eh-HSP10 and Eh-HSP40 are highest in the heart under normal conditions, followed by the hepatopancreas, gills, and epidermis, similar to the expression of other crustacean aquatic animals. For example, HSP10 of S. paramamosain is most abundantly expressed in the heart [38], and HSP40 in M. japonicus is highly expressed in the hepatopancreas and heart [37]. The heart is an important organ for blood circulation, playing an important role in metabolism. The hepatopancreas, as the main metabolic center responsible for the production of reactive oxygen species, is the main site for the digestion and absorption of nutrients and is an important tissue for the immune defense of crustaceans [39]. High expression of the Eh-HSP10 and Eh-HSP40 genes in the heart and hepatopancreas were recorded, possibly due to the rapid response of immune organs such as the heart and hepatopancreas, which help protect E. hepuensis against foreign pathogens and from bodily injury.
As a common pathogen in aquaculture, V. parahaemolyticus has strong pathogenicity to aquatic organisms. In this study, under the stress of V. parahaemolyticus, Eh-HSP10 in the hepatopancreas increased rapidly at 6 h and then gradually decreased, similar to the expression results of HSP10 in other crustaceans. For example, the expression of PmHSP10 was rapidly upregulated after 6 h under stress [40], and HSP10 in Penaeus vannamei was rapidly increased under low-temperature induction [41]. It was hypothesized that hepatopancreas might the most vulnerable organ to be damaged under stress conditions [42], indicating that the HSP10 protein could rapidly synthesize and maintain body cells when E. hepuensis is under the V. parahaemolyticus stress, in addition to repairing or refolding damaged proteins. In the gills, the expression levels of Eh-HSP10 and Eh-HSP40 both showed a double peak at 12 h, similar to that of the HSP40 gene of Pinctada fucata martensii under the Vibrio alginolyticus stress [14]. The results show that both the HSP10 and HSP40 genes can be induced between species and participate in immune responses. In the heart, the Eh-HSP10 and Eh-HSP40 genes showed distinct expression trends, indicating that the difference in the protein structure of the two proteins might lead to different mechanisms in resisting Vibrio infections. Our findings suggest that Eh-HSP10 and Eh-HSP40 genes perform an essential role in fighting against Vibrio infections.
5. Conclusions
In this study, the HSP10 and HSP40 genes of Eriocheir hepuensis were cloned with a coding sequence length of 309 bp and 1191 bp, encoding 102 and 396 amino acids, respectively. Eh-HSP10 and Eh-HSP40 genes were expressed in all tested tissues, and the highest expression was observed in the heart. Under Vibrio parahaemolyticus stress, the highest expression level of Eh-HSP10 in the hepatopancreas, gills, and heart was 1.68, 3.33, and 2.37 times that in the control group at 6 h, 24 h, and 12 h, respectively. The highest expression level of Eh-HSP40 in the hepatopancreas, gills, and heart was 4.92, 1.95, and 3.7 times that in the control group at 9 h, 12 h, and 48 h, respectively, indicating that Eh-HSP10 and Eh-HSP40 play a role in crab antibacterial immunity.
Author Contributions
Y.L., P.Z. and J.L. designed this project. Q.F. cultured the experimental Eriocheir hepuensis. T.R., Z.Z. and Z.M. collected samples and extracted RNA. P.Z. and Q.F. performed data analysis and drafted the manuscript. Z.L. (Zhenyu Lan), Y.D. and Z.L. (Ziwei Liang) participated in the sequence alignment, B.C. and Y.Z. performed the statistical analysis. J.L. and Y.L. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Fund (grant no. NSFC 32260918), the Guangxi Natural Science Foundation (grant no. 2019GXNSFBA245081), and the Marine Science Guangxi First-Class Subject, Beibu Gulf University (grant nos. DRB005 and DTB001).
Institutional Review Board Statement
This article does not contain any studies with human participants. All animals used in this study were purchased from the market. The experimental protocols adhere to the Guidelines of Animal Care and were approved by the Animal Management and Ethics Committee of BeiBu Gulf University (approval code BBgulf20210005, approved on 12 September 2021).
