Transcriptome-Based Analysis of the Response Mechanism of Leopard Coralgrouper Liver at Different Flow Velocities
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Enzyme Activity Analysis
2.3. RNA Isolation and Transcriptome Sequencing
2.4. Data Assembly, Annotation, and Variance Analysis
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Data Processing
3. Results and Analysis
3.1. Growth Analysis
3.2. Enzyme Activity Analysis
3.3. Transcriptome Sequencing
3.4. Differentially Expressed Gene Analysis
3.5. Quantitative Real-Time PCR (qRT-PCR)
3.6. GO and KEGG Analysis
4. Discussion
4.1. The Effect of Flow Velocity on Fish Growth
4.2. Effects of Various Flow Velocities on P. leopardus
4.3. Regulatory Pathway
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, D.S.; Kim, S.Y.; Chung, J.H.; Kim, K.H.; Eun, H.C.; Park, K.C. Delayed ERK activation by ceramide reduces melanin synthesis in human melanocytes. Cell. Signal. 2002, 14, 779–785. [Google Scholar] [CrossRef]
- Wang, R.; Chen, T.; Zhao, B.; Fan, R.; Ji, K.; Yu, X.; Wang, X.; Dong, C. FGF21 regulates melanogenesis in alpaca melanocytes, via, ERK1/2-mediated MITF downregulation. Biochem. Biophys. Res. Commun. 2017, 490, 466–471. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, S.Y.; Lee, J.E.; Kwon, S.B.; Joo, Y.H. Sphingosine-1-phosphate-induced ERK activation protects human melanocytes from UVB-induced apoptosis. Arch. Pharmacal Res. 2003, 26, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Mayrhofer, M.; Gourain, V.; Reischl, M.; Affaticati, P.; Jenett, A.; Joly, J.S.; Benelli, M.; Demichelis, F.; Poliani, P.L.; Sieger, D.; et al. A novel brain tumour model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. Dis. Models Mech. 2017, 10, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.; Roberts, C.; Hawkins, J. The threatened status of groupers (Epinephelinae). Biodivers. Conserv. 2000, 9, 919–942. [Google Scholar] [CrossRef]
- Carter, A.B.; Russ, G.R.; Tobin, A.J.; Williams, A.J.; Davies, C.R.; Mapstone, B.D. Spatial variation in the effects of size and age on reproductive dynamics of common coral trout Plectropomus leopardus. J. Fish Biol. 2014, 84, 1074–1098. [Google Scholar] [CrossRef]
