Footprints of Natural Selection in North Atlantic Eels: A Review
Abstract
1. Introduction
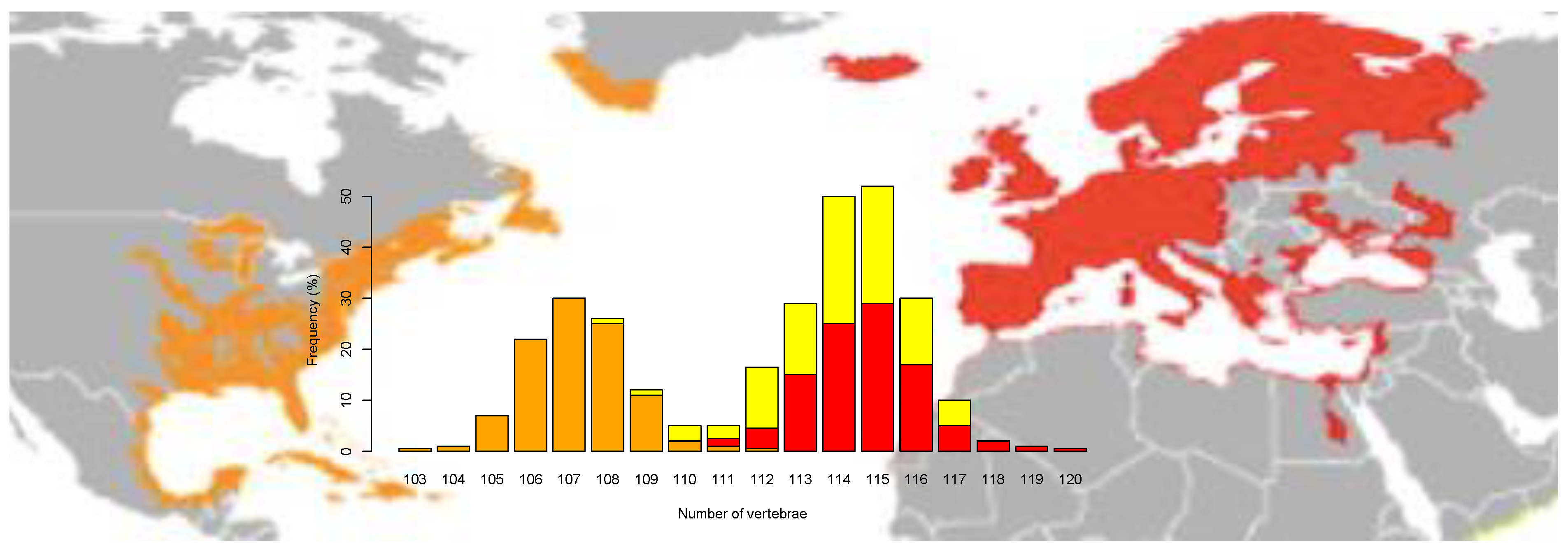
2. Excellent Species for the Study of Footprints of Selection
3. How Panmixia Affects the Detection of Signatures of Selection
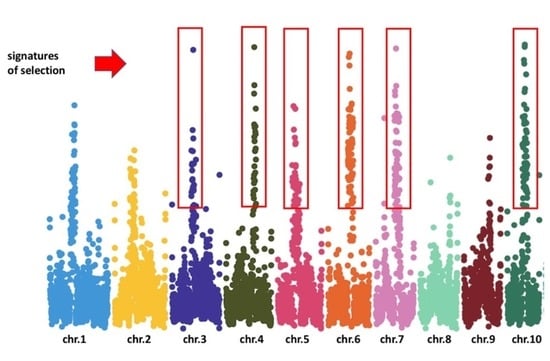
4. Studies of Selection in American Eel
5. Studies of Selection in European Eel
6. Signature of Selection between Life-Cycle Stages
7. Signatures of Selection between European and American Eel
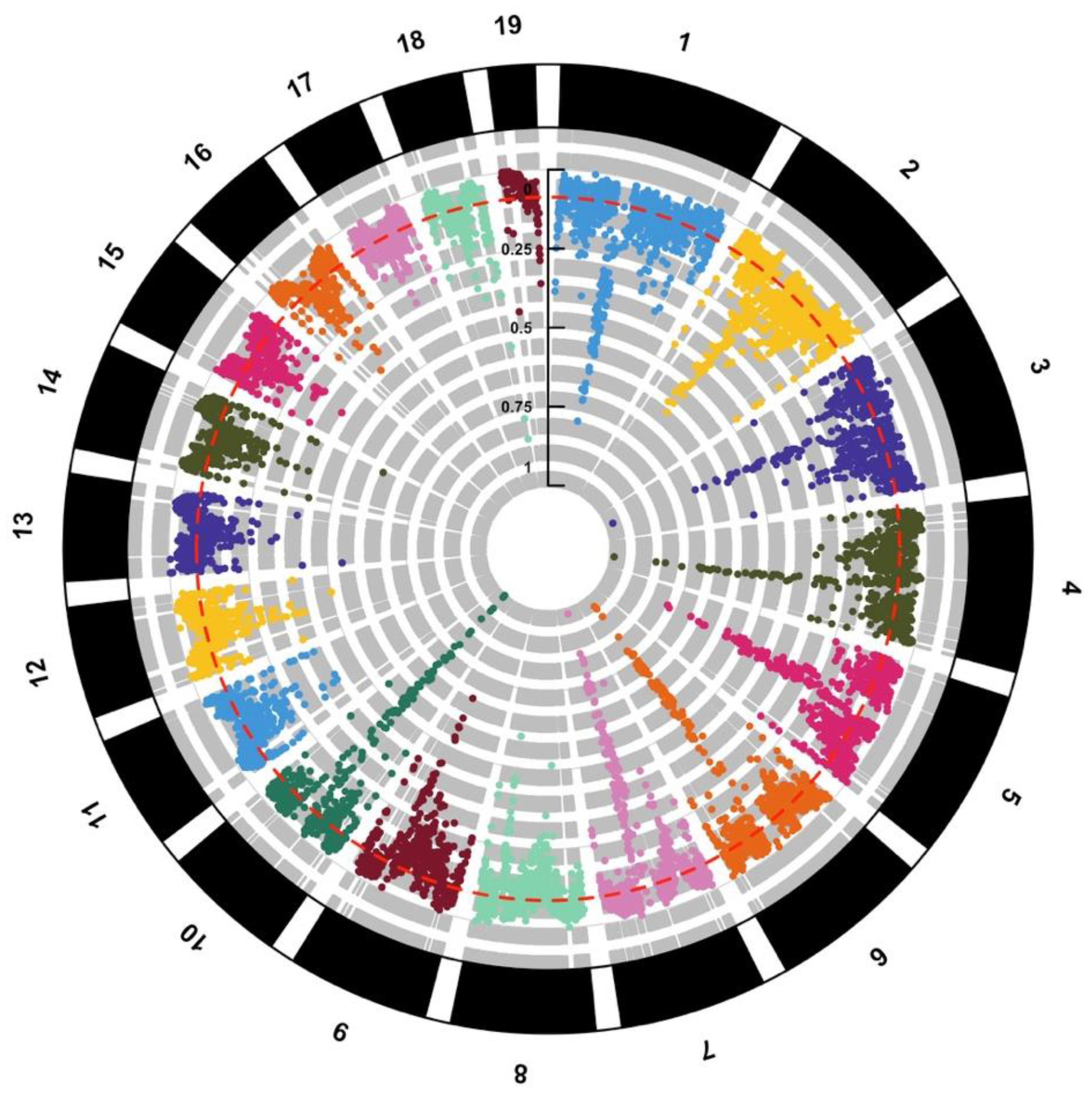
8. Genomic Islands of Divergence
9. Making Sense of Genomic Islands of Divergence
Author Contributions
Funding
Conflicts of Interest
References
- Stapley, J.; Reger, J.; Feulner, P.G.D.; Smadja, C.; Galindo, J.; Ekblom, R.; Bennison, C.; Ball, A.D.; Beckerman, A.P.; Slate, J. Adaptation genomics: The next generation. Trends Ecol. Evol. 2010, 25, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Bourret, V.; Dionne, M.; Kent, M.P.; Lien, S.; Bernatchez, L. Landscape genomics in Atlantic salmon (Salmo salar): Searching for gene-environment interactions driving local adaptation. Evolution 2013, 67, 3469–3487. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Fraser, D.J.; Weir, L.K.; Bernatchez, L.; Hansen, M.M.; Taylor, E.B. Extend and scale of local adaptation in salmonid fishes: Review and meta-analysis. Heredity 2011, 106, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Feder, J.L.; Nosil, P. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 2010, 64, 1729–1747. [Google Scholar] [CrossRef] [PubMed]
- Maynard Smith, J.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974, 23, 23–35. [Google Scholar] [CrossRef]
- Stephan, W. Selective sweeps. Genetics 2019, 211, 5–13. [Google Scholar] [CrossRef]
- Nosil, P.; Egan, S.P.; Funk, D.J. Heterogenous genomic differentiation between walking-stick ecotypes: “Isolation by adaptation” and multiple roles for divergent selection. Evolution 2008, 62, 316–336. [Google Scholar] [CrossRef]
- Hoffer, T.; Foll, M.; Excoffier, L. Evolutionary forces shaping genomic islands of population differentiation in humans. BMC Genomics 2012, 13, 107. [Google Scholar] [CrossRef]
- Hohenlohe, P.A.; Basshan, S.; Etter, P.D.; Stiffler, N.; Johnson, E.A.; Cresko, W.A. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010, 6, e1000862. [Google Scholar] [CrossRef]
