Skin Mucus Proteome Analysis Reveals Disease-Resistant Biomarker Signatures in Hybrid Grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) against Vibrio alginolyticus
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Bacterial Infection, Mucus Sample Collection and Preparation
2.2. Liquid Chromatography–Mass Spectrometry (Hybrid Quadrupole-Orbitrap Technology)
2.3. Data Processing and Protein Annotation
2.4. Statistics and Protein Enrichment
3. Results
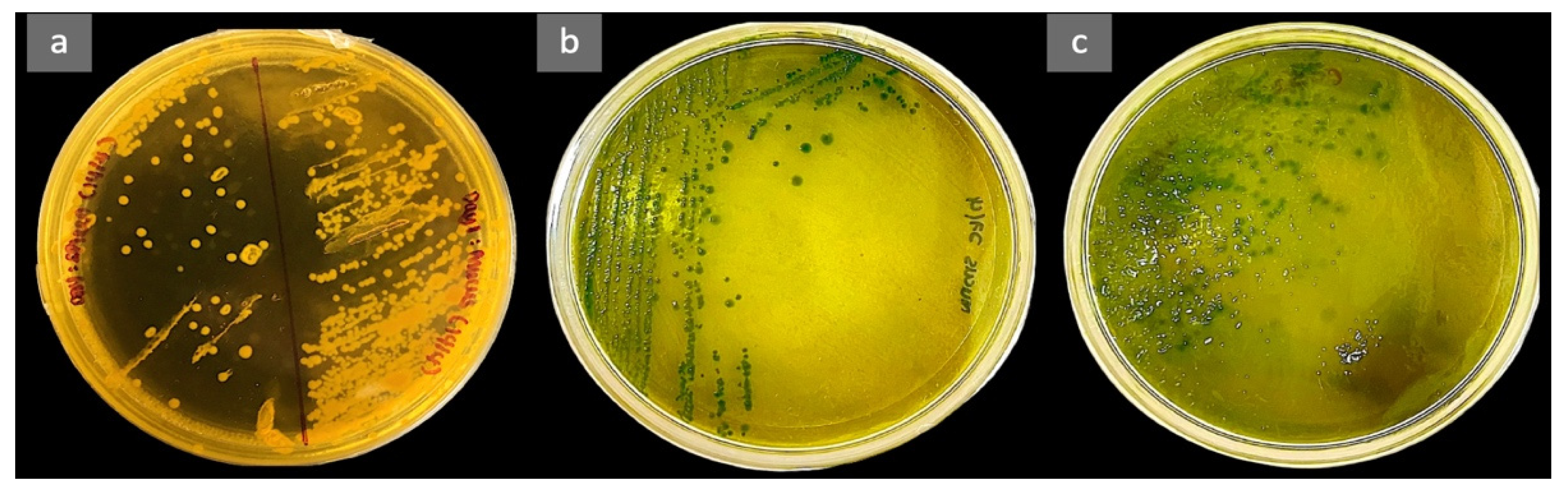
3.1. Experimental Infection of Hybrid Grouper with Vibrio alginolyticus
3.2. Skin Mucus Proteome Profiling and Differential Expression Analysis
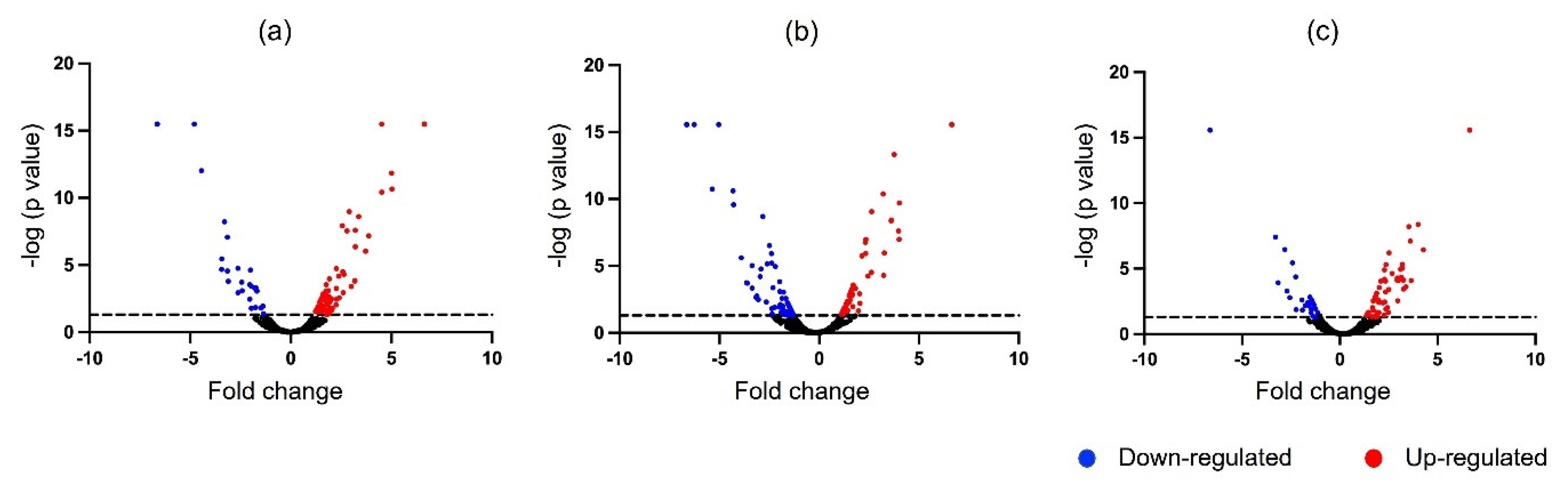
3.3. Differentially Expressed Proteins (DEPs) in Response to V. alginolyticus Experimental Infection
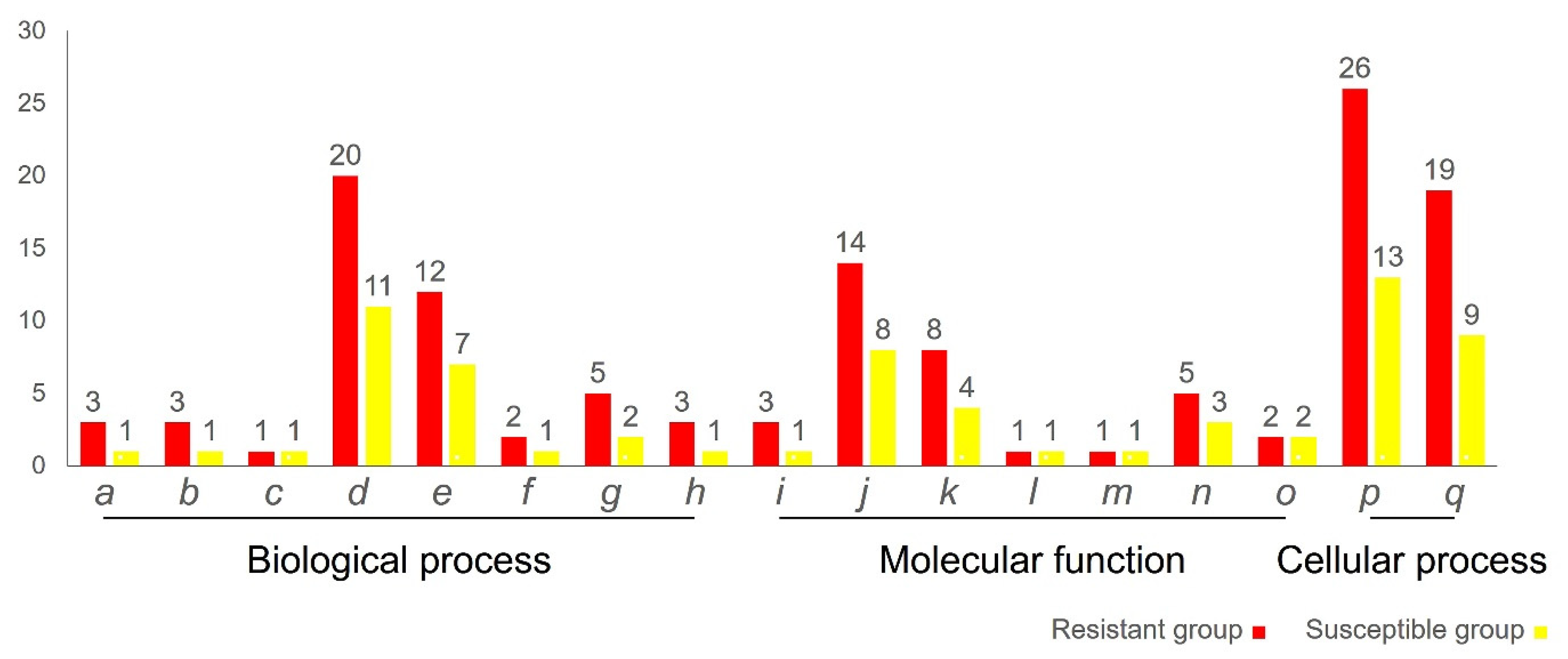
3.4. Gene Ontology Analysis of DEPs
3.5. KEGG Enrichment
3.6. Identification of Potential Biomarker Candidates (PBCs)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Pathway ID | Description | Protein Count | p-Value | Protein | |
|---|---|---|---|---|---|
| Accession | Name | ||||
| 1100 | Metabolic pathways | 30 | 0.000 | 834906005 | glyceraldehyde-3-phosphate dehydrogenase |
| 295792242 | nucleoside diphosphate kinase B | ||||
| 328677247 | glyceraldehyde 3-phosphate dehydrogenase isoform, partial | ||||
| 221048061 | fructose-bisphosphate aldolase | ||||
| 834904058 | malate dehydrogenase | ||||
| 1832677686 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | ||||
| 834904842 | phosphoglycerate kinase | ||||
| 1832683648 | nucleolin | ||||
| 1832687225 | 3-hydroxybutyrate dehydrogenase type 2 | ||||
| 1832613275 | cytochrome c oxidase subunit 5A, mitochondrial | ||||
| 197725770 | mitochondrial cytochrome C oxidase subunit Vb precursor | ||||
| 1832617105 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | ||||
| 1832614309 | very-long-chain 3-oxoacyl-CoA reductase-A | ||||
| 1832627296 | isovaleryl-CoA dehydrogenase, mitochondrial | ||||
| 1832669039 | aldo-keto reductase family 1 member B1 isoform X1 | ||||
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | ||||
| 343459175 | ATP synthase, H+ transporting, mitochondrial F0 complex, partial | ||||
| 295792326 | mitochondrial cytochrome c oxidase subunit 7C | ||||
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | ||||
| 1832641526 | uroporphyrinogen decarboxylase | ||||
| 1832664977 | cytochrome c oxidase subunit 6B1 | ||||
| 1832644368 | peroxiredoxin-6 | ||||
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | ||||
| 1832671044 | deoxyribose-phosphate aldolase | ||||
| 1832640714 | L-threonine dehydrogenase | ||||
| 1832608120 | cytochrome c oxidase subunit 4 isoform 1, mitochondrial | ||||
| 2067150115 | Thioredoxin domain-containing protein 12 | ||||
| 343459105 | NADH dehydrogenase 1 alpha, partial | ||||
| 1832680021 | betaine--homocysteine S-methyltransferase 1 | ||||
| 295792244 | fructose-bisphosphate aldolase A | ||||
| 190 | Oxidative phosphorylation | 9 | 0.000 | 1832613275 | cytochrome c oxidase subunit 5A, mitochondrial |
| 197725770 | mitochondrial cytochrome C oxidase subunit Vb precursor | ||||
| 1832608120 | cytochrome c oxidase subunit 4 isoform 1, mitochondrial | ||||
| 343459105 | NADH dehydrogenase 1 alpha, partial | ||||
| 343459175 | ATP synthase, H+ transporting, mitochondrial F0 complex, partial | ||||
| 295792326 | mitochondrial cytochrome c oxidase subunit 7C | ||||
| 1832617105 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | ||||
| 1832664977 | cytochrome c oxidase subunit 6B1 | ||||
| 1832677686 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | ||||
| 10 | Glycolysis/Gluconeogenesis | 7 | 0.000 | 834906005 | glyceraldehyde-3-phosphate dehydrogenase |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | ||||
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | ||||
| 295792244 | fructose-bisphosphate aldolase A | ||||
| 328677247 | glyceraldehyde 3-phosphate dehydrogenase isoform, partial | ||||
| 221048061 | fructose-bisphosphate aldolase | ||||
| 834904842 | phosphoglycerate kinase | ||||
| 1200 | Carbon metabolism | 8 | 0.000 | 834906005 | glyceraldehyde-3-phosphate dehydrogenase |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | ||||
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | ||||
| 295792244 | fructose-bisphosphate aldolase A | ||||
| 221048061 | fructose-bisphosphate aldolase | ||||
| 834904058 | malate dehydrogenase | ||||
| 834904842 | phosphoglycerate kinase | ||||
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | ||||
| 1230 | Biosynthesis of amino acids | 6 | 0.000 | 834906005 | glyceraldehyde-3-phosphate dehydrogenase |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | ||||
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | ||||
| 295792244 | fructose-bisphosphate aldolase A | ||||
| 221048061 | fructose-bisphosphate aldolase | ||||
| 834904842 | phosphoglycerate kinase | ||||
| 4260 | Cardiac muscle contraction | 6 | 0.000 | 1832606995 | tropomyosin alpha-4 chain-like isoform X4 |
| 1832613275 | cytochrome c oxidase subunit 5A, mitochondrial | ||||
| 197725770 | mitochondrial cytochrome C oxidase subunit Vb precursor | ||||
| 1832608120 | cytochrome c oxidase subunit 4 isoform 1, mitochondrial | ||||