- Tu, Z.G.; Jiang, Y.F.; Qiu, M.Y. Current Situation and Development Suggestions of Breeding of Plectropomus leopardus in Yazhou District, Sanya. China Fish. 2009, 2009, 54–55. [Google Scholar]
- Tang, X.M.; Lee, X.M.; Chen, J.M. The advantages and industrialization status of Hainan’s development of deep-water anti-wind and wave cage fish culture. In Proceedings of the 2009 National Symposium on Marine Aquaculture, Lianyungang, China, 6 November 2009; pp. 51–59. [Google Scholar]
- Hockley, A.F.; Wilson, C.A.M.E.; Brew, A.; Cable, J. Fish responses to flow velocity and turbulence in relation to size, sex and parasite load. J. R. Soc. Interface 2014, 8, 20130814. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Y.B.; Liu, J.Y.; Gu, Y.L.; Fu, S.Y. Transcriptome analysis of Plectropomus leopardus liver under different flow velocity. S. China Fish. Sci. 2022, 18, 109–117. [Google Scholar]
- Li, D.; Lin, X.T.; Zhu, Z.M.; Yi, M.M. Effect of Flow Rate on Swimming States and Activity Metabolism in Juvenile Hybrid Sturgeon. Acta Hydrobiol. Sin. 2011, 35, 578–585. [Google Scholar]
- Ogata, H.Y.; Oku, H. Effects of water velocity on growth performance of juvenile Japanese flounder Paralichthys olivaceus. J. World Aquac. Soc. 2000, 31, 225–231. [Google Scholar] [CrossRef]
- German, E.M.; Piedrahita, R.H.; Conklin, D.E. Effect of water velocity on the growth of California halibut (Paralichthys californicus) juveniles. Aquaculture 2007, 271, 206–215. [Google Scholar]
- Sun, G.X.; Li, M.; Wang, J.; Liu, Y. Effects of flow rate on growth performance and welfare of juvenile turbot (Scophthalmus maximus L.) in recirculating aquaculture systems. Aquac. Res. 2014, 47, 1341–1352. [Google Scholar] [CrossRef]
- Huang, N.N.; Cheng, Q.Q.; Gao, L.L.; Me, Z.L.; Xia, L.J. Effect of water current and temperature on growth of juvenile Acipenser baaeri. J. Fish. China 2007, 1, 31–37. [Google Scholar]
- Simonato, J.D.; Mela, M.; Doria, H.B.; Guiloski, I.C.; Randi, M.A.F.; Carvalho, P.S.M.; Meletti, P.C.; Assis, H.C.S.D.; Bianchini, A.; Martinez, C.B.R. Biomarkers of waterborne copper exposure in the Neotropical fish Prochilodus lineatus. Aquat. Toxicol. 2016, 170, 31–41. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, Z.; Ji, M.; Zhou, F.; Zhao, J. Effects of flow velocity on growth and physiology of juvenile largemouth bass (Micropterus salmoides) in recirculating aquaculture systems. Aquac. Res. 2021, 52, 3093–3100. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef]