- Jones, F.C.; Grabherr, M.G.; Chan, Y.F.; Russell, P.; Mauceli, E.; Johnson, J.; Swofford, R.; Pirun, M.; Zody, M.C.; White, S.; et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 2012, 484, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Hemmer-Hansen, J.; Nielsen, E.E.; Therkildsen, N.J.; Taylor, M.I.; Ogden, R.; Geffen, A.J.; Bekkevold, D.; Helyar, S.; Pampoulie, C.; Johansen, T.; et al. A genomic island linked to ecotype divergence in Atlantic cod. Mol. Ecol. 2013, 22, 2653–2667. [Google Scholar] [CrossRef]
- Rodríguez-Ramilo, S.T.; Baranski, M.; Moghadam, H.; Grove, H.; Lien, S.; Goddard, M.E.; Meuwissen, T.H.E.; Sonesson, A.K. Strong selection pressures maintain divergence on genomic islands in Atlantic cod (Gadus morhua) populations. Genet. Sel. Evol. 2019, 51, 61. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, P.A.; Pavey, S.A.; Normandeau, E.; Bernatchez, L. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assessed by RAD sequencing. Evolution 2013, 67, 2483–2497. [Google Scholar] [CrossRef] [PubMed]
- Als, T.D.; Hansen, M.M.; Maes, G.E.; Castonguay, M.; Riemann, L.; Aerestrup, K.; Munk, P.; Sparholt, T.; Hanel, R.; Bernatchez, L. All roads lead to home: Panmixia of European eel in the Sargasso Sea. Mol. Ecol. 2011, 20, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Côté, C.; Gagnaire, P.A.; Bourret, V.; Verrault, G.; Castonguay, M.; Bernatchez, L. Population genetics of the American eel (Anguilla rostrata): FST = 0 and North Atlantic Oscillation effects on demographic fluctuations of a panmictic species. Mol. Ecol. 2013, 22, 1763–1776. [Google Scholar] [CrossRef]
- Pujolar, J.M.; Jacobsen, M.W.; Als, T.D.; Frydenberg, J.; Munch, K.; Jónsson, B.; Jiang, X.; Cheng, L.; Maes, G.E.; Bernatchez, L.; et al. Genome-wide signatures of within-generation local selection in the panmictic European eel. Mol. Ecol. 2014, 23, 2514–2528. [Google Scholar] [CrossRef]
- Enbody, E.D.; Petterson, M.E.; Sprehn, C.G.; Palm, S.; Wickstrom, H.; Andersson, L. Ecological adaptation in European eels is based on phenotypic plasticity. Proc. Natl. Acad. Sci. USA 2021, 118, e2022620118. [Google Scholar] [CrossRef]
- Nikolic, N.; Liu, S.; Jacobsen, M.W.; Jónsson, B.; Bernatchez, L.; Gagnaire, P.A.; Hansen, M.M. Speciation history of European (Anguilla Anguilla) and American eel (A. rostrata), analysed using genomic data. Mol. Ecol. 2020, 29, 565–577. [Google Scholar] [CrossRef]
- Daverat, F.; Limburg, K.E.; Thibaut, I.; Shiao, J.C.; Dodson, J.J.; Caron, F.; Tzeng, W.N.; Iizuka, Y.; Wickstrom, H. Phenotypic plasticity of habitat use by three temperate eel species Anguilla anguilla, A. japonica and A. rostrata. Mar. Ecol. Progr. Ser. 2006, 308, 231–241. [Google Scholar] [CrossRef]