| 295792326 | mitochondrial cytochrome c oxidase subunit 7C | ||||
| 1832664977 | cytochrome c oxidase subunit 6B1 | ||||
| 3010 | Ribosome | 5 | 0.001 | 1832612579 | 60S ribosomal protein L27a |
| 1832640175 | FAU ubiquitin-like and ribosomal protein S30 fusion a | ||||
| 1832673253 | 60S ribosomal protein L8 | ||||
| 1832610610 | 60S ribosomal protein L39 | ||||
| 157929900 | ribosomal protein L23 | ||||
| 30 | Pentose phosphate pathway | 3 | 0.001 | 295792244 | fructose-bisphosphate aldolase A |
| 1832671044 | deoxyribose-phosphate aldolase | ||||
| 221048061 | fructose-bisphosphate aldolase | ||||
| 4672 | Intestinal immune network for IgA production | 3 | 0.001 | 380006108 | MHC class II antigen |
| 326632479 | MHC class II antigen | ||||
| 62255674 | immunoglobulin mu heavy chain | ||||
| 51 | Fructose and mannose metabolism | 3 | 0.001 | 295792244 | fructose-bisphosphate aldolase A |
| 221048061 | fructose-bisphosphate aldolase | ||||
| 1832669039 | aldo-keto reductase family 1 member B1 isoform X1 | ||||
| 260 | Glycine, serine and threonine metabolism | 3 | 0.002 | 1832680021 | betaine--homocysteine S-methyltransferase 1 |
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | ||||
| 1832640714 | L-threonine dehydrogenase | ||||
| 4514 | Cell adhesion molecules (CAMs) | 4 | 0.005 | 380006108 | MHC class II antigen |
| 161935902 | MHC class I alpha antigen | ||||
| 326632479 | MHC class II antigen | ||||
| 1832656308 | neural cell adhesion molecule 1a isoform X4 | ||||
| 4145 | Phagosome | 4 | 0.007 | 380006108 | MHC class II antigen |
| 161935902 | MHC class I alpha antigen | ||||
| 326632479 | MHC class II antigen | ||||
| 62255674 | immunoglobulin mu heavy chain | ||||
| 5168 | Herpes simplex virus 1 infection | 4 | 0.014 | 380006108 | MHC class II antigen |
| 161935902 | MHC class I alpha antigen | ||||
| 1832633330 | protein phosphatase 1, catalytic subunit, alpha isozyme a | ||||
| 326632479 | MHC class II antigen | ||||
| 630 | Glyoxylate and dicarboxylate metabolism | 2 | 0.015 | 834904058 | malate dehydrogenase |
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | ||||
| 270 | Cysteine and methionine metabolism | 2 | 0.029 | 834904058 | malate dehydrogenase |
| 1832680021 | betaine--homocysteine S-methyltransferase 1 | ||||
| 72 | Synthesis and degradation of ketone bodies | 1 | 0.047 | 1832687225 | 3-hydroxybutyrate dehydrogenase type 2 |
| Accession | Name | KEGG Orthology |
|---|---|---|
| 1832702676 | von Willebrand factor A domain-containing protein 5A-like isoform X1 | K24510 |
| 1832606995 | tropomyosin alpha-4 chain-like isoform X4 | K10373 |
| 1832633330 | protein phosphatase 1, catalytic subunit, alpha isozyme a | K06269 |
| 1832613275 | cytochrome c oxidase subunit 5A, mitochondrial | K02264 |
| 1832681058 | antithrombin-III | K03911 |
| 1832628613 | EH domain-containing protein 4 | K12477 |
| 1832617553 | small nuclear ribonucleoprotein Sm D1 | K11087 |
| 1832634797 | transmembrane emp24 domain-containing protein 2 isoform X1 | K20347 |
| 1832661391 | complement factor I | K01333 |
| 1832671044 | deoxyribose-phosphate aldolase | K01619 |
| 1832645324 | arginine--tRNA ligase, cytoplasmic | K01887 |
| 1832640714 | L-threonine dehydrogenase | K15789 |
| 1832610610 | 60S ribosomal protein L39 | K02924 |
| 1832685492 | 39S ribosomal protein L45, mitochondrial | K17426 |
| 1832700793 | TATA-binding protein-associated factor 2N isoform X1 | K13098 |
| 1832687225 | 3-hydroxybutyrate dehydrogenase type 2 | K25939 |
| 1832690325 | uncharacterized protein LOC117256382 isoform X3 | K08870 |
| 1832685958 | arachidonate 15-lipoxygenase B-like isoform X1 | K18684 |
| 1832683648 | nucleolin | K11294 |
| 145105480 | interleukin enhancer-binding factor 2 | K13089 |
| 161935902 | MHC class I alpha antigen | K06751 |
| 1832684689 | trypsin-3 | K23011 |
| 1832608120 | cytochrome c oxidase subunit 4 isoform 1, mitochondrial | K02263 |
| 1832640175 | FAU ubiquitin-like and ribosomal protein S30 fusion a | K02983 |
| 343459105 | NADH dehydrogenase 1 alpha, partial | K03948 |
| 1832688344 | ADP/ATP translocase 1 | K05863 |
| 1832620903 | cystatin-B | K13907 |
| 1832667755 | elongation factor 1-alpha 1a | K03231 |
| 327239696 | complement component C6, partial | K03995 |
| 1832674019 | deleted in malignant brain tumors 1 protein-like isoform X1 | K13912 |
| 1832691617 | ubiquinol-cytochrome-c reductase complex assembly factor 2 | K17682 |
| 1832614309 | very-long-chain 3-oxoacyl-CoA reductase-A | K10251 |
| 1832627296 | isovaleryl-CoA dehydrogenase, mitochondrial | K00253 |
| 1832635569 | annexin A1 | K17091 |
| 1832617105 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | K03939 |
| 1832631564 | E3 ubiquitin-protein ligase RBX1 | K03868 |
| 197725770 | mitochondrial cytochrome C oxidase subunit Vb precursor | K02265 |
| 1832626150 | C-reactive protein | K16143 |
| 2067150115 | Thioredoxin domain-containing protein 12 | K05360 |
| 1832680021 | betaine--homocysteine S-methyltransferase 1-like | K00544 |
| 1832680525 | ubiquitin-conjugating enzyme E2 L3 | K04552 |
| 1832606043 | ras-related protein Rab-27A | K07885 |
| 1710371347 | electron transfer flavoprotein subunit beta | K03522 |
| 1832702451 | LOW-QUALITY PROTEIN: apolipoprotein(a)- | K01315 |