Data Availability Statement
The data supporting these findings of this study are included in the manuscript.
Conflicts of Interest
None of the authors has any conflict of interest to declare.
References
- Li, F.; Liao, Y.Y.; Li, Z.G.; Giang, Y.J.; Yang, B.H. Research progress of Eriocheir hepuensis. Curr. Fish. 2021, 46, 72–74. [Google Scholar]
- Tang, B.; Zhou, K.; Song, D.; Yang, G.; Dai, A. Molecular systematics of the Asian mitten crabs, genus Eriocheir (Crustacea: Brachyura). Mol. Phylogenetics Evol. 2003, 29, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Zeng, H. Study on Morphology of Eriocheir hepuensis. J. Zhejiang Ocean. Univ. (Nat. Sci. Ed.) 2000, 19, 327–332. [Google Scholar] [CrossRef]
- Wang, J.; Huang, L.; Cheng, Q.X.; Lu, G.Q.; Wang, C.H. Complete mitochondrial genomes of three mitten crabs, Eriocheir sinensis, E. hepuensis, and E. japonica. Mitochondrial DNA A 2016, 27, 1175–1176. [Google Scholar] [CrossRef]
- Srivastava, P. Roles of heat-shock proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2002, 2, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.T.; Liu, B.; Xie, J.; Zhou, Q.L.; Ge, X.P.; Xu, P. Research progress of heat shock protein and its research prospect in aquatic animals. Jiangsu Agric. Sci. 2011, 39, 303–306. [Google Scholar]
- Zininga, T.; Ramatsui, L.; Shonhai, A. Heat Shock Proteins as Immunomodulants. Molecules 2018, 23, 2846. [Google Scholar] [CrossRef]
- Mallouk, Y.; Vayssier-Taussat, M.; Bonventre, J.V.; Polla, B.S. Heat shock protein 70 and ATP as partners in cell homeostasis (Review). Int. J. Mol. Med. 1999, 4, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.L.; Marzano, N.R.; van Oijen, A.M.; Ecroyd, H. Using Single-Molecule Approaches to Understand the Molecular Mechanisms of Heat-Shock Protein Chaperone Function. J. Mol. Biol. 2018, 430, 4525–4546. [Google Scholar] [CrossRef]
- Morimoto, R.I.; Sarge, K.D.; Abravaya, K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J. Biol. Chem. 1992, 267, 21987–21990. [Google Scholar] [CrossRef]
- Georgopoulos, C.P.; Lundquist-Heil, A.; Yochem, J.; Feiss, M. Identification of the E. coli dnaJ gene product. Mol. Gen. Genet. 1980, 178, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Zylicz, M.; Yamamoto, T.; Mckittrick, N.; Sell, S.; Georgopoulos, C. Purification and properties of the dnaJ replication protein of Escherichia coli. J. Biol. Chem. 1985, 260, 7591–7598. [Google Scholar] [CrossRef] [PubMed]
- Kästle, M.; Grune, T. Chapter 4—Interactions of the Proteasomal System with Chaperones: Protein Triage and Protein Quality control. In Progress in Molecular Biology and Translational Science; Grune, T., Ed.; Academic Press: Cambridge, MA, USA, 2012; pp. 113–160. [Google Scholar]
- Li, J.; Zhang, Y.H.; Liu, Y.; Zhang, Y.; Xiao, S.; Yu, Z.N. Co-expression of heat shock protein (HSP) 40 and HSP70 in Pinctada martensii response to thermal, low salinity and bacterial challenges. Fish Shellfish. Immunol. 2016, 48, 239–243. [Google Scholar] [CrossRef]
- Knox, C.; Luke, G.A.; Blatch, G.L.; Pesce, E.R. Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle. Virus Res. 2011, 160, 15–24. [Google Scholar] [CrossRef]