- Tesch, F. The Eel; Blackwell Science Ltd.: Oxford, UK, 2003. [Google Scholar]
- Bonhommeau, S.; Blanke, B.; Tréguier, A.M.; Grima, N.; Rivot, E.; Vermand, Y.; Greiner, E.; Le Pape, O. How fast can the European eel (Anguilla anguilla) larvae cross the Atlantic Ocean? Fish. Oceanogr. 2009, 18, 371–385. [Google Scholar] [CrossRef]
- Åström, M.; Dekker, W. When will the eel recover? A full life cycle model. ICES J. Mar. Sci. 2007, 64, 1491–1498. [Google Scholar] [CrossRef]
- Van den Thillart, G.; Rankin, J.C.; Dufour, S. Spawning Migration of the European Eel: Reproduction Index, a Useful Tool for Conservation Management; Springer: Dordecht, The Netherlands, 2009. [Google Scholar]
- Wirth, T.; Bernatchez, L. Decline of Atlantic eels: A fatal synergy? Proc. Biol. Sci. 2003, 270, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Dannewitz, J.; Maes, G.E.; Johansson, L.; Wickström, H.; Volckaert, F.A.M.; Jarvi, T. Panmixia in the European eel: A matter of time. Proc. Biol. Sci. 2005, 272, 1129–1137. [Google Scholar] [CrossRef]
- Maes, G.E.; Pujolar, J.M.; Hellemans, B.; Volckaert, F.A.M. Evidence for isolation by time in the European eel (Anguilla Anguilla). Mol. Ecol. 2006, 15, 2095–2107. [Google Scholar] [CrossRef]
- Palm, S.; Dannewitz, J.; Prestegaard, T.; Wickström, H. Panmixia in European eel revisited: No genetic difference between maturing adults from southern and northern Europe. Heredity 2009, 103, 82–89. [Google Scholar] [CrossRef]
- Baltazar-Soares, M.; Biastoch, A.; Harrod, C.; Hanel, R.; Marohn, L.; Prigge, E.; Evans, D.; Bodles, K.; Behrens, E.; Böning, C.W.; et al. Recruitment collapse and population structure of the European eel shaped by local ocean current dynamics. Curr. Biol. 2014, 24, 104–108. [Google Scholar] [CrossRef]
- Jacobsen, M.W.; Pujolar, J.M.; Gilbert, M.T.P.; Moreno-Mayar, J.V.; Bernatchez, L.; Als, T.D.; Lobón-Cervià, J.; Hansen, M.M. Speciation and demographic history of Atlantic eels (Anguilla anguilla and A. rostrata) revealed by mitogenome sequencing. Heredity 2014, 113, 432–442. [Google Scholar] [CrossRef]
- Gagnaire, P.A.; Normandeau, E.; Côté, C.; Hansen, M.M.; Bernatchez, L. The genetic consequences of spatially varying selection in the panmictic American eel (Anguilla rostrata). Genetics 2012, 190, 725–736. [Google Scholar] [CrossRef]
- Ulrik, M.G.; Pujolar, J.M.; Ferchaud, A.L.; Jacobsen, M.W.; Als, T.D.; Gagnaire, P.A.; Frydenberg, J.; Bøcher, P.K.; Jónsson, B.; Bernatchez, L.; et al. Do North Atlantic eels show parallel patterns of spatially varying selection? BMC Evol. Biol. 2014, 14, 138. [Google Scholar] [CrossRef]