- Cloonan, N.; Grimmond, S.M. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008, 9, 234. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.J.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Lin, M.D.; Chen, G.; Ma, J.; Wang, Z.L.; Zhang, J.D.; Xie, R.T.; Tang, B.G.; Huang, J.S.; Zhou, H.; Shi, G.; et al. Differentially Expressed Genes Analysis of Hybrid Grouper and Female Tiger Grouper Based on Transcriptome Sequencing. Fisheies Coll. Guangdong Ocean. Univ. 2019, 39, 15–23. [Google Scholar]
- Juliana, C.S.; Douglas, D.; Martins, L.F.; Wei, Z. RNA-Seq differential expression analysis: An extended review and a software tool. PLoS ONE 2017, 12, e0190152. [Google Scholar]
- Xi, Q.; Yi, B.; Zhuang, Q.; Zhong, G. RNA-Seq technology and its application in fish transcriptomics. Omics 2014, 18, 98–110. [Google Scholar]
- Luo, H.; Ye, H.; Xiao, S.J.; Zheng, S.M.; Wang, X.Q.; Wang, Z.Y. Application of transcriptomics technology to aquatic animal research. J. Fish. China 2015, 39, 598–607. [Google Scholar]
- Meng, W.; Xu, K.D.; Li, Z.H.; Zhou, Y.D. Transcriptome analysis of Nibea japonica under acute salinity stress. J. Fish. China 2021, 45, 649–660. [Google Scholar]
- Wang, C.H.; Wachholtz, M.; Wang, J.; Liao, X.L.; Lu, G.Q. Analysis of the skin transcriptome in two Oujiang color varieties of common carp. PLoS ONE 2014, 9, e90074. [Google Scholar] [CrossRef]
- Hu, J.R.; Yan, X.C.; Hang, W.; Li, G.L.; Zhang, Y.F.; Wang, G.X.; Zhao, H.X.; Hang, Y.H. Analysis of Different Selenium Sources on Regulation Signal Pathways and Key Gene of Pelteobrus fulvidraco under Low Temperature Stress Based on Transcriptome. Chin. J. Anim. Nutr. 2021, 33, 4662–4674. [Google Scholar]
- Wang, H.M.; Ding, X.J.; Chen, L.; Song, S.G.; Tan, B.P. Effect of Dietary Manman-Oligosaccharides on Growth Performace, Serum Immune Indices, Transcriptome and Intestinal Microflora of Epinephelus fuscoguttatus♀×Epinephelus lanceolatus♂. Chin. J. Anim. Nutr. 2021, 33, 6982–6998. [Google Scholar]