| 1832645146 | pigment epithelium-derived factor | K19614 |
| 1832639949 | collagen alpha-1(X) chain | K19479 |
| 1832701092 | latexin | K23594 |
| 1832662114 | parvalbumin alpha | K23926 |
| 1832699882 | peptidyl-prolyl cis-trans isomerase FKBP10 | K09575 |
| 1832670505 | coatomer subunit gamma-2 | K17267 |
| 209981964 | alpha-1-antitrypsin | K04525 |
| 1832702196 | vasodilator-stimulated phosphoprotein isoform X1 | K06274 |
| 834904058 | malate dehydrogenase | K00026 |
| 1832656308 | neural cell adhesion molecule 1a isoform X4 | K06491 |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | K00134 |
| 306008591 | heat shock protein | K04077 |
| 295792244 | fructose-bisphosphate aldolase A | K01623 |
| 328677149 | hypothetical protein, partial | K01315 |
| 343459175 | ATP synthase, H+ transporting, mitochondrial F0 complex, partial | K02138 |
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | K00134 |
| 1832630592 | glucosidase 2 subunit beta | K08288 |
| 1832669039 | aldo-keto reductase family 1 member B1 isoform X1 | K00011 |
| 1832632244 | parvalbumin beta | K23926 |
| 1832620905 | stefin-C | K13907 |
| 1832677686 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | K03937 |
| 1832638111 | hyaluronan and proteoglycan link protein 1-like | K06848 |
| 1832619571 | tetranectin-like | K17520 |
| 1832616138 | testin | K24270 |
| 1832604382 | histone H1 | K11275 |
| 834904842 | phosphoglycerate kinase | K00927 |
| 1832645835 | ruvB-like 2 | K11338 |
| 1832635997 | proteasome subunit beta type-7 isoform X1 | K02739 |
| 1832641526 | uroporphyrinogen decarboxylase | K01599 |
| 1832690337 | chitinase, acidic.1 | K01183 |
| 1832673253 | 60S ribosomal protein L8 | K02938 |
| 1832638138 | transcription factor BTF3 | K01527 |
| 1832643512 | rho GTPase-activating protein 12-like isoform X1 | K20636 |
| 1832644997 | coatomer subunit beta’ isoform X1 | K17302 |
| 1832642084 | neurogenic differentiation factor 6-B | K09080 |
| 1832685063 | hydroperoxide isomerase ALOXE3-like | K18684 |
| 1832644368 | peroxiredoxin-6 | K11188 |
| 1832670971 | LSM8 homolog, U6 small nuclear RNA associated | K12627 |
| 1832638785 | leucine-rich alpha-2-glycoprotein-like | K25431 |
| 1832664977 | cytochrome c oxidase subunit 6B1 | K02267 |
| 1832612090 | heat shock 70 kDa protein 4b | K09489 |
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | K00281 |
| 1832622572 | acyl-CoA thioesterase 9, tandem duplicate 1 isoform X1 | K17361 |
| 1832656769 | vesicle-associated membrane protein 2 | K13504 |
| 328677247 | glyceraldehyde 3-phosphate dehydrogenase isoform, partial | K10705 |
| 1832679851 | drebrin-like b isoform X1 | K20520 |
| 1832650400 | prefoldin subunit 1 isoform X1 | K09548 |
| 221048061 | fructose-bisphosphate aldolase | K01623 |
| 1832658469 | fibrinogen beta chain | K03904 |
| 1832612579 | 60S ribosomal protein L27a | K02900 |
| 222087999 | fibrinogen beta chain precursor | K03904 |
| 1832689402 | fibrinogen gamma chain | K03905 |
| 1832662038 | leukocyte cell-derived chemotaxin-2 | K25755 |
| 1832685624 | microfibril-associated glycoprotein 4 | K25409 |
| 1832634516 | keratin, type I cytoskeletal 13 | K07604 |
| 1832685241 | beta-2-glycoprotein 1 | K17305 |
| 1832623025 | apolipoprotein Eb | K04524 |
| 405790938 | MHC class II antigen, partial | K06752 |
| 295792326 | mitochondrial cytochrome c oxidase subunit 7C | K02272 |
| 1832626146 | allograft inflammatory factor 1-like | K18617 |
| 1832610590 | DNA-directed RNA polymerase I subunit RPA1 | K02999 |
| 295792242 | nucleoside diphosphate kinase B | K00940 |
| 1832702726 | serpin H1b | K09501 |
| 1832665474 | sulfotransferase 2B1 | K01015 |
| 1832641369 | phosphotriesterase-related protein | K07048 |
| 1832686354 | puromycin-sensitive aminopeptidase | K08776 |
| 1832695641 | N-alpha-acetyltransferase 10 isoform X1 | K20791 |
| 1832682447 | dedicator of cytokinesis protein 7 isoform X1 | K21852 |
| 1832667938 | sideroflexin-3 | K23500 |
| 1832608362 | AP-1 complex subunit gamma-1 isoform X1 | K12391 |
| 1832621671 | rRNA 2’-O-methyltransferase fibrillarin | K14563 |
| 1832611868 | transmembrane 9 superfamily member 2 isoform X1 | K17086 |
| 1832671468 | coagulation factor VIIi | K01320 |
| 1832618423 | class I histocompatibility antigen, F10 alpha chain-like isoform X1 | K06751 |
| 62255674 | immunoglobulin mu heavy chain | K06856 |
| 157929900 | ribosomal protein L23 | K02894 |
| 2022331196 | immunoglobulin M heavy-chain constant mu variant 1 | K06856 |
| 834906005 | glyceraldehyde-3-phosphate dehydrogenase | K00134 |
| 1832604432 | erythroblast NAD(P)(+)--arginine ADP-ribosyltransferase | K19977 |
| 1832697311 | nucleobindin-2a isoform X1 | K20371 |
| 1832604652 | fibrinogen alpha chain-like | K03903 |
| 326632479 | MHC class II antigen | K06752 |
| 1832660271 | macrosialin | K06501 |
| 380006108 | MHC class II antigen | K06752 |
| 2022331198 | immunoglobulin M heavy-chain constant mu variant 2 | K06856 |
References
- Che Cob, Z.; Kumar Das, S. Optimum temperature for the growth form of Tiger grouper (Epinephelus fuscoguttatus ♀) × Giant grouper (Epinephelus lanceolatus ♂) hybrid. Sains Malays. 2016, 45, 541–549. [Google Scholar]