- Cheetham, M.E.; Caplan, A.J. Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones 1998, 3, 28–36. [Google Scholar] [CrossRef]
- Cappello, F. HSP60 and HSP10 as diagnostic and prognostic tools in the management of exocervical carcinoma. Gynecol. Oncol. 2003, 91, 661. [Google Scholar] [CrossRef]
- Magen, D.; Georgopoulos, C.; Bross, P.; Ang, D.; Segev, Y.; Goldsher, D.; Nemirovski, A.; Shahar, E.; Ravid, S.; Luder, A.; et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am. J. Hum. Genet. 2008, 83, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, Y.; Huo, Z.J.; Zhou, X.R.; Pang, B.P. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs from Galeruca daurica (Coleoptera: Chrysomelidae) and their expression analysis. Bull. Entomol. Res. 2018, 108, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.K.; Cui, Y.G.; Liu, J.Y. Research Progress of Heat Shock Protein 10. Med. Recapitul. 2018, 108, 510–522. [Google Scholar]
- Corrao, S.; Campanella, C.; Anzalone, R.; Farina, F.; Zummo, G.; Conway de Macario, E.; Macario, A.J.; Cappello, F.; La Rocca, G. Human Hsp10 and Early Pregnancy Factor (EPF) and their relationship and involvement in cancer and immunity: Current knowledge and perspectives. Life Sci. 2010, 86, 145–152. [Google Scholar] [CrossRef]
- Zhang, F.Y.; Jiang, K.J.; Sun, M.N.; Zhang, D.; Ma, L.B. Multiplex immune- related genes expression analysis response to bacterial challenge in mud crab, Scylla paramamosain. Fish Shellfish. Immunol. 2013, 34, 712–716. [Google Scholar] [CrossRef]
- Lu, S.J.; Jiao, J.B.; Yuan, X.M.; Yu, Z.; Zhang, H.Q.; Hang, X.Y.; Shi, W.D.; Wu, Y.L. Research progress of Eriocheir sinensis immunostimulants. Jiangsu Agric. Sci. 2021, 49, 19–23. [Google Scholar]
- Luo, J.M.; Luo, J.X. Research Progress of the Immunity of Heat Shock Proteins. Med. Recapitul. 2013, 19, 1179–1181. [Google Scholar]
- Jung, H.; Lyons, R.E.; Li, Y.; Thanh, N.M.; Dinh, H.; Hurwood, D.A.; Salin, K.R.; Mather, P.B. A candidate gene association study for growth performance in an improved giant freshwater prawn (Macrobrachium rosenbergii) culture line. Mar. Biotechnol. 2014, 16, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Liu, J.X.; Ren, T.J.; Xu, Y.H.; Lu, M.; Fang, H.Y.; Zhang, Y.; Liao, Y.Y.; Zhu, P. Cloning and analysis of three glutathione S-transferases in Eriocheir hepuensis and their expression in response to azadirachtin stress. Aquac. Rep. 2021, 19, 1–11. [Google Scholar] [CrossRef]
- Zhang, L.; Pang, Y.Y.; Yang, H.; Yang, Z.G. Cloning and Expression Analysis of Aquaporin 11 Gene in Eriocheir sinensis. Genom. Appl. Biol. 2021, 40, 1497–1504. [Google Scholar]
- Liao, Y.Y.; Liu, K.; Ren, T.J.; Zhang, Z.N.; Ma, Z.H.; Dan, S.F.; Lan, Z.Y.; Lu, M.; Fang, H.Y.; Zhang, Y.; et al. The characterization, expression and activity analysis of three superoxide dismutases in Eriocheir hepuensis under azadirachtin stress. Fish Shellfish. Immunol. 2021, 117, 228–239. [Google Scholar] [CrossRef]