- Jacobsen, M.W.; Pujolar, J.M.; Bernatchez, L.; Munch, K.; Jian, J.; Niu, Y.; Hansen, M.M. Genomic footprints of speciation in Atlantic eels Anguilla anguilla and A. rostrata. Mol. Ecol. 2014, 23, 4785–4798. [Google Scholar] [CrossRef] [PubMed]
- Pavey, S.A.; Gaudin, J.; Normandeau, E.; Dionne, M.; Castonguay, M.; Audet, C.; Bernatchez, L. RAD sequencing highlights polygenic discrimination of habitat ecotypes in the panmictic American eel. Curr. Biol. 2015, 25, 1666–1671. [Google Scholar] [CrossRef] [PubMed]
- Pujolar, J.M.; Jacobsen, M.W.; Bekkevold, D.; Lobon-Cervià, J.; Jónsson, B.; Bernatchez, L.; Hansen, M.M. Signatures of natural selection between life cycles stages separated by metamorphosis in European eel. BMC Genom. 2015, 16, 600. [Google Scholar] [CrossRef] [PubMed]
- Pujolar, J.M.; Jacobsen, M.W.; Bertolini, F. Comparative genomics and signatures of selection in North Atlantic eels. Mar. Genom. 2022, 62, 100933. [Google Scholar] [CrossRef]
- Ling, S.; Tengstedt, A.N.B.; Jacobsen, M.W.; Pujolar, J.M.; Jónsson, B.; Lobón-Cervià, J.; Bernatchez, L.; Hansen, M.M. Genome-wide methylation in the panmictic European eel (Anguilla anguilla). Mol. Ecol. 2022, 31, 4286–4306. [Google Scholar] [CrossRef]
- Somero, G.N. Adaptation of enzymes to temperature: Searching for basic “strategies”. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 139, 321–333. [Google Scholar] [CrossRef]
- Nyman, L. Some effects of temperature on eel (Anguilla) behaviour. Rep. Inst. Freshw. Res. Drottingholm 1972, 52, 90–102. [Google Scholar]
- Walsh, P.J.; Foster, G.D.; Moon, T.W. The effects of temperature and metabolism of the American eel Anguilla rostrata: Compensation in the summer and torpor in the winter. Physiol. Zool. 1983, 56, 532–540. [Google Scholar] [CrossRef]
- Linton, E.D.; Jónsson, B.; Noakes, D.L.G. Effects of water temperature on the swimming and climbing behavior of glass eels Anguilla spp. Environ. Biol. Fishes 2007, 78, 189–192. [Google Scholar] [CrossRef]
- Heyland, A.; Moroz, L.L. Signalling mechanisms underlying metamorphic transitions in animals. Integr. Comp. Biol. 2006, 46, 743–759. [Google Scholar] [CrossRef]
- Moran, N.A. Adaptation and constraint in the complex life cycles of animals. Annu. Rev. Ecol. Syst. 1994, 25, 573–600. [Google Scholar] [CrossRef]
- Avise, J.C.; Nelson, W.S.; Arnold, J.; Koehn, R.K.; Williams, G.C.; Thorsteinsson, V. The evolutionary genetic status of Icelandic eels. Evolution 1990, 44, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Lecomte-Finiger, R. The genus Anguilla: Current state of knowledge and questions. Rev. Fish. Biol. Fish. 2003, 13, 265–279. [Google Scholar] [CrossRef]