- Mo, Z.Q.; Wu, H.C.; Hu, Y.T.; Lu, Z.J.; Lai, X.L.; Chen, H.P.; He, Z.; Luo, X.; Li, Y.; Dan, X. Transcriptomic analysis reveals innate immune mechanisms of an underlying parasite-resistant grouper hybrid (Epinephelus fuscogutatus × Epinephelus lanceolatus). Fish Shellfish. Immunol. 2021, 119, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Glamuzina, B.; Rimmer, M.A. Grouper Aquaculture World Status and Perspectives. In Biology and Ecology of Groupers; CRC Press: Boca Raton, FL, USA, 2022; pp. 166–190. [Google Scholar]
- Rahman, M.A.; Lee, S.-G.; Yusoff, F.; Rafiquzzaman, S. Hybridization and Its Application in Aquaculture. In Sex Control in Aquaculture; John Wiley & Sons, Ltd.: Chichester, UK, 2018; pp. 163–178. [Google Scholar] [CrossRef]
- Ch ’ng, C.L.; Senoo, S. Egg and Larval Development of a New Hybrid Grouper, Tiger Grouper (Epinephelus fuscoguttatus) and Giant Grouper (Epinephelus lanceolatus). Aquac. Sci. 2008, 56, 505–512. [Google Scholar]
- Rimmer, M.A.; Glamuzina, B. A review of grouper (Family Serranidae: Subfamily Epinephelinae) aquaculture from a sustainability science perspective. Rev. Aquac. 2017, 11, 58–87. [Google Scholar] [CrossRef]
- Harikrishnan, R.; Balasundaram, C.; Heo, M.-S. Molecular studies, disease status and prophylactic measures in grouper aquaculture: Economic importance, diseases and immunology. Aquaculture 2010, 309, 1–14. [Google Scholar] [CrossRef]
- Petersen, E.H.; My Chinh, D.T.; Diu, N.T.; Phuoc, V.V.; Phuong, T.H.; Dung, N.V.; Dat, N.K.; Giang, P.T.; Glencross, B.D. Bioeconomics of grouper, Serranidae: Epinephelinae, culture in Vietnam. Rev. Fish. Sci. 2013, 21, 49–57. [Google Scholar] [CrossRef]
- Bunlipatanon, P.; U-taynapun, K. Growth performance and disease resistance against Vibrio vulnificus infection of novel hybrid grouper (Epinephelus lanceolatus × Epinephelus fuscoguttatus). Aquac. Res. 2017, 48, 1711–1723. [Google Scholar] [CrossRef]
- Ye, G.; Dong, X.; Yang, Q.; Chi, S.; Liu, H.; Zhang, H.; Tan, B.; Zhang, S. Low-gossypol cottonseed protein concentrate used as a replacement of fish meal for juvenile hybrid grouper (Epinephelus fuscoguttatus♀ × Epinephelus lanceolatus♂): Effects on growth performance, immune responses and intestinal microbiota. Aquaculture 2020, 524, 735309. [Google Scholar] [CrossRef]
- Liao, J.; Cai, Y.; Wang, X.; Shang, C.; Zhang, Q.; Shi, H.; Wang, S.; Zhang, D.; Zhou, Y. Effects of a potential host gut-derived probiotic, Bacillus subtilis 6-3-1, on the growth, non-specific immune response and disease resistance of hybrid grouper (Epinephelus fuscoguttatus × Epinephelus lanceolatus♂). Probiotics Antimicrob. Proteins 2021, 13, 1119–1137. [Google Scholar] [CrossRef]
- Xiao, H.; Liu, M.; Li, S.; Shi, D.; Zhu, D.; Ke, K.; Xu, Y.; Dong, D.; Zhu, L.; Yu, Q. Isolation and characterization of a ranavirus associated with disease outbreaks in cultured hybrid grouper (♀ Tiger Grouper Epinephelus fuscoguttatus × giant grouper Epinephelus lanceolatus) in Guangxi, China. J. Aquat. Anim. Health 2019, 31, 364–370. [Google Scholar] [CrossRef]
- Zhu, Z.M.; Dong, C.F.; Weng, S.P.; He, J.G. The high prevalence of pathogenic Vibrio harveyi with multiple antibiotic resistance in scale drop and muscle necrosis disease of the hybrid grouper, Epinephelus fuscoguttatus (♀) × Epinephelus lanceolatus (♂), in China. J. Fish Dis. 2018, 41, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Suyanti, E.; Mahasri, G.; Lokapirnasari, W.P. Marine Leech Zeylanicobdella arugamensis Infestation As A Predisposing Factor For Vibrio alginolyticus Infection On The Hybrid Grouper “Cantang”(Epinephelus fuscoguttatus × Epinephelus lanceolatus) From Traditional Ponds In The Kampung Kerapu Lamongan Eas. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2021; p. 12035. [Google Scholar]
- Lee, K.-K. Pathogenesis studies on Vibrio alginolyticus in the grouper, Epinephelus malabaricus, Bloch et Schneider. Microb. Pathog. 1995, 19, 39–48. [Google Scholar] [CrossRef]
- Mohamed Alipiah, N.; Ramli, N.; Low, C.; Shamsudin, M.; Yusoff, F. Protective effects of sea cucumber surface-associated bacteria against Vibrio harveyi in brown-marbled grouper fingerlings. Aquac. Environ. Interact. 2016, 8, 147–155. [Google Scholar] [CrossRef]
- Mohamad, A.; Mursidi, F.-A.; Zamri-Saad, M.; Amal, M.N.A.; Annas, S.; Monir, S.; Loqman, M.; Hairudin, F.; Al-Saari, N.; Ina-Salwany, Y. Laboratory and Field Assessments of Oral Vibrio Vaccine Indicate the Potential for Protection against Vibriosis in Cultured Marine Fishes. Animals 2022, 12, 133. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Martin, S.A.; Silva, T.S.; Boonanuntanasarn, S.; Schrama, D.; Moreira, M.; Raposo, C. Proteomics in fish and aquaculture research. In Proteomics in Domestic Animals: From Farm to Systems Biology; Springer International Publishing: Cham, Switzerland, 2018; pp. 311–338. [Google Scholar]
- Mischak, H.; Ioannidis, J.P.A.; Argiles, A.; Attwood, T.K.; Bongcam-Rudloff, E.; Broenstrup, M. Implementation of proteomic biomarkers: Making it work. Eur. J. Clin. Investig. 2012, 42, 1027–1036. [Google Scholar] [CrossRef]
- Braceland, M.; McLoughlin, M.F.; Tinsley, J.; Wallace, C.; Cockerill, D.; McLaughlin, M.; Eckersall, P.D. Serum enolase: A non-destructive biomarker of white skeletal myopathy during pancreas disease (PD) in Atlantic salmon Salmo salar L. J. Fish Dis. 2015, 38, 821–831. [Google Scholar] [CrossRef]
