Genomic Analysis of the Proteasome Subunit Gene Family and Their Response to High Density and Saline-Alkali Stresses in Grass Carp
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Classification of the PS Genes in Grass Carp
2.2. Phylogenetic and Conserved Motif Analysis of the PS Gene Family
2.3. Chromosomal Localization and Gene Duplication Analysis of the PS Gene Family
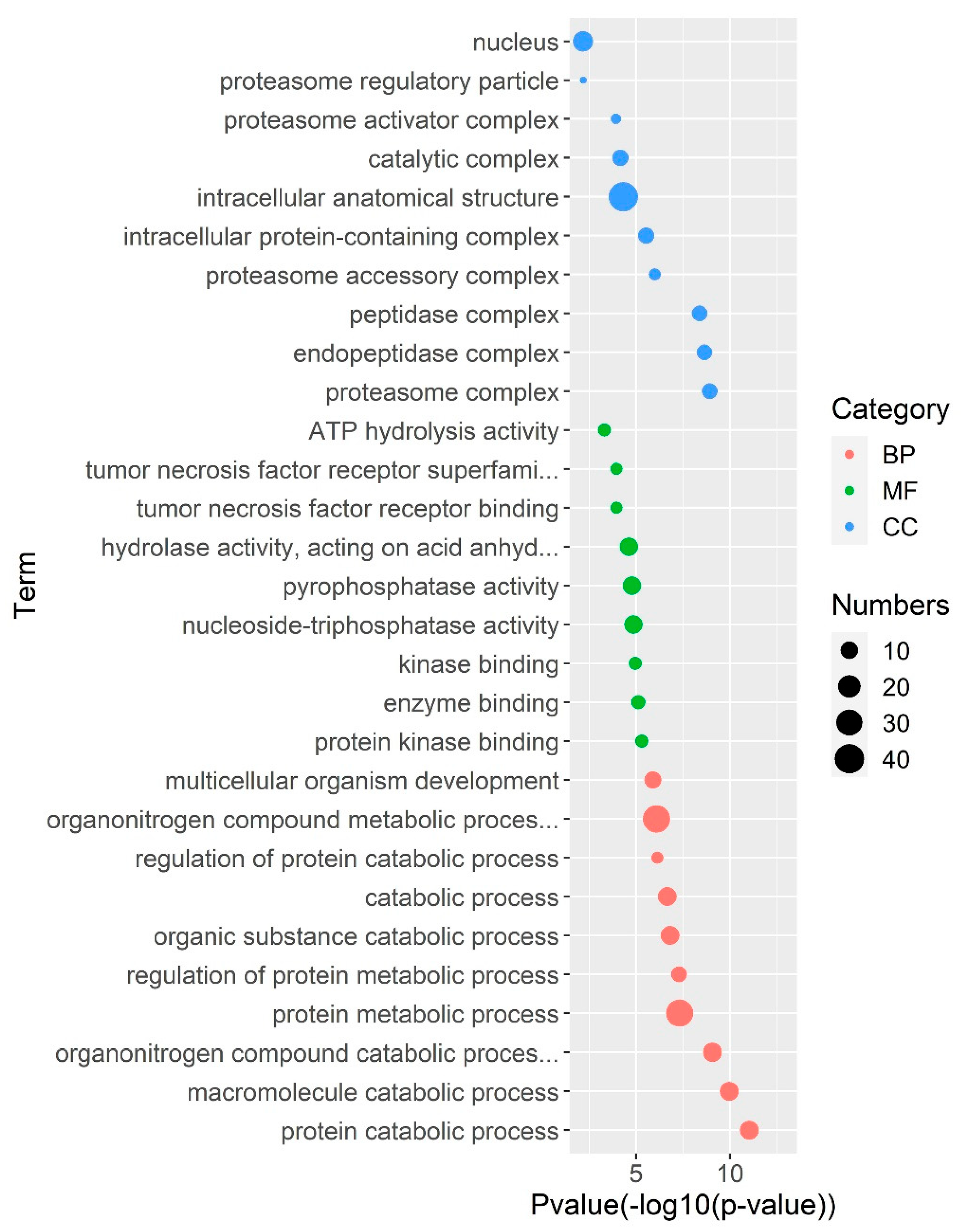
2.4. Gene Regulatory Network Analysis of PS Genes in Grass Carp
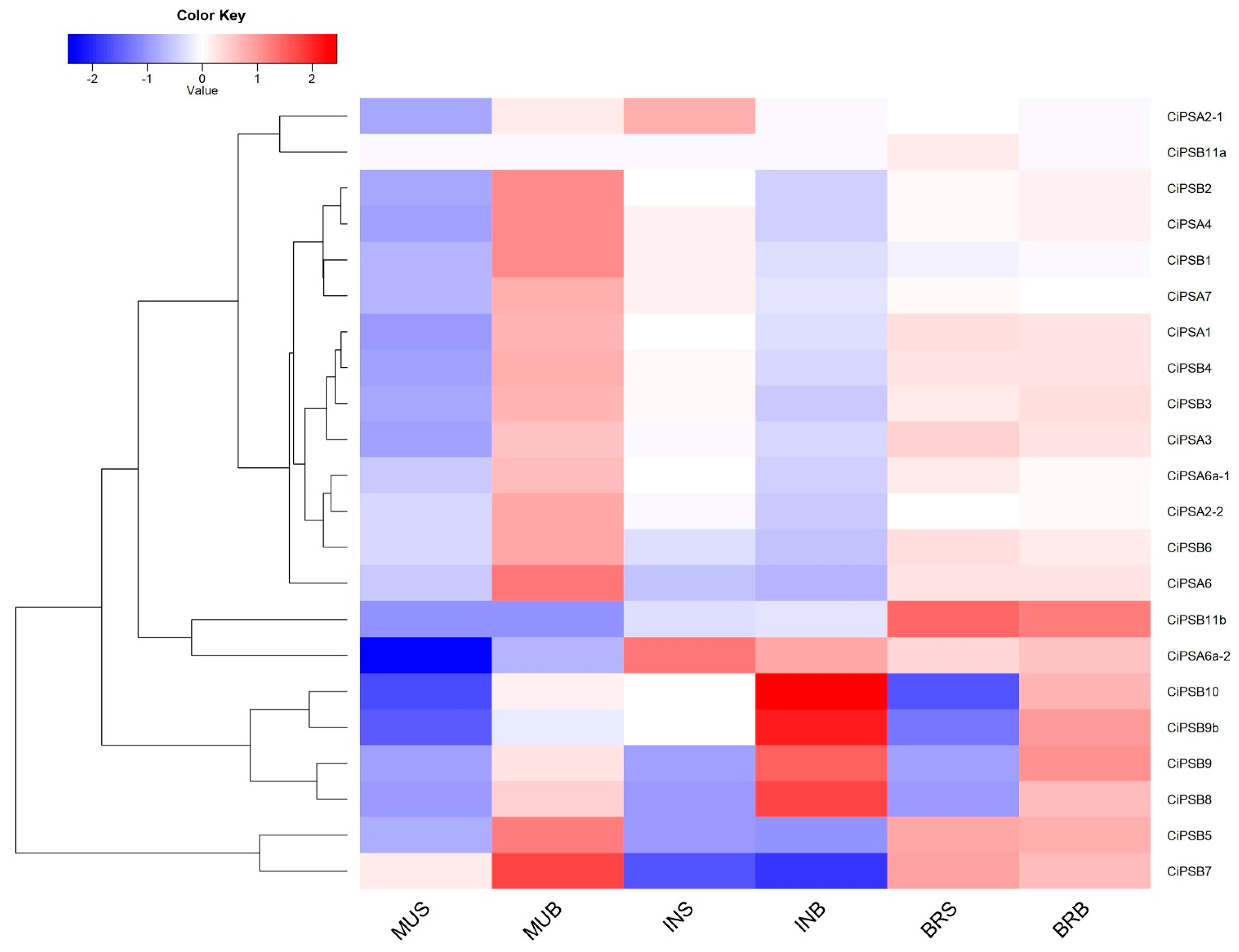
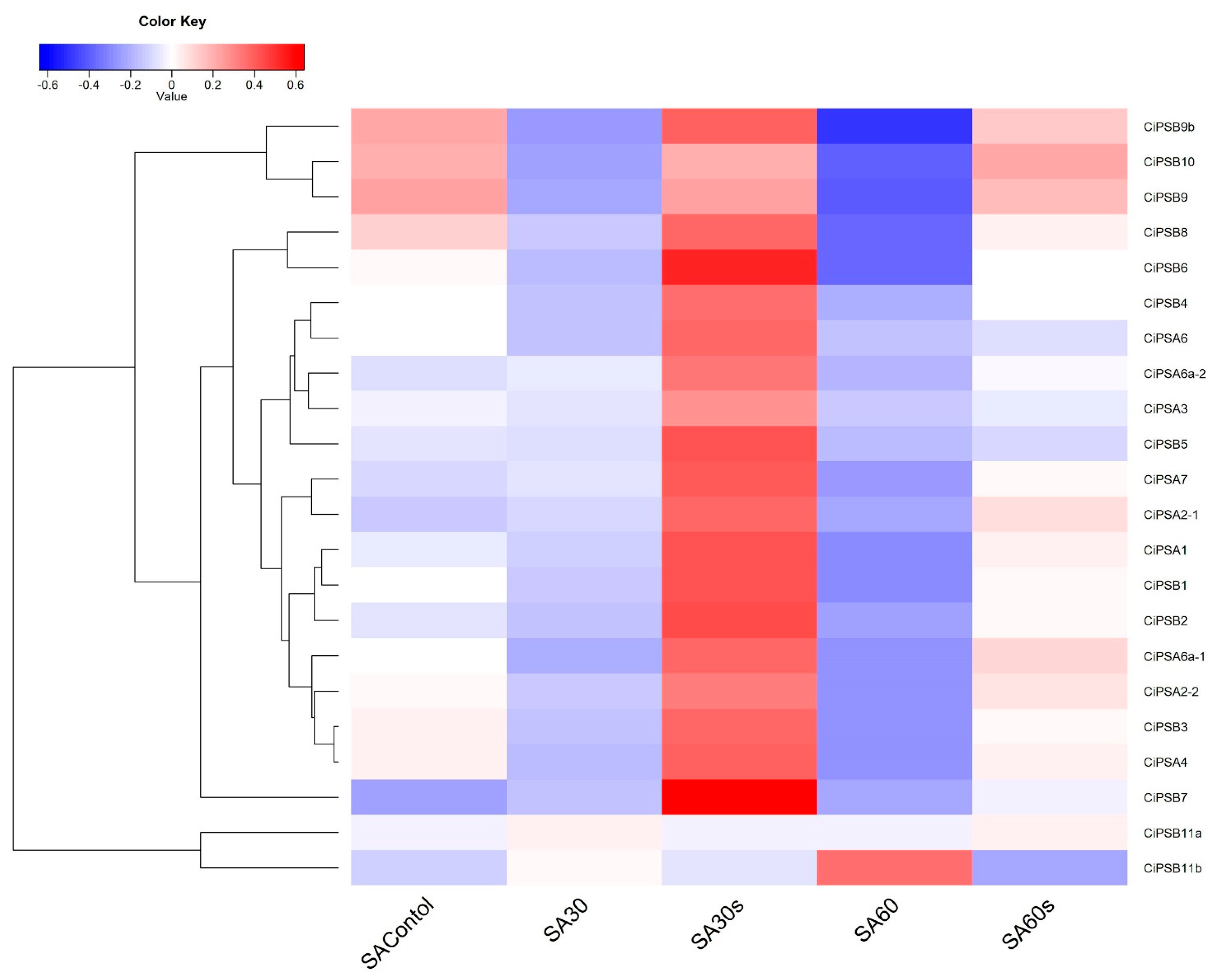
2.5. Transcriptomic Analysis of the Response of PS Genes to Environmental Stresses
3. Results
3.1. Identification and Phylogenetic Analysis of the PS Genes in Grass Carp
3.2. Conserved Motif Analysis of the PS Genes
3.3. Chromosomal Localization and Gene Duplication Analysis of the PS Genes
3.4. Gene Regulatory Network Analysis of PS Genes
3.5. Expression Analysis of CiPS Genes in Response to Environmental Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fishery Administration of Ministry of Agriculture and Rural Affairs. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2022.