- Liu, M.; Yan, J.L.; Lian, Q.P.; Ni, M.; Gu, Z.M. Effects of Different Water Flow Rates on the Growth Performance, Antioxidant Capacity, Energy Metabolism and Tissue Structure of Micropterus Salmoides under An In-Pone Recirculating Aquanculture System. Acta Hydrobiol. Sin. 2022, 1–27. [Google Scholar]
- Wei, X.L.; Yu, S.; Yang, Y.; Xiao, Y.; Li, C. Effects of exercise intensity on growth, blood innate immunity, hepatic antioxidant capacity, and HSPs70 mRNA expression of Epinephelus coioides. J. Fish. Sci. China 2017, 24, 1055–1064. [Google Scholar] [CrossRef]
- Young, P.S.; Cech, J.J., Jr. Optimum exercise conditioning speed for growth, muscular development, and swimming performance in young-of-the-year striped bass (Morone saxatilis). Can. J. Fish. Aquat. Sci. 1994, 51, 1519–1527. [Google Scholar] [CrossRef]
- Young, P.S.; Cech, J.J., Jr. Improved growth, swimming performance, and muscular development in exercise-conditioned young-of-the-year Striped Bass (Morone saxatilis). Can. J. Fish. Aquat. Sci. 1993, 50, 703–707. [Google Scholar] [CrossRef]
- Xu, Y.Q. Effects of Flow Rate on Growth, Nonspecific Immunity and Fatty Acid Composition in Juvenile Phoxinus lagowskii Dybowskii. Master’s Thesis, Dalian Ocean University, Dalian, China, 2020; pp. 8–19. [Google Scholar]
- Wen, X.; Hu, Y.; Zhang, X.; Wei, X.Z.; Wang, T.; Yin, S.W. Integrated application of multi-omics provides insights into cold stress responses in pufferfish Takifugu fasciatus. BMC Genomices 2019, 20, 563. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhang, X.; Hu, Y.; Wang, T.; Yin, S. iTRAQ-based quantitative proteomic analysis of Takifugu fasciatus liver in response to low-temperature stress. J. Proteom. 2019, 201, 27–36. [Google Scholar] [CrossRef]
- Azuma, T.; Noda, S.; Yada, T.; Ototake, M.; Nagoya, H.; Moriyama, S.; Yamada, H.; Nakanishi, T.; Iwata, M. Profiles in growth, smoltification, immune function and swimming performance of 1-year-old masu salmon (Oncorhynchus masou) reared in water flow. Jpn. Soc. Fish. Sci. 2002, 68, 1282–1294. [Google Scholar] [CrossRef]
- Song, B.L. Effects of Water Current Onswimming Activity, Growth and Ecophysiologocal Aspect of Young Barbodes Schwanenfeldi. Ph.D. Thesis, Jinan University, Guangzhou, China, 2008; pp. 8–101. [Google Scholar]