- Coates, C.J.; Decker, H. Immunological properties of oxygen-transport proteins: Hemoglobin, hemocyanin and hemerythrin. Cell. Mol. Life Sci. 2016, 74, 293–317. [Google Scholar] [CrossRef]
- Hua, Y.; Qingsong, L.; Hui, L. Applications of transcriptomics and proteomics in understanding fish immunity. Fish Shellfish. Immunol. 2018, 77, 319–327. [Google Scholar]
- Leng, W.; Wu, X.; Shi, T.; Xiong, Z.; Yuan, L.; Jin, W.; Gao, R. Untargeted Metabolomics on Skin Mucus Extract of Channa argus against Staphylococcus aureus: Antimicrobial Activity and Mechanism. Foods 2021, 10, 2995. [Google Scholar] [CrossRef]
- Leng, W.; Wu, X.; Xiong, Z.; Shi, T.; Sun, Q.; Yuan, L.; Gao, R. Study on antibacterial properties of mucus extract of snakehead (Channa argus) against Escherichia coli and its application in chilled fish fillets preservation. LWT 2022, 167, 113840. [Google Scholar] [CrossRef]
- Lau, B.Y.C.; Amiruddin, M.D.; Othman, A. Proteomics analysis on lipid metabolism in Elaeis guineensis and Elaeis oleifera. Data Brief 2020, 31, 105714. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting significant changes in protein abundance. EuPA Open Proteom. 2015, 7, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Das, S.; Samal, J.; Thatoi, H.N. Epidermal mucus, a major determinant in fish health: A review. Iran. J. Vet. Res. 2018, 19, 72–81. [Google Scholar] [CrossRef]
- Valdenegro-Vega, V.A.; Crosbie, P.; Bridle, A.; Leef, M.; Wilson, R.; Nowak, B.F. Differentially expressed proteins in gill and skin mucus of Atlantic salmon (Salmo salar) affected by amoebic gill disease. Fish Shellfish Immunol. 2014, 40, 69–77. [Google Scholar] [CrossRef]
- Raposo de Magalhães, C.S.F.; Cerqueira, M.A.C.; Schrama, D.; Moreira, M.J.V.; Boonanuntanasarn, S.; Rodrigues, P.M.L. A Proteomics and other Omics approach in the context of farmed fish welfare and biomarker discovery. Rev. Aquac. 2020, 12, 122–144. [Google Scholar] [CrossRef]
- Canellas, A.L.B.; Costa, W.F.; Freitas-Silva, J.; Lopes, I.R.; de Oliveira, B.F.R.; Laport, M.S. In sickness and in health: Insights into the application of omics in aquaculture settings under a microbiological perspective. Aquaculture 2022, 554, 738132. [Google Scholar] [CrossRef]
- Drago, F.; Sautière, P.E.; le Marrec-Croq, F.; Accorsi, A.; van Camp, C.; Salzet, M.; Lefebvre, C.; Vizioli, J. Microglia of medicinal leech (Hirudo medicinalis) express a specific activation marker homologous to vertebrate ionized calcium-binding adapter molecule 1 (Iba1/alias aif-1). Dev. Neurobiol. 2014, 74, 987–1001. [Google Scholar] [CrossRef]
- Lü, A.; Hu, X.; Wang, Y.; Shen, X.; Li, X.; Zhu, A.; Tian, J.; Ming, Q.; Feng, Z. iTRAQ analysis of gill proteins from the zebrafish (Danio rerio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2014, 36, 229–239. [Google Scholar] [CrossRef]
- Mierziak, J.; Burgberger, M.; Wojtasik, W. 3-Hydroxybutyrate as a Metabolite and a Signal Molecule Regulating Processes of Living Organisms. Biomolecules 2021, 11, 402. [Google Scholar] [CrossRef]
- Stubbs, B.J.; Koutnik, A.P.; Goldberg, E.L.; Upadhyay, V.; Turnbaugh, P.J.; Verdin, E.; Newman, J.C. Investigating Ketone Bodies as Immunometabolic Countermeasures against Respiratory Viral Infections. Med 2020, 1, 43–65. [Google Scholar] [CrossRef] [PubMed]
- Dedkova, E.N.; Blatter, L.A. Role of Î2-hydroxybutyrate, its polymer poly-Î2-hydroxybutyrate and inorganic polyphosphate in mammalian health and disease. Front. Physiol. 2014, 17, 5. [Google Scholar] [CrossRef] [PubMed]
- Graff, E.C.; Fang, H.; Wanders, D.; Judd, R.L. Anti-inflammatory effects of the hydroxycarboxylic acid receptor 2. Metabolism 2016, 65, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Ballarin, L.; Cammarata, M.; Franchi, N.; Parrinello, N. Routes in Innate Immunity Evolution: Galectins and Rhamnose-binding Lectins in Ascidians. In Marine Protein and Peptides. Biological Activities and Applications; Wiley-Blackwell: Chichester, UK, 2013; pp. 185–205. [Google Scholar] [CrossRef]
- Shiina, N.; Tateno, H.; Ogawa, T.; Muramoto, K.; Saneyoshi, M.; Kamiya, H. Isolation and characterization of L-rhamnose-binding lectins from chum salmon (Oncorhynchus keta) eggs. Fish. Sci. 2002, 68, 1352–1366. [Google Scholar] [CrossRef]
- Cammarata, M.; Parisi, M.; Benenati, G.; Vasta, G.; Parrinello, N. A rhamnose-binding lectin from sea bass (Dicentrarchus labrax) plasma agglutinates and opsonizes pathogenic bacteria. Dev. Comp. Immunol. 2014, 44, 332–340. [Google Scholar] [CrossRef]
- Okamoto, M.; Tsutsui, S.; Tasumi, S.; Suetake, H.; Kikuchi, K.; Suzuki, Y. Tandem repeat l-rhamnose-binding lectin from the skin mucus of ponyfish, Leiognathus nuchalis. Biochem. Biophys. Res. Commun. 2005, 333, 463–469. [Google Scholar] [CrossRef]
- Vibhute, P.; Radhakrishnan, A.; Sivakamavalli, J.; Chellapandian, H.; Selvin, J. Antimicrobial and Immunomodulatory Role of Fish Lectins. In Aquat. Lectins; Springer: Singapore, 2022; pp. 257–286. [Google Scholar] [CrossRef]
- Maher, K.; Kokelj, B.J.; Butinar, M.; Mikhaylov, G.; Manček-Keber, M.; Stoka, V.; Vasiljeva, O.; Turk, B.; Grigoryev, S.A.; Kopitar-Jerala, N. A Role for Stefin B (Cystatin B) in Inflammation and Endotoxemia. J. Biol. Chem. 2014, 289, 31736–31750. [Google Scholar] [CrossRef]