- Xie, C.; Li, J.; Li, D.; Shen, Y.; Gao, Y.; Zhang, Z. Grass Carp: The Fish that Feeds Half of China. In Aquaculture in China; Gui, J., Tang, Q., Li, Z., Liu, J., De Silva, S.S., Eds.; John Wiley & Sons Press: Oxford, UK, 2018; pp. 93–115. [Google Scholar]
- Zhao, Y.; Zhang, L.; Wang, C.; Xie, C. Biology and Ecology of Grass Carp in China: A Review and Synthesis. North Am. J. Fish. Manag. 2020, 40, 1379–1399. [Google Scholar] [CrossRef]
- Petitjean, Q.; Jean, S.; Gandar, A.; Côte, J.; Laffaille, P.; Jacquin, L. Stress responses in fish: From molecular to evolutionary processes. Sci. Total Environ. 2019, 684, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Pankhurst, N.W. The endocrinology of stress in fish: An environmental perspective. Gen. Comp. Endocrinol. 2011, 170, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Schulte, P.M. What is environmental stress? Insights from fish living in a variable environment. J. Exp. Biol. 2014, 217, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y. Structure, Dynamics and Function of the 26S Proteasome. Subcell. Biochem. 2021, 96, 1–151. [Google Scholar] [PubMed]
- Gomes, A.V. Genetics of proteasome diseases. Sci. Cairo 2013, 2013, 637629. [Google Scholar] [CrossRef]