- Van Ginneken, V.J.T.; Maes, G.E. The European eel (Anguilla Anguilla), its life cycle, evolution and reproduction: A literature review. Rev. Fish. Biol. Fish. 2005, 15, 367–398. [Google Scholar] [CrossRef]
- Mineguishi, Y.; Aoyama, J.; Inoue, J.G.; Miya, M.; Nishida, M.; Tsukamoto, K. Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Mol. Phylogenet. Evol. 2005, 34, 134–146. [Google Scholar] [CrossRef]
- Mank, J.E.; Avise, J.C. Microsatellite variation and differentiation in North Atlantic eels. J. Hered. 2003, 94, 30–34. [Google Scholar] [CrossRef][Green Version]
- Gagnaire, P.A.; Albert, V.; Jónsson, B.; Bernatchez, L. Natural selection influences AFLP intraspecific variability and introgression patterns in Atlantic eels. Mol. Ecol. 2009, 18, 1678–1691. [Google Scholar] [CrossRef]
- Miller, M.J.; Bonhommeau, S.; Munk, P.; Castonguay, M.; Hanel, R.; McCleave, J.D. A century of research on the larval distribution of the Atlantic eels: A re-examination of the data. Biol. Rev. Camb. Philos. Soc. 2015, 90, 1035–1064. [Google Scholar] [CrossRef]
- Albert, V.; Jónsson, B.; Bernatchez, L. Natural hybrids in Atlantic eels (Anguilla anguilla, A. rostrata): Evidence for successful reproduction and fluctuating abundance in space and time. Mol. Ecol. 2006, 15, 1903–1916. [Google Scholar] [CrossRef]
- Pujolar, J.M.; Jacobsen, M.W.; Als, T.D.; Frydenberg, J.; Magnussen, E.; Jónsson, B.; Jiang, X.; Cheng, L.; Bekkevold, D.; Maes, G.E.; et al. Assessing patterns of hybridization between North Atlantic eels using diagnostic single nucleotide polymorphisms. Heredity 2014, 112, 627–637. [Google Scholar] [CrossRef]
- Jacobsen, M.; Smedegaard, L.; Sørensen, S.; Pujolar, J.M.; Munk, P.; Jónsson, B.; Magnussen, E.; Hansen, M.M. Assessing pre- and post-zygotic barriers between North Atlantic eels (Anguilla anguilla and A. rostrata). Heredity 2017, 118, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, D.M.P.; Casselman, J.M.; Crooks, V.; Delucia, M.B.; Ahn, H.; Kaifu, K.; Kurwie, T.; Sasal, P.; Silfvergrip, A.M.C.; Smith, K.; et al. Synergistic patterns of threat and the challenges facing global anguillid eel conservation. Glob. Ecol. Conserv. 2015, 4, 321–333. [Google Scholar] [CrossRef]
- Henkel, C.V.; Burgerhout, E.; de Wijze, D.L.; Dirks, R.P.; Minegishi, Y.; Jansen, H.J.; Spaink, H.P.; Dufour, S.; Weltzien, F.A.; Tsukamoto, K.; et al. Primitive duplicate Hox clusters in the European eel’s genome. PLoS ONE 2012, 7, e32231. [Google Scholar] [CrossRef] [PubMed]