- Rivera-Rivera, L.; Perez-Laspiur, J.; Colón, K.; Meléndez, L.M. Inhibition of interferon response by cystatin B: Implication in HIV replication of macrophage reservoirs. J. Neurovirol. 2012, 18, 20–29. [Google Scholar] [CrossRef][Green Version]
- Wickramasinghe, P.; Kwon, H.; Elvitigala, D.A.S.; Wan, Q.; Lee, J. Identification and characterization of cystatin B from black rockfish, Sebastes schlegelii, indicating its potent immunological importance. Fish Shellfish Immunol. 2020, 104, 497–505. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Wang, G. Response of complement expression to challenge with lipopolysaccharide in embryos/larvae of zebrafish Danio rerio: Acquisition of immunocompetent complement. Fish Shellfish Immunol. 2008, 25, 264–270. [Google Scholar] [CrossRef]
- Kimura, A.; Nonaka, M. Molecular cloning of the terminal complement components C6 and C8β of cartilaginous fish. Fish Shellfish Immunol. 2009, 27, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, X. Identification of differentially expressed genes in lipopolysaccharide-stimulated yellow grouper Epinephelus awoara spleen. Fish Shellfish Immunol. 2007, 23, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Schorn, T. Identification and Modulation of the Allograft Inflammatory Factor-1 (AIF-1) Homologous in the Medicinal Leech Hirudo Medicinalis. Doctoral Dissertation, Universita degli Studi dell’lnsubria, Varese, Italy, 2014. [Google Scholar]
- De Zoysa, M.; Nikapitiya, C.; Kim, Y.; Oh, C.; Kang, D.H.; Whang, I.; Kim, S.-J.; Lee, J.-S.; Choi, C.Y.; Lee, J. Allograft inflammatory factor-1 in disk abalone (Haliotis discus): Molecular cloning, transcriptional regulation against immune challenge and tissue injury. Fish Shellfish Immunol. 2010, 29, 319–326. [Google Scholar] [CrossRef]
- Miyata, M.; Iinuma, K.; Miyazaki, T. DNA cloning and characterization of an allograft inflammatory factor-1 homologue in red sea bream (Chrysophrys major). Aquaculture 2001, 194, 63–74. [Google Scholar] [CrossRef]
- Ying, Z.Y.; Qiu, L.Y.; You, X. Functional Identification of Allograft Inflammatory Factor 1-Like Gene in Luning Chicken. Anim. Biotechnol. 2018, 29, 234–240. [Google Scholar]
- Ligtenberg, A.J.M.; Karlsson, N.G.; Veerman, E.C.I. Deleted in Malignant Brain Tumors-1 Protein (DMBT1): A Pattern Recognition Receptor with Multiple Binding Sites. Int. J. Mol. Sci. 2010, 11, 5212–5233. [Google Scholar] [CrossRef]
- Müller, H.; Nagel, C.; Weiss, C.; Mollenhauer, J.; Poeschl, J. Deleted in malignant brain tumors 1 (DMBT1) elicits increased VEGF and decreased IL-6 production in type II lung epithelial cells. BMC Pulm. Med. 2015, 15, 32. [Google Scholar] [CrossRef]
- Stenger, S.; Modlin, R.L. Cytotoxic T cell responses to intracellular pathogens. Curr. Opin. Immunol. 1998, 10, 471–477. [Google Scholar] [CrossRef]
- Grimholt, U. MHC and Evolution in Teleosts. Biology 2016, 5, 6. [Google Scholar] [CrossRef]
- Balebona, M.C.; Andreu, M.J.; Bordas, M.A.; Zorrilla, I.; Moriñigo, M.A.; Borrego, J.J. Pathogenicity of Vibrio alginolyticus for Cultured Gilt-Head Sea Bream (Sparus aurata L.). Appl. Environ. Microbiol. 1998, 64, 4269–4275. [Google Scholar] [CrossRef]
- Solito, E.; De Coupade, C.; Canaider, S.; Goulding, N.J.; Perretti, M. Transfection of annexin 1 in monocytic cells produces a high degree of spontaneous and stimulated apoptosis associated with caspase-3 activation. J. Cereb. Blood Flow Metab. 2001, 133, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Barton, G.J.; Newman, R.H.; Freemont, P.S.; Crumpton, M.J. Amino acid sequence analysis of the annexin super-gene family of proteins. JBIC J. Biol. Inorg. Chem. 1991, 198, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Linke, B.; Abeler-Dörner, L.; Jahndel, V.; Kurz, A.; Mahr, A.; Pfrang, S.; Linke, L.; Krammer, P.H.; Weyd, H. The Tolerogenic Function of Annexins on Apoptotic Cells Is Mediated by the Annexin Core Domain. J. Immunol. 2015, 194, 5233–5242. [Google Scholar] [CrossRef]
- Carson, D.A.; Ribeiro, J.M. Apoptosis and disease. Lancet 1993, 341, 1251–1254. [Google Scholar] [CrossRef]
- Wang, H.-G.; Pathan, N.; Ethell, I.M.; Krajewski, S.; Yamaguchi, Y.; Shibasaki, F.; McKeon, F.; Bobo, T.; Franke, T.F.; Reed, J.C. Ca 2+ -Induced Apoptosis Through Calcineurin Dephosphorylation of BAD. Science 1999, 284, 339–343. [Google Scholar] [CrossRef]
- Solito, E.; Kamal, A.; Russo-Marie, F.; Buckingham, J.C.; Marullo, S.; Perretti, M. A novel calcium-dependent proapoptotic effect of annexin 1 on human neutrophils. FASEB J. 2003, 17, 1–27. [Google Scholar] [CrossRef]
- Roh, J.S.; Sohn, D.H. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef] [PubMed]
- Maderna, P.; Yona, S.; Perretti, M.; Godson, C. Modulation of Phagocytosis of Apoptotic Neutrophils by Supernatant from Dexamethasone-Treated Macrophages and Annexin-Derived Peptide Ac2–26. J. Immunol. 2005, 174, 3727–3733. [Google Scholar] [CrossRef]
- Young, N.; Cooper, G.; Nowak, B.; Koop, B.; Morrison, R. Coordinated down-regulation of the antigen processing machinery in the gills of amoebic gill disease-affected Atlantic salmon (Salmo salar L.). Mol. Immunol. 2008, 45, 2581–2597. [Google Scholar] [CrossRef]

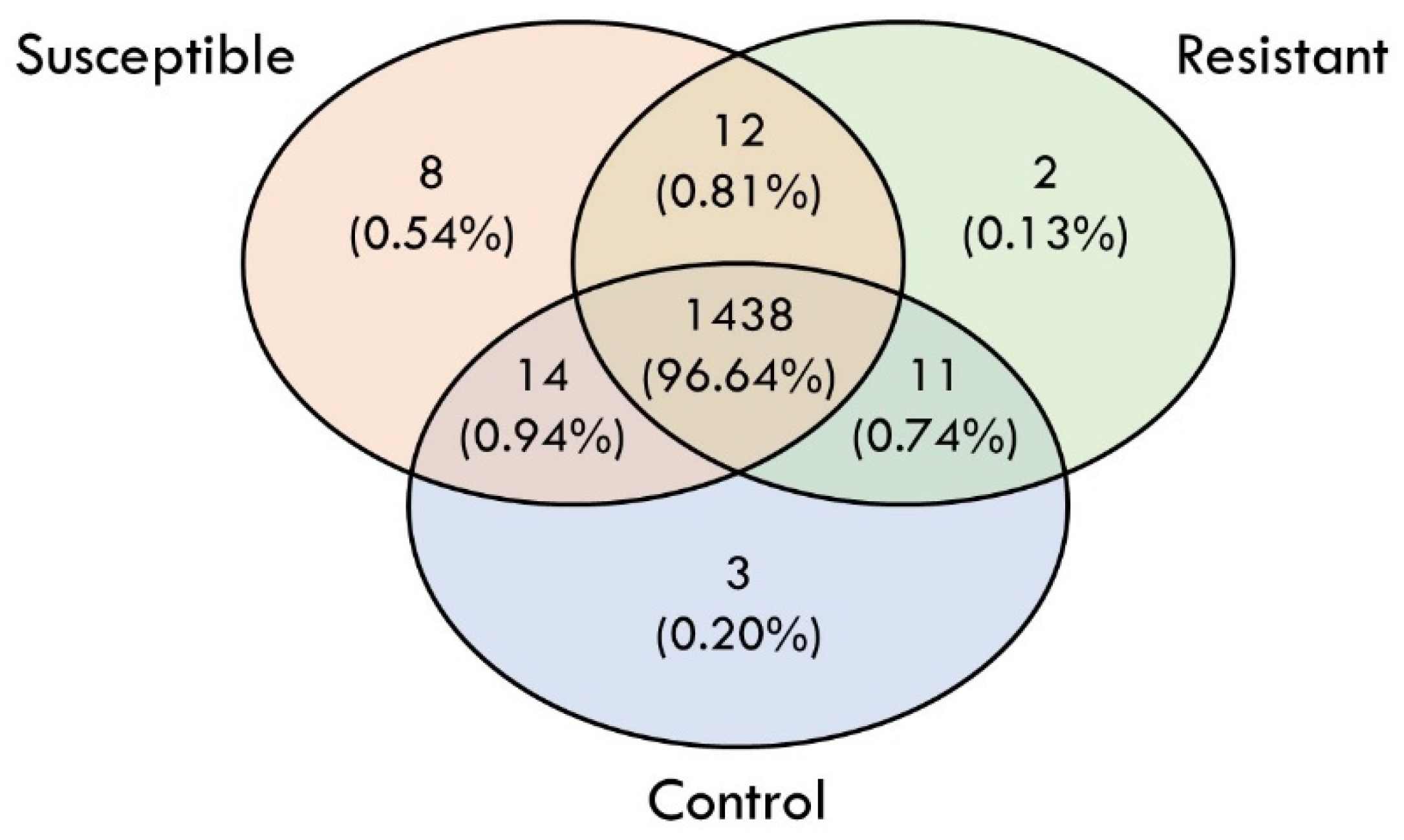