- Adams, J. The proteasome: Structure, function, and role in the cell. Cancer Treat Rev. 2003, 29, 3–9. [Google Scholar] [CrossRef]
- Adams, J. The proteasome: A suitable antineoplastic target. Nat. Rev. Cancer 2004, 4, 349–360. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, X.; Chen, J.; Tang, R.; Li, L.; Li, D. Change in Ubiquitin Proteasome System of Grass Carp Ctenopharyngodon idellus Reared in the Different Stocking Densities. Front Physiol. 2018, 9, 837. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Y.; Zhang, Y.; Ning, Z.; Li, Y.; Zhao, Q.; Lu, H.; Huang, R.; Xia, X.; Feng, Q.; et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015, 47, 625–631. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment analysis for gene ontology. R Package 2019, version 2.38.1.
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- He, Y.; Yu, H.; Zhao, H.; Zhu, H.; Zhang, Q.; Wang, A.; Shen, Y.; Xu, X.; Li, J. Transcriptomic analysis to elucidate the effects of high stocking density on grass carp (Ctenopharyngodon idella). BMC Genom. 2021, 22, 620. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Khalil, R. Ubiquitin-Proteasome Pathway and Muscle Atrophy. Adv. Exp. Med. Biol. 2018, 1088, 235–248. [Google Scholar] [PubMed]