- Zhu, Z.M. Research of Carbohydrate and Lipid Metabolism in Muscles and Liver in Barbodes Schwanenfeldi during Exercise Training. Ph.D. Thesis, Jinan University, Guangzhou, China, 2014; pp. 28–55. [Google Scholar]
- Fam, B.C.; Joannides, C.N.; Andrikopoulos, S. The liver: Key in regulating appetite and body weight. Adipocyte 2012, 1, 259–264. [Google Scholar] [CrossRef]
- Wu, Q.Y.; Zeng, L.Q.; Cao, Z.D.; Peng, J.L.; Fu, S.J. The preferred water velocity behavior of six Cyprinids with different feeding habits. J. Fish. China 2014, 39, 1807–1816. [Google Scholar]
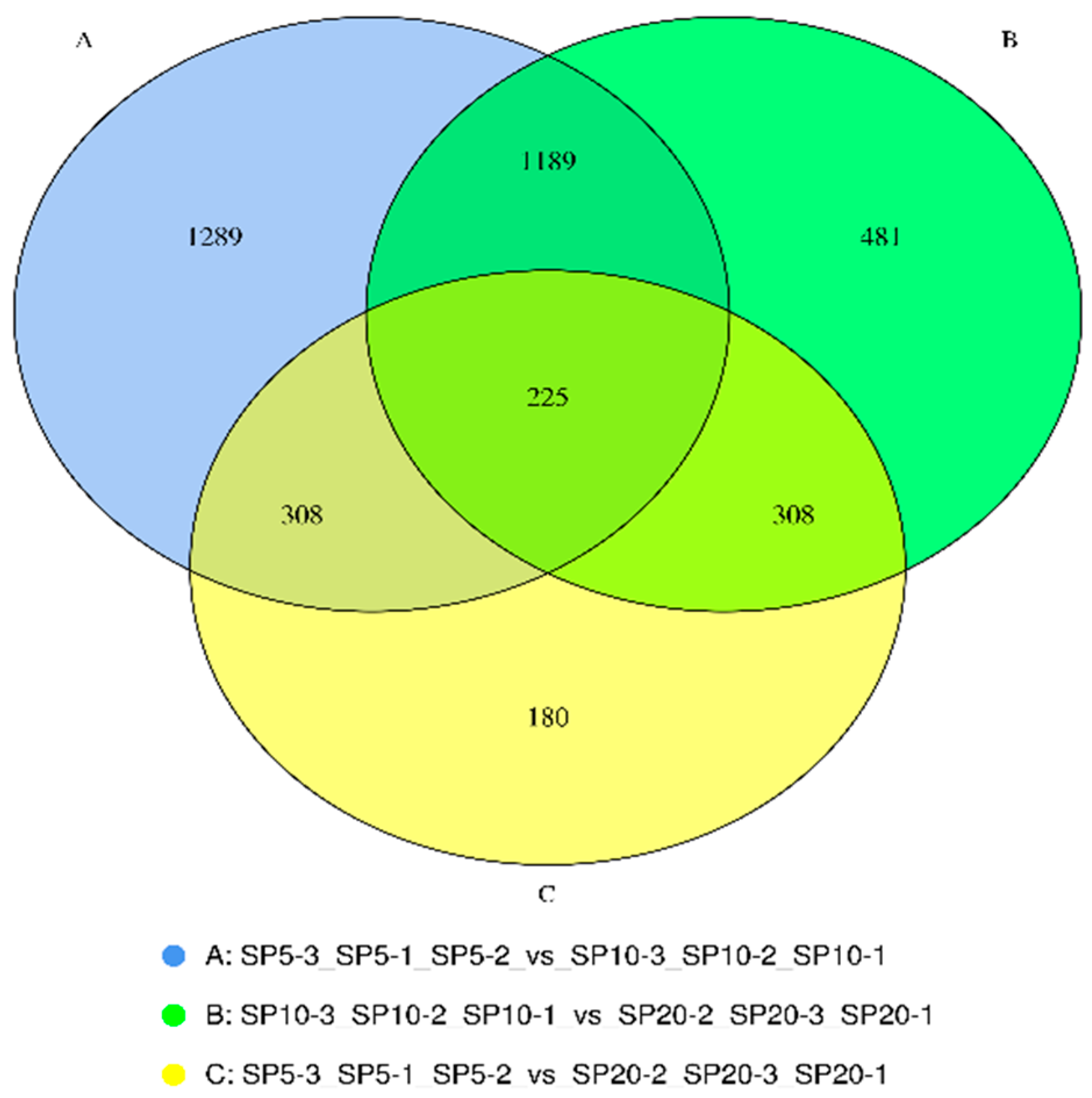
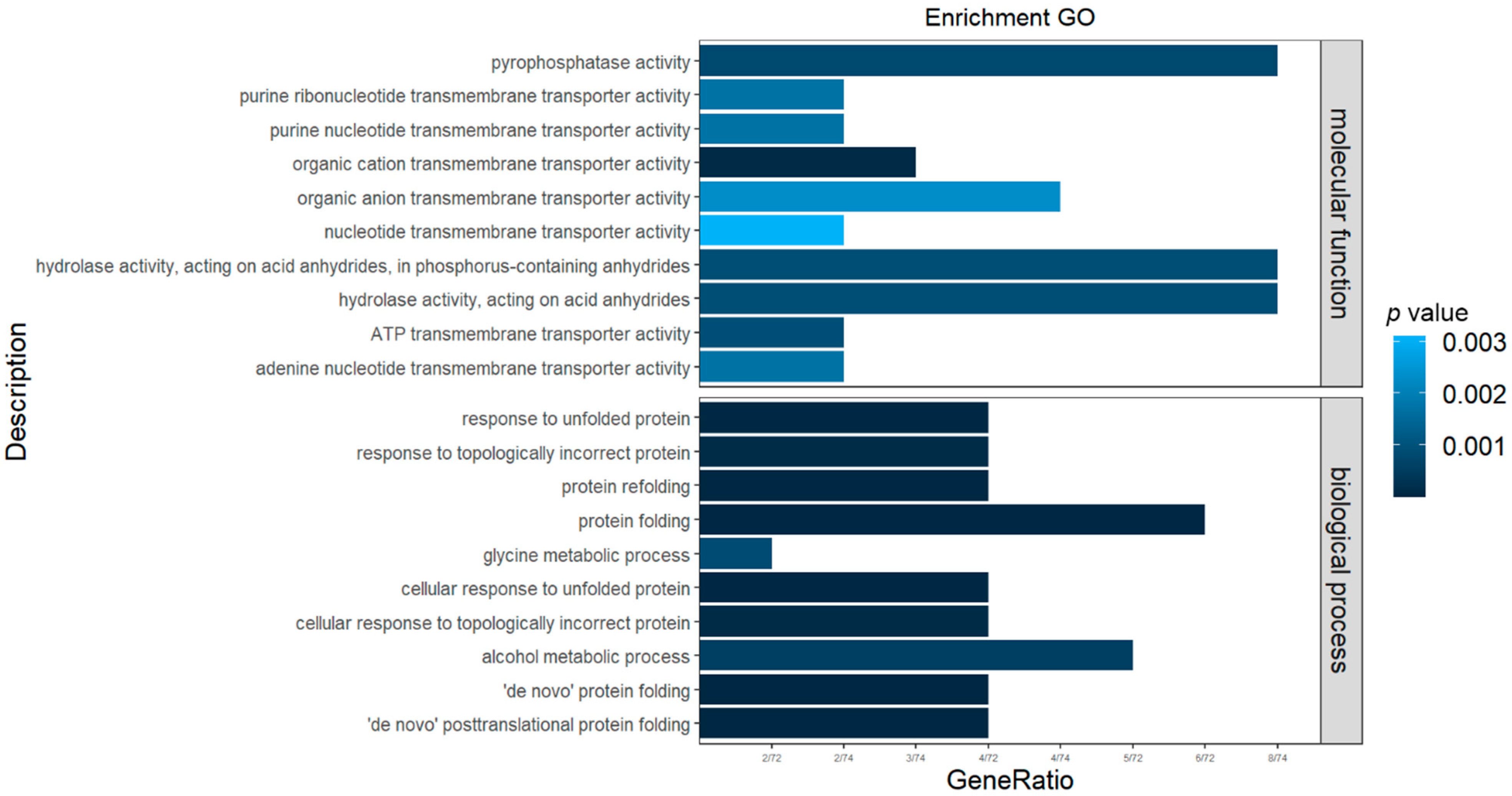

| 20 cm/s | 10 cm/s | 5 cm/s | ||||
|---|---|---|---|---|---|---|
| Average Body Length (cm) | Average Weight (g) | Average Body Length(cm) | Average Weight (g) | Average Body Length(cm) | Average Weight (g) | |
| Month 1 | 19.30 ± 1.3 | 104.18 ± 2.6 | 19.47 ± 1.2 | 105.22 ± 2.3 | 19.40 ± 0.9 | 104.16 ± 2.3 |
| Month 2 | 22.96 ± 1.3 | 174.57 ± 2.4 | 22.65 ± 1.1 | 179.38 ± 2.8 | 22.75 ± 1.1 | 166.41 ± 3.6 |
| Month 3 | 24.81 ± 1.4 | 227.92 ± 3.7 | 24.35 ± 0.9 | 226.38 ± 3.2 | 23.95 ± 1.2 | 225.05 ± 3.9 |
| Month 4 | 26.47 ± 1.4 | 263.68 ± 3.9 | 25.82 ± 1.2 | 288.75 ± 2.4 | 26.03 ± 0.8 | 262.07 ± 3.1 |
| Month 5 | 27.83 ± 1.5 | 344.25 ± 4.6 | 28.03 ± 1.1 | 358.97 ± 3.3 | 27.60 ± 1.3 | 330.73 ± 4.9 |
| Gene Name | Forward Sequences | Reverse Sequences |
|---|---|---|
| cyp7b1 | ACTTCATCGCCCTCTACCCTC | TGAGCCTCTGACCGTCTTTG |
| cpt1a | AGCACCTGACTGACCGTAAGC | GCATCTCAAGTTCACTGGGTAAG |
| irs2 | TGACATCAGCGACCCTTGTG | CGCCACTACTCTCTGTTGACG |
| Kmt5c | GCAGCAAAGACTGGAGCAAG | TCGGTGAACTCATCTGGCAC |
| acod | AGCAATGTTCTCCCTGAGGC | CCAAAGCAAGGTCAAAGGATG |
| cyp2j2 | GGCAACTTATTCTCTGTGGATTTC | GCTGTCTCCCTGATTTACCAGTG |
| acsbg2 | GCAGCAGAAGAGCCTGACCTAC | TAGATGCCAACAGCAAACCC |
| β-actin | CACCACAGCCGAGAGGGA | TCTGGGCAACGGAACCTCT |
| Enzyme Activity (U/g) | Flow Velocities | ||
|---|---|---|---|
| 20 cm/s | 10 cm/s | 5 cm/s | |
| alanine aminotransferase(ALT) | 9.62 b | 12.17 a | 10.72 b |
| superoxide dismutase(SOD) | 560.30 b | 586.02 a | 444.79 c |
| glutathione peroxidase(GPX) | 48.28 b | 62.10 a | 5.70 c |