| Group | Accession | Description |
|---|---|---|
| Control | 834906564 | cysteine desulfhydrase |
| 1832645835 | ruvB-like 2 | |
| 1832690337 | chitinase | |
| Resistant | 1832634797 | transmembrane emp24 domain-containing protein 2 isoform X1 |
| 1832622809 | RWD domain-containing protein 1 isoform X2 | |
| Susceptible | 1832685063 | hydroperoxide isomerase ALOXE3 |
| 1832644368 | peroxiredoxin-6 | |
| 1832670971 | LSM8 homolog, U6 small nuclear RNA associated | |
| 1832638785 | leucine-rich alpha-2-glycoprotein | |
| 1832664977 | cytochrome c oxidase subunit 6B1 | |
| 1832612090 | heat shock 70 kDa protein 4b | |
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | |
| 1832627111 | nesprin-2-like isoform X4 |
| Accession | Description | MW (kDa) | Fold Change | p-Value | Abundances Control | Abundances Resistant |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| 1832619927 | elongation factor 1-alpha | 50.3 | −6.64 | 0.000 | 31.5 | |
| 1832702676 | von Willebrand factor A domain-containing protein 5A-like isoform X1 | 69.9 | −6.64 | 0.000 | 102.0 | |
| 1832606995 | tropomyosin alpha-4 chain-like isoform X4 | 28.4 | −6.64 | 0.000 | 172.3 | |
| 1832633330 | protein phosphatase 1, catalytic subunit, alpha isozyme a | 37.5 | −6.64 | 0.000 | 24.9 | 125.1 |
| 1832671158 | 23 kDa integral membrane protein | 26.7 | −6.64 | 0.000 | 49.9 | |
| 1832663676 | catechol O-methyltransferase domain-containing protein 1-like isoform X1 | 28.6 | −6.64 | 0.000 | 86.7 | |
| 1832613275 | cytochrome c oxidase subunit 5A, mitochondrial | 16 | −6.64 | 0.000 | 118.8 | |
| 1832681058 | antithrombin-III | 51.2 | −6.64 | 0.000 | 44.5 | 92.2 |
| 1832628613 | EH domain-containing protein 4 | 62.3 | −6.64 | 0.000 | 98.1 | |
| 1832617553 | small nuclear ribonucleoprotein Sm D1 | 13.3 | −6.64 | 0.000 | 146.4 | |
| 1832651057 | uncharacterized protein si:ch1073-126c3.2 | 23.2 | −6.64 | 0.000 | 194.3 | |
| 1832634797 | transmembrane emp24 domain-containing protein 2 isoform X1 | 23.3 | −6.64 | 0.000 | 300.0 | |
| 1832661391 | complement factor I | 77.7 | −6.64 | 0.000 | 89.6 | |
| 1832671044 | deoxyribose-phosphate aldolase | 35 | −6.64 | 0.000 | 34.0 | 134.8 |
| 1832645324 | Arginine-tRNA ligase, cytoplasmic | 75.6 | −6.64 | 0.000 | 108.9 | |
| 1832640714 | L-threonine dehydrogenase | 41.7 | −6.64 | 0.000 | 33.9 | 137.4 |
| 1832610610 | 60S ribosomal protein L39 | 6.4 | −6.64 | 0.000 | 126.1 | |
| 1832685492 | 39S ribosomal protein L45, mitochondrial | 35.9 | −6.64 | 0.000 | 72.9 | 168.9 |
| 1832700793 | TATA-binding protein-associated factor 2N isoform X1 | 43.2 | −6.64 | 0.000 | 19.6 | 147.4 |
| 1832687225 | 3-hydroxybutyrate dehydrogenase type 2 | 26.4 | −6.64 | 0.000 | 121.5 | 178.5 |
| 1832690325 | uncharacterized protein LOC117256382 isoform X3 | 72.6 | −6.64 | 0.000 | 130.4 | 169.6 |
| 1832622809 | RWD domain-containing protein 1 isoform X2 | 23.2 | −6.64 | 0.000 | 300.0 | |
| 1996377847 | DUF2850 domain-containing protein | 16.9 | −4.8 | 0.000 | 41.2 | 206.2 |
| 1832685958 | arachidonate 15-lipoxygenase B-like isoform X1 | 76.9 | −4.44 | 0.000 | 6.9 | 45.6 |
| 1832683648 | nucleolin | 74.2 | −3.43 | 0.000 | 71.9 | 105.2 |
| 1832680261 | low molecular weight neuronal intermediate filament | 59.2 | −3.3 | 0.000 | 66.6 | 152.2 |
| 1832613023 | transmembrane protein 258 | 9.1 | −3.15 | 0.000 | 90.0 | 104.2 |
| 1832671502 | basic leucine zipper and W2 domain-containing protein 1-A | 48.1 | −3.15 | 0.000 | 103.3 | 116.1 |
| 145105480 | interleukin enhancer-binding factor 2 | 42.8 | −2.63 | 0.001 | 62.4 | 174.9 |
| 161935902 | MHC class I alpha antigen | 41.6 | −2.63 | 0.000 | 23.1 | 176.2 |
| 1832684689 | trypsin-3 | 30.1 | −2.44 | 0.000 | 88.5 | 163.5 |
| 1832608120 | cytochrome c oxidase subunit 4 isoform 1, mitochondrial | 19.4 | −2.42 | 0.001 | 30.0 | 194.5 |
| 1832640175 | FAU ubiquitin-like and ribosomal protein S30 fusion a | 14.5 | −2.04 | 0.004 | 37.7 | 171.2 |
| 1832678052 | CD59B glycoprotein-like isoform X1 | 11.5 | −2.04 | 0.000 | 31.5 | 105.5 |
| 343459105 | NADH dehydrogenase 1 alpha, partial | 9.5 | −2.01 | 0.000 | 81.3 | 139.3 |
| 1832688344 | ADP/ATP translocase 1 | 32.9 | 2.26 | 0.001 | 28.7 | 215.2 |
| 1832676130 | brain acid soluble protein 1 | 36.1 | 2.27 | 0.004 | 76.4 | 137.1 |
| 1832620903 | cystatin-B | 11.9 | 2.41 | 0.003 | 17.5 | 89.8 |
| 1832667755 | elongation factor 1-alpha 1a | 50.5 | 3.18 | 0.000 | 54.3 | 99.7 |
| 327239696 | complement component C6, partial | 11.4 | 3.18 | 0.000 | 14.4 | 19.4 |
| 1832625921 | uncharacterized protein LOC117266161 | 62.4 | 3.2 | 0.000 | 64.4 | 120.6 |
| 1832674019 | deleted in malignant brain tumors 1 protein-like isoform X1 | 141.3 | 3.72 | 0.000 | 122.9 | 135.4 |
| 1832691617 | ubiquinol-cytochrome-c reductase complex assembly factor 2 | 15 | 6.64 | 0.000 | 104.7 | 113.7 |
| 1832614309 | very-long-chain 3-oxoacyl-CoA reductase-A | 37.2 | 6.64 | 0.000 | 48.9 | 187.2 |
| 1832627296 | isovaleryl-CoA dehydrogenase, mitochondrial | 45.9 | 6.64 | 0.000 | 95.8 | 116.4 |
| 1832635569 | annexin A1 | 39.2 | 6.64 | 0.000 | 58.0 | 125.4 |
| 1832617105 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 6, mitochondrial | 14.1 | 6.64 | 0.000 | 114.8 | 117.4 |
| Downregulated | ||||||
| 1832631564 | E3 ubiquitin-protein ligase RBX1 | 12.3 | −6.64 | 0.000 | 102.9 | 83.5 |