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, X.; Qi, X.; Fu, X.; Ghimire, S.; Ma, R.; Li, S.; Zhang, N.; Si, H. The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System. Int. J. Mol. Sci. 2020, 21, 2894. [Google Scholar] [CrossRef]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Seiliez, I.; Panserat, S.; Skiba-Cassy, S.; Fricot, A.; Vachot, C.; Kaushik, S.; Tesseraud, S. Feeding status regulates the polyubiquitination step of the ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss) muscle. J. Nutr. 2008, 138, 487–491. [Google Scholar] [CrossRef]
- Ahongo, Y.D.; Le Cam, A.; Montfort, J.; Bugeon, J.; Lefèvre, F.; Rescan, P.Y. Gene expression profiling of trout muscle during flesh quality recovery following spawning. BMC Genom. 2022, 23, 9. [Google Scholar] [CrossRef]
- Li, Z.; Du, X.; Wen, L.; Li, Y.; Qin, J.; Chen, Z.; Huang, Y.; Wu, X.; Luo, H.; Lin, Y.; et al. Transcriptome analysis reveals the involvement of ubiquitin-proteasome pathway in the regulation of muscle growth of rice flower carp. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 41, 100948. [Google Scholar] [CrossRef]
- Li, Y.; Xiao, T.; Zou, J. Fish TNF and TNF receptors. Sci. China Life Sci. 2021, 64, 196–220. [Google Scholar] [CrossRef]
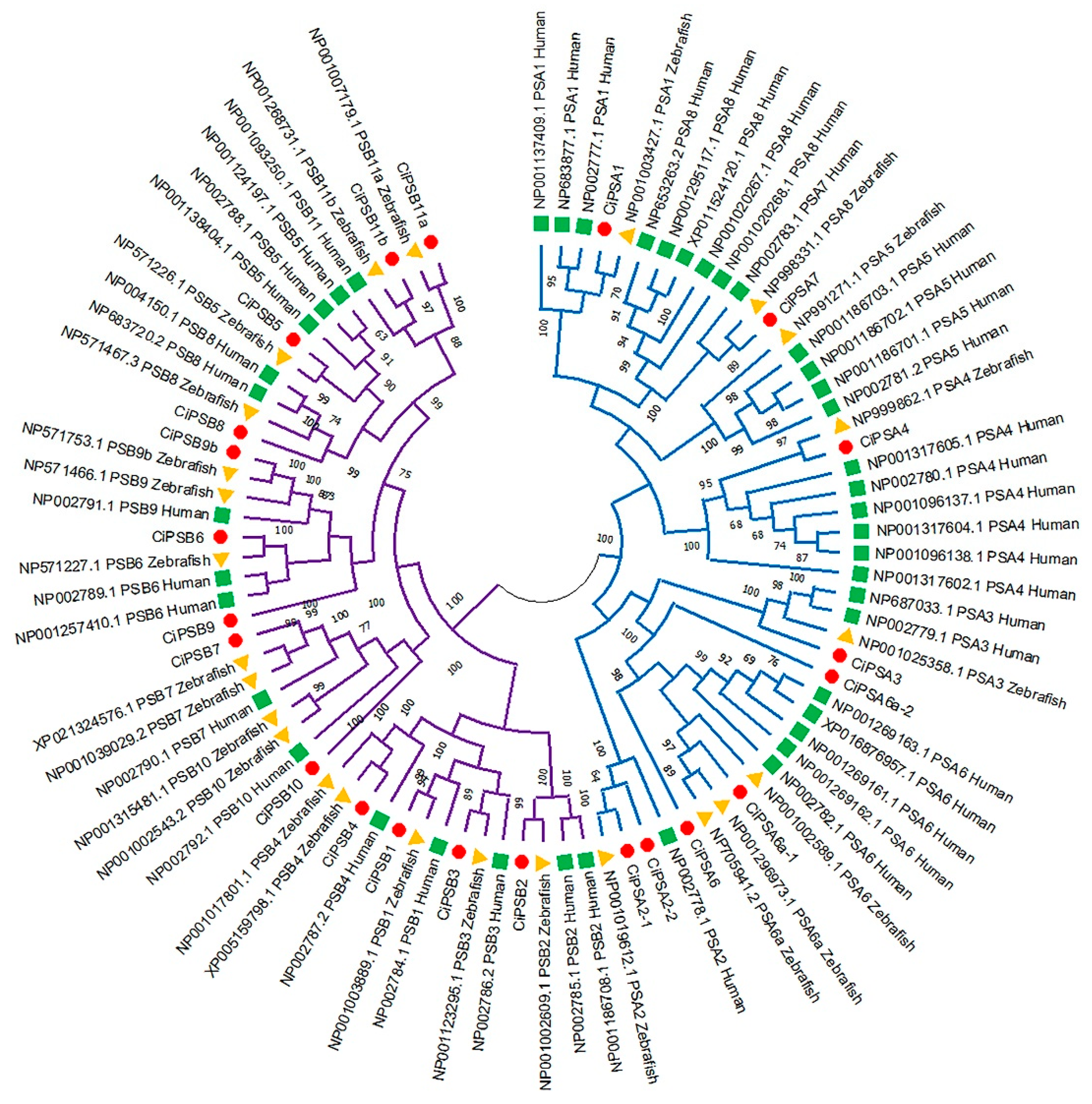
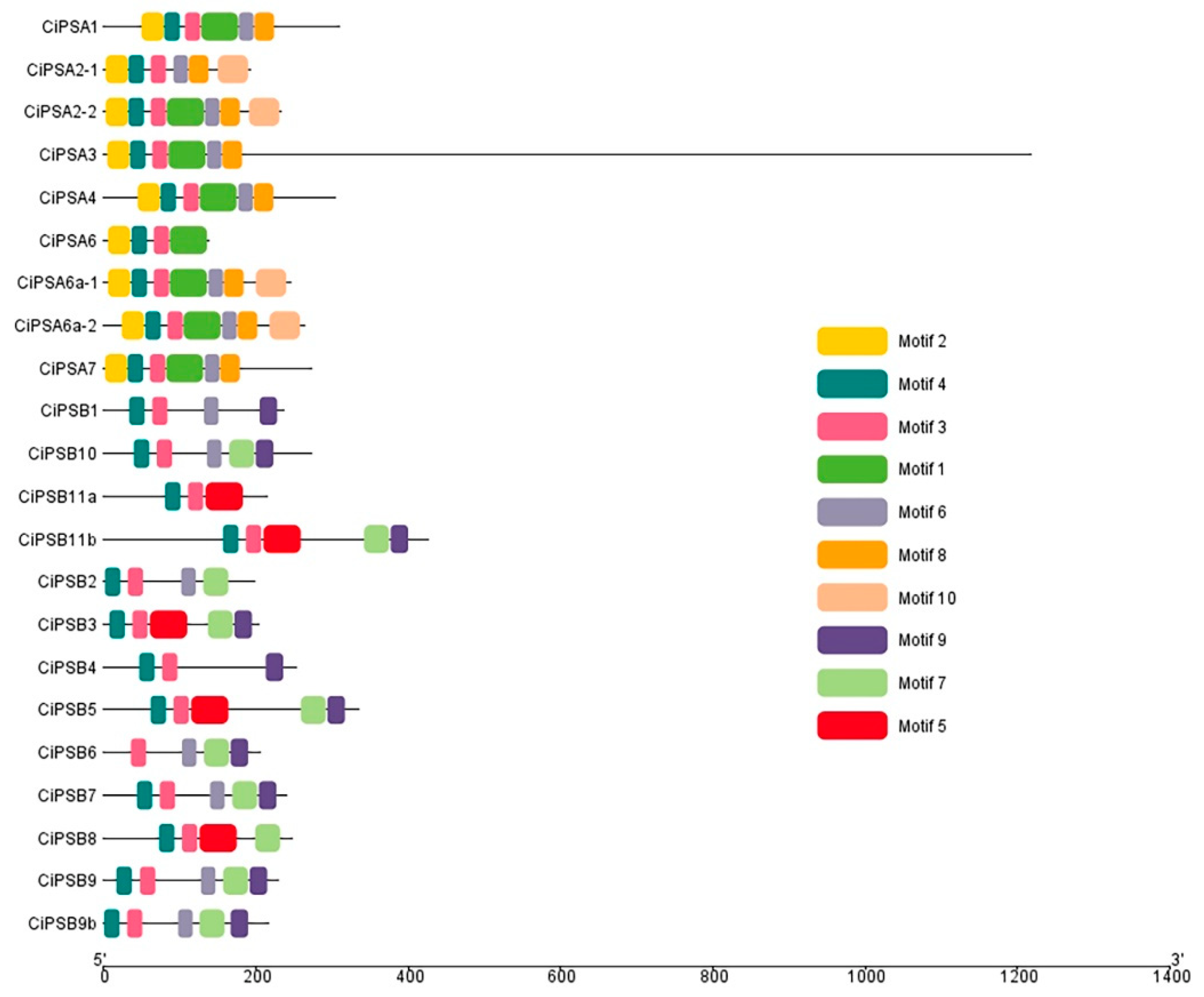

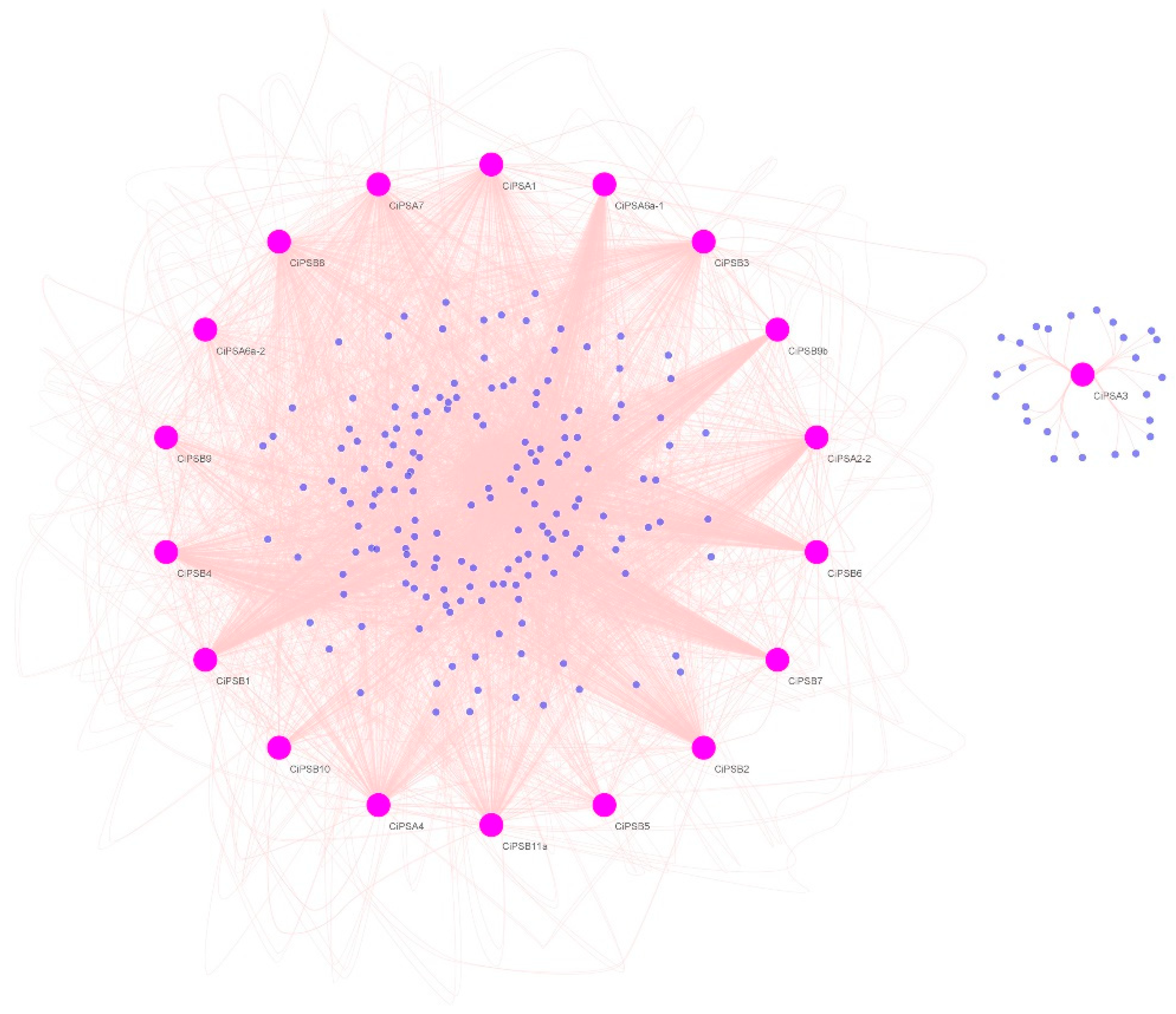