- Rhie, A.; McCarthy, S.A.; Fedrigo, O.; Damas, J.; Formenti, G.; Koren, S.; Uliano-Silva, M.; Chow, W.; Fungtammasan, A.; Kim, J.; et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 2021, 592, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ding, X.; Qanbari, S.; Weigend, S.; Zhang, Q.; Simianer, H. Properties of different selection signature statistics and a new strategy to combine them. Heredity 2015, 115, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Cadzow, M.; Boocock, J.; Nguyen, H.T.; Wilcox, P.; Merriman, T.R.; Black, M.A. A bioinformatics workflow for detecting signatures of selection in genomic data. Front. Genet. 2014, 5, 293. [Google Scholar] [CrossRef]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef]
- Hermisson, J.; Pennings, P.S. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics 2005, 169, 2335–2352. [Google Scholar] [CrossRef]
- Via, S.; West, J. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol. Ecol. 2008, 17, 4334–4345. [Google Scholar] [CrossRef]
- Storz, J.F. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 2005, 14, 671–688. [Google Scholar] [CrossRef]
- Bernatchez, L.; St-Cyr, J.; Normandeau, E.; Maes, G.E.; Als, T.D.; Kalujnaia, S.; Cramb, G.; Castonguay, M.; Hansen, M.M. Differential timing of gene expression between leptocephali of the two Anguilla eel species in the Sargasso Sea. Ecol. Evol. 2011, 1, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Dupond, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef]
- Srikant, T.; Drost, H.H. How stress facilitates phenotypic innovation through epigenetic diversity. Front. Plant Sci. 2020, 11, 606800. [Google Scholar] [CrossRef] [PubMed]
| Study | Species | Marker | Sampling Details | Main Results |
|---|---|---|---|---|
| Gagnaire et al. [31] | American eel | 100 candidate SNPs | A total of 992 individuals from 16 sampling sites from Florida to Quebec | Positive correlations at 8 loci with environmental variables, mostly related to energy production and metabolism |
| Ulrik et al. [32] | European eel | 80 candidate SNPs | A total of 321 glass eels collected at 8 sampling sites from Iceland to the Mediterranean | Positive associations at 11 loci with environmental variables, mostly related to metabolism |
| Pujolar et al. [17] | European eel | RAD sequencing | A total of 259 glass eels from 8 sampling sites ranging from Iceland to Morocco | A total of 754 loci under selection with a variety of functions including calcium signaling and circadian rhythms |