- Liang, H.Y.; Wang, Z.X.; Lei, Q.N.; Huang, R.L.; Deng, Y.W.; Wang, Q.H.; Jiao, Y.; Du, X.D. Molecular cloning and expression analysis of a pearl oyster (Pinctada martensii) heat shock protein 90 (HSP90). Genet. Mol. Res. 2015, 14, 18778–18791. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Jin, Z.X.; Huang, H.Y.; Ye, H.H.; Li, S.J. Cloning and expression analysis of the regulatory subunit B gene of PP2A in the mud crab Scylla paramamosain. Oceanol. Limnol. Sin. 2012, 43, 932–937. [Google Scholar]
- Zhou, S.-M.; Tao, Z.; Shen, C.; Qian, D.; Wang, C.-L.; Zhou, Q.-C.; Jin, S. β-actin gene expression is variable among individuals and not suitable for normalizing mRNA levels in Portunus trituberculatus. Fish Shellfish. Immunol. 2018, 81, 338–342. [Google Scholar] [CrossRef]
- Nitnavare, R.B.; Yeshvekar, R.K.; Sharma, K.K.; Vadez, V.; Reddy, M.K.; Reddy, P.S. Molecular cloning, characterization and expression analysis of a heat shock protein 10 (Hsp10) from Pennisetum glaucum (L.), a C-4 cereal plant from the semi-arid tropics. Mol. Biol. Rep. 2016, 43, 861–870. [Google Scholar] [CrossRef]
- David, S.; Bucchieri, F.; Corrao, S.; Czarnecka, A.M.; Campanella, C.; Farina, F.; Peri, G.; Tomasello, G.; Sciumè, C.; Modica, G.; et al. Hsp10: Anatomic distribution, functions, and involvement in human disease. Front. Biosci. 2013, 5, 768–778. [Google Scholar] [CrossRef]
- Martinez-Yamout, M.; Legge, G.B.; Zhang, O.; Wright, P.E.; Dyson, H.J. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J. Mol. Biol. 2000, 300, 805–818. [Google Scholar] [CrossRef]
- Lu, Z.; Cyr, D.M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 1998, 273, 5970–5978. [Google Scholar] [CrossRef]
- Cunnea, P.M.; Miranda-Vizuete, A.; Bertoli, G.; Simmen, T.; Damdimopoulos, A.E.; Hermann, S.; Leinonen, S.; Huikko, M.P.; Gustafsson, J.A.; Sitia, R.; et al. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 2003, 278, 1059–1066. [Google Scholar] [CrossRef]
- Kiyun, P.; Ihn-Sil, K. Salinity and bisphenol A alter cellular homeostasis and immune defense by heat shock proteins in the intertidal crab Macrophthalmus japonicus—ScienceDirect. Estuar. Coast. Shelf Sci. 2019, 229, 106381. [Google Scholar]
- Ding, J.; Chen, F.Y.; Ren, S.Y.; Qiao, K.; Chen, B.; Wang, K.J. Molecular characterization and promoter analysis of crustacean heat shock protein 10 in Scylla paramemosain. Genome 2013, 56, 273–281. [Google Scholar] [CrossRef]
- Söderhäll, K.; Cerenius, L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 1998, 10, 23–28. [Google Scholar] [CrossRef]
- Shi, J.X.; Fu, M.J.; Zhao, C.; Zhou, F.L.; Yang, Q.B.; Qiu, L.B. Characterization and function analysis of Hsp60 and Hsp10 under different acute stresses in black tiger shrimp, Penaeus monodon. Cell Stress Chaperones 2016, 21, 295–312. [Google Scholar] [CrossRef]
- Peng, J.X.; Wei, B.Y.; Yin, Q.; Jiang, X.Z.; Li, M.; Chen, X.H. Sequence of HSP10 gene in Litopenaeus vannmei and its low temperature expression analysis. J. South. Agric. 2013, 44, 838–843. [Google Scholar]
- Shi, H.N.; Liu, Z.; Zhang, J.P.; Kang, Y.J.; Wang, J.F.; Huang, J.Q.; Wang, W.M. Short Communication: Effect of heat stress on heat-shock protein (Hsp60) mRNA expression in rainbow trout Oncorhynchus mykiss. Genet. Molecilar Res. 2015, 14, 5280–5286. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).