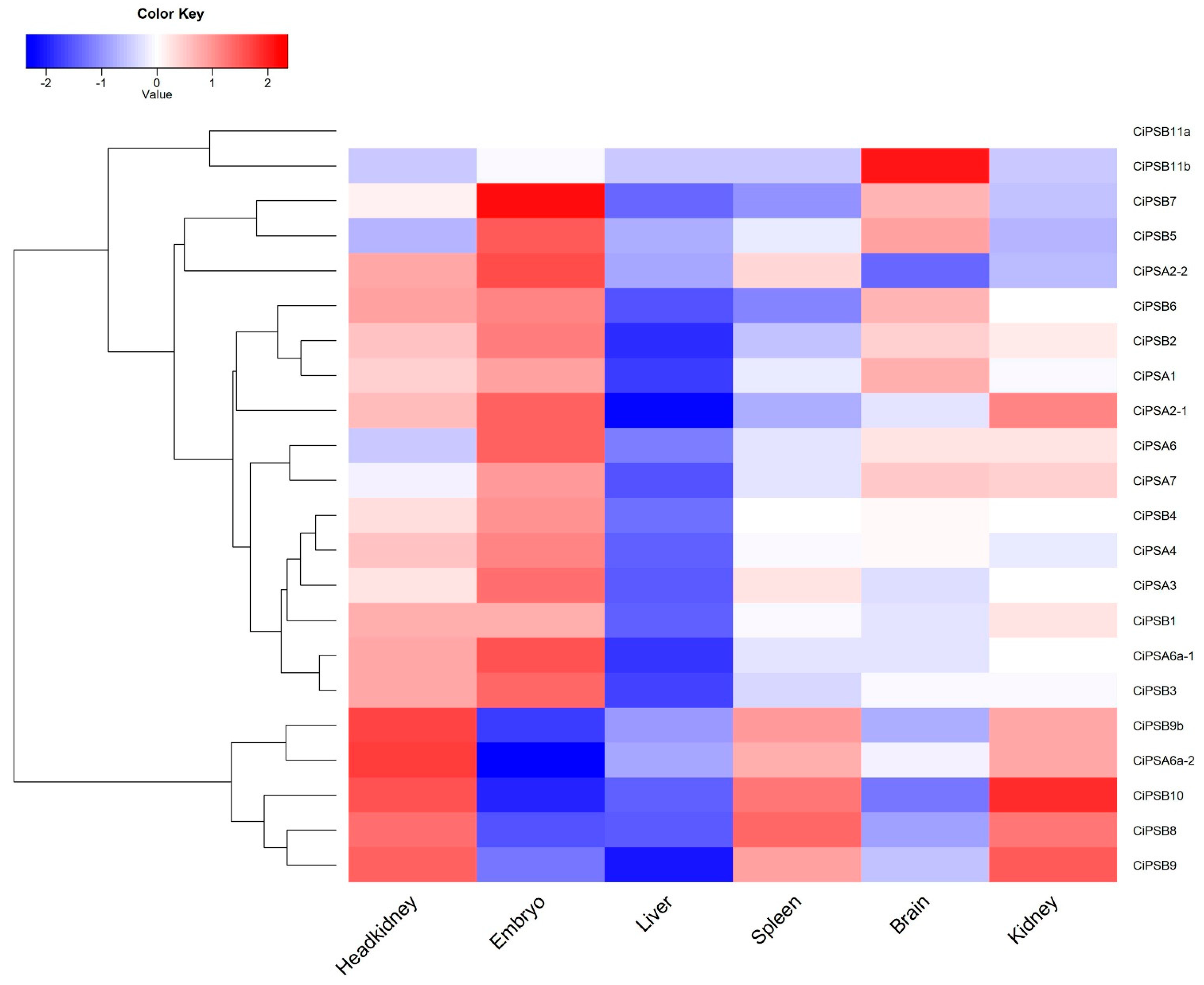
| Gene Name | Chromosome Segment | Gene Locus | Exon | Amino Acid | Subfamily |
|---|---|---|---|---|---|
| CiPSA1 | CI01000191 | 00591202_00594485 | 10 | 309 | A |
| CiPSA2-1 | CI01000027 | 02991231_02994560 | 7 | 193 | A |
| CiPSA2-2 | CI01000054 | 05825039_05827325 | 8 | 234 | A |
| CiPSA3 | CI01000310 | 04428945_04459038 | 30 | 1218 | A |
| CiPSA4 | CI01000139 | 00003689_00012005 | 10 | 304 | A |
| CiPSA6 | CI01000051 | 04165819_04167331 | 4 | 138 | A |
| CiPSA6a-1 | CI01000311 | 00675876_00680433 | 7 | 246 | A |
| CiPSA6a-2 | CI01000468 | 00039181_00047704 | 8 | 264 | A |
| CiPSA7 | CI01000051 | 00775546_00780498 | 7 | 273 | A |
| CiPSB1 | CI01000005 | 04829696_04837047 | 6 | 237 | B |
| CiPSB2 | CI01000223 | 00398156_00405578 | 6 | 199 | B |
| CiPSB3 | CI01000024 | 00161629_00165522 | 5 | 204 | B |
| CiPSB4 | CI01000054 | 01613638_01617674 | 6 | 254 | B |
| CiPSB5 | CI01000004 | 16221780_16232426 | 4 | 336 | B |
| CiPSB6 | CI01000129 | 01661645_01663621 | 5 | 205 | B |
| CiPSB7 | CI01000008 | 01141528_01145824 | 7 | 240 | B |
| CiPSB8 | CI01000319 | 05816948_05819499 | 5 | 247 | B |
| CiPSB9 | CI01000319 | 05808908_05812498 | 6 | 230 | B |
| CiPSB9b | CI01000319 | 05804236_05806955 | 5 | 216 | B |