| Sample | Clean Reads | Clean Bases | Q30 (%) | GC Content (%) | Total Mapped |
|---|---|---|---|---|---|
| SP20-1 | 32,524,608 | 9.06 G | 95.11 | 51.68 | 30,229,755 (92.94%) |
| SP20-2 | 24,959,181 | 6.95 G | 95.16 | 50.72 | 23,119,553 (92.63%) |
| SP20-3 | 29,608,098 | 8.26 G | 94.88 | 51.56 | 27,452,439 (92.72%) |
| SP10-1 | 26,861,072 | 7.50 G | 95.30 | 51.42 | 25,190,298 (93.78%) |
| SP10-2 | 35,829,110 | 9.99 G | 95.05 | 51.51 | 33,509,319 (93.53%) |
| SP10-3 | 23,503,733 | 6.56 G | 94.98 | 50.44 | 22,011,276 (93.65%) |
| SP5-1 | 27,742,612 | 7.74 G | 95.33 | 50.71 | 25,838,704 (93.14%) |
| SP5-2 | 30,041,710 | 8.38 G | 95.12 | 51.02 | 27,966,268 (93.09%) |
| SP5-3 | 21,989,239 | 6.14 G | 95.44 | 50.82 | 20,452,207 (93.01%) |
| Gene Name | Gene ID | log2fold Change | log2fold Change |
|---|---|---|---|
| cyp7b1 | utg000043l-1.624 | −2.87 | −1.96 * |
| cpt1a | utg000134l-0.116 | −3.66 | −1.87 * |
| irs2 | utg000150l-1.275 | −4.32 | −2.35 * |
| kmt5c | utg000519l-0.406 | −2.35 | −2.89 * |
| cyp2j2 | utg000129l-0.32 | 3.96 | 5.22 * |
| acsbg2 | utg000537l-0.141 | 4.31 | 3.87 * |
| acod | utg000003l-2.243 | 3.11 | 4.21 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Gao, J.; Ke, H.; Wang, Y.; Liu, J.; Wen, X.; Fu, S.; Wang, J. Transcriptome-Based Analysis of the Response Mechanism of Leopard Coralgrouper Liver at Different Flow Velocities. Fishes 2022, 7, 279. https://doi.org/10.3390/fishes7050279
Yang M, Gao J, Ke H, Wang Y, Liu J, Wen X, Fu S, Wang J. Transcriptome-Based Analysis of the Response Mechanism of Leopard Coralgrouper Liver at Different Flow Velocities. Fishes. 2022; 7(5):279. https://doi.org/10.3390/fishes7050279
Chicago/Turabian StyleYang, Min, Jin Gao, Hongji Ke, Yongbo Wang, Jinye Liu, Xin Wen, Shuyuan Fu, and Jiang Wang. 2022. "Transcriptome-Based Analysis of the Response Mechanism of Leopard Coralgrouper Liver at Different Flow Velocities" Fishes 7, no. 5: 279. https://doi.org/10.3390/fishes7050279
APA StyleYang, M., Gao, J., Ke, H., Wang, Y., Liu, J., Wen, X., Fu, S., & Wang, J. (2022). Transcriptome-Based Analysis of the Response Mechanism of Leopard Coralgrouper Liver at Different Flow Velocities. Fishes, 7(5), 279. https://doi.org/10.3390/fishes7050279