| Jacobsen et al. [33] | European and American eel | RAD sequencing | A total of 30 American eel and 30 European eel individuals either glass or yellow eels | A total of 3757 highly differentiated candidate SNPs, located in genes mostly related to development and energy production |
| Pavey et al. [34] | American eel | RAD sequencing | A total of 379 individuals from 8 locations each of freshwater and brackish/marine habitats from the Atlantic Canada and St. Lawrence river region | A total of 331 loci under selection associated with environmental variables showing differences between ecotypes (freshwater vs. brackish/marine) |
| Pujolar et al. [35] | European eel | 80 candidate SNPs + RAD sequencing | A total of 123 glass eels and 113 silver eels collected in Iceland, Ireland and Spain | A total of 1413 potentially selected SNPs, located in genes related to growth among other functions |
| Enbody et al. [18] | European eel | Whole-genome sequencing | A total of 417 Europeam eel samples from 10 locations across Europe | When comparing Baltic vs. Mid-Atlantic samples only a small region under selection located on chromosome 1 and two other regions on chromosomes 13 and 15 were found |
| Pujolar et al. [36] | European and American eel | RAD sequencing | Re-analysis of 359 samples retrieved from Genbank, including 254 European eel and 105 American eel | Islands of divergence were detected at 7 chromosomes, with candidate genes involved in energy production, development and regulation |
| Liu et al. [37] | European eel | DNA methylation | A total of 50 individuals were analysed representing 7 localities in Europe and northern Africa | Differentially methylated regions were reported including genes involved in development, in particular Hox genes. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pujolar, J.M.; Bertolini, F.; Jacobsen, M.W. Footprints of Natural Selection in North Atlantic Eels: A Review. Fishes 2022, 7, 311. https://doi.org/10.3390/fishes7060311
Pujolar JM, Bertolini F, Jacobsen MW. Footprints of Natural Selection in North Atlantic Eels: A Review. Fishes. 2022; 7(6):311. https://doi.org/10.3390/fishes7060311
Chicago/Turabian StylePujolar, José Martin, Francesca Bertolini, and Magnus W. Jacobsen. 2022. "Footprints of Natural Selection in North Atlantic Eels: A Review" Fishes 7, no. 6: 311. https://doi.org/10.3390/fishes7060311
APA StylePujolar, J. M., Bertolini, F., & Jacobsen, M. W. (2022). Footprints of Natural Selection in North Atlantic Eels: A Review. Fishes, 7(6), 311. https://doi.org/10.3390/fishes7060311