| CiPSB10 | CI01000319 | 05812788_05816097 | 8 | 273 | B |
| CiPSB11a | CI01000004 | 16217935_16218580 | 1 | 215 | B |
| CiPSB11b | CI01000021 | 06402127_06406030 | 3 | 426 | B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.; Shu, Y.; Luan, P.; Zhang, T.; Chen, F.; Zheng, X. Genomic Analysis of the Proteasome Subunit Gene Family and Their Response to High Density and Saline-Alkali Stresses in Grass Carp. Fishes 2022, 7, 350. https://doi.org/10.3390/fishes7060350
Hu G, Shu Y, Luan P, Zhang T, Chen F, Zheng X. Genomic Analysis of the Proteasome Subunit Gene Family and Their Response to High Density and Saline-Alkali Stresses in Grass Carp. Fishes. 2022; 7(6):350. https://doi.org/10.3390/fishes7060350
Chicago/Turabian StyleHu, Guo, Yongjun Shu, Peixian Luan, Tianxiang Zhang, Feng Chen, and Xianhu Zheng. 2022. "Genomic Analysis of the Proteasome Subunit Gene Family and Their Response to High Density and Saline-Alkali Stresses in Grass Carp" Fishes 7, no. 6: 350. https://doi.org/10.3390/fishes7060350
APA StyleHu, G., Shu, Y., Luan, P., Zhang, T., Chen, F., & Zheng, X. (2022). Genomic Analysis of the Proteasome Subunit Gene Family and Their Response to High Density and Saline-Alkali Stresses in Grass Carp. Fishes, 7(6), 350. https://doi.org/10.3390/fishes7060350