| 197725770 | mitochondrial cytochrome C oxidase subunit Vb precursor | 14.9 | −3.44 | 0.000 | 74.5 | 73.5 |
| 1832626150 | C-reactive protein | 25.5 | −3.11 | 0.000 | 65.4 | 53.7 |
| 2067150115 | Thioredoxin domain-containing protein 12 | 22.7 | 2.01 | 0.021 | 125.9 | 60.8 |
| 1832680021 | Betaine-homocysteine S-methyltransferase 1 | 44.1 | 2.02 | 0.003 | 207.7 | 86.8 |
| 1832680525 | ubiquitin-conjugating enzyme E2 L3 | 17.8 | 2.04 | 0.022 | 95.7 | 27.5 |
| 1832606043 | ras-related protein Rab-27A | 24.8 | 2.06 | 0.017 | 103.4 | 83.7 |
| 1710371347 | electron transfer flavoprotein subunit beta | 31.4 | 2.25 | 0.009 | 130.4 | 54.0 |
| 1832702451 | LOW-QUALITY PROTEIN: apolipoprotein(a)- | 122.9 | 2.26 | 0.000 | 132.7 | 27.3 |
| 1832645146 | pigment epithelium-derived factor | 44.7 | 2.38 | 0.000 | 228.9 | 46.5 |
| 1832639949 | collagen alpha-1(X) chain | 62.9 | 2.56 | 0.000 | 200.3 | 42.5 |
| 1832701092 | latexin | 33 | 2.58 | 0.000 | 136.3 | 24.0 |
| 1832662114 | parvalbumin alpha | 11.5 | 2.6 | 0.001 | 269.9 | 13.3 |
| 1832699882 | peptidyl-prolyl cis-trans isomerase FKBP10 | 62.4 | 2.64 | 0.000 | 241.4 | 32.8 |
| 343458999 | nattectin, partial | 18.1 | 2.79 | 0.000 | 229.8 | 50.1 |
| 1832696854 | protein bassoon | 431.2 | 2.9 | 0.000 | 159.2 | 43.0 |
| 1832670505 | coatomer subunit gamma-2 | 97.4 | 3 | 0.000 | 22.2 | 17.8 |
| 1379027526 | ribonuclease G | 55.4 | 3.21 | 0.000 | 142.9 | 15.5 |
| 209981964 | alpha-1-antitrypsin | 46.1 | 3.37 | 0.000 | 199.3 | 22.4 |
| 1832702196 | vasodilator-stimulated phosphoprotein isoform X1 | 44.6 | 3.87 | 0.000 | 177.5 | 122.5 |
| 834904058 | malate dehydrogenase | 32.4 | 4.52 | 0.000 | 200.3 | 54.0 |
| 1832627647 | PTTG1-interacting protein a | 18.8 | 4.52 | 0.000 | 147.0 | 30.5 |
| 1832656308 | neural cell adhesion molecule 1a isoform X4 | 109.2 | 5 | 0.000 | 155.3 | 14.2 |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | 36 | 5.03 | 0.000 | 130.0 | 65.7 |
| 306008591 | heat shock protein | 61.2 | 6.64 | 0.000 | 84.3 | |
| 295792244 | fructose-bisphosphate aldolase A | 39.6 | 6.64 | 0.000 | 221.9 | 29.6 |
| 328677149 | hypothetical protein, partial | 33.1 | 6.64 | 0.000 | 132.9 | 18.3 |
| 343459175 | ATP synthase, H+ transporting, mitochondrial F0 complex, partial | 18.2 | 6.64 | 0.000 | 156.9 | |
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | 36 | 6.64 | 0.000 | 188.4 | 87.4 |
| 1832630592 | glucosidase 2 subunit beta | 59.5 | 6.64 | 0.000 | 151.8 | |
| 1832669039 | aldo-keto reductase family 1 member B1 isoform X1 | 35.7 | 6.64 | 0.000 | 105.8 | |
| 1832632244 | parvalbumin beta | 11.7 | 6.64 | 0.000 | 180.6 | 96.3 |
| 1832620905 | stefin-C | 11.7 | 6.64 | 0.000 | 171.2 | 51.2 |
| 1832677686 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | 19 | 6.64 | 0.000 | 89.3 | 12.3 |
| 1832638111 | hyaluronan and proteoglycan link protein 1-like | 40 | 6.64 | 0.000 | 271.7 | |
| 1832619571 | tetranectin | 22.4 | 6.64 | 0.000 | 193.6 | 84.8 |
| 1832616138 | testin | 62.2 | 6.64 | 0.000 | 72.2 | |
| 1832604382 | histone H1 | 27.4 | 6.64 | 0.000 | 48.3 | |
| 1832692739 | protein SSUH2 homolog | 39.7 | 6.64 | 0.000 | 66.0 | |
| 834904842 | phosphoglycerate kinase | 41 | 6.64 | 0.000 | 286.0 | |
| 1832645835 | ruvB-like 2 | 50.7 | 6.64 | 0.000 | 300.0 | |
| 1832635997 | proteasome subunit beta type-7 isoform X1 | 32.9 | 6.64 | 0.000 | 300.0 | |
| 1832640826 | papilin b, proteoglycan-like sulfated glycoprotein | 63 | 6.64 | 0.000 | 161.6 | 54.5 |
| 1832641526 | uroporphyrinogen decarboxylase | 41.3 | 6.64 | 0.000 | 157.7 | |
| 1832690337 | chitinase | 50.6 | 6.64 | 0.000 | 300.0 | |
| 1832673253 | 60S ribosomal protein L8 | 28 | 6.64 | 0.000 | 101.8 | |
| 834904288 | TDP-fucosamine acetyltransferase | 25.4 | 6.64 | 0.000 | 199.5 | 6.5 |
| 1832638138 | transcription factor BTF3 | 17.7 | 6.64 | 0.000 | 79.7 | 14.6 |
| 1832643512 | rho GTPase-activating protein 12-like isoform X1 | 100.9 | 6.64 | 0.000 | 129.7 | |
| 1832644997 | coatomer subunit beta’ isoform X1 | 105.9 | 6.64 | 0.000 | 70.5 | |
| 1832642084 | neurogenic differentiation factor 6-B | 37.7 | 6.64 | 0.000 | 193.0 | 107.0 |
| 834906564 | cysteine desulfhydrase | 52.8 | 6.64 | 0.000 | 300.0 |
| Accession | Description | MW (kDa) | Fold Change | p-Value | Abundances Control | Abundances Susceptible |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| 306008591 | heat shock protein | 61.2 | −6.64 | 0.000 | 84.3 | 215.7 |
| 1832619927 | elongation factor 1-alpha | 50.3 | −6.64 | 0.000 | 268.5 | |
| 1832702676 | von Willebrand factor A domain-containing protein 5A-like isoform X1 | 69.9 | −6.64 | 0.000 | 198.0 | |
| 1832606995 | tropomyosin alpha-4 chain-like isoform X4 | 28.4 | −6.64 | 0.000 | 127.7 | |
| 1832685063 | hydroperoxide isomerase ALOXE3 | 82.7 | −6.64 | 0.000 | 300.0 | |
| 1832633330 | protein phosphatase 1, catalytic subunit, alpha isozyme a | 37.5 | −6.64 | 0.000 | 24.9 | 150.0 |
| 1832683648 | nucleolin | 74.2 | −6.64 | 0.000 | 71.9 | 122.8 |
| 1832671158 | 23 kDa integral membrane protein | 26.7 | −6.64 | 0.000 | 250.1 | |
| 1832663676 | catechol O-methyltransferase domain-containing protein 1-like isoform X1 | 28.6 | −6.64 | 0.000 | 213.3 | |
| 1832644368 | peroxiredoxin-6 | 24.6 | −6.64 | 0.000 | 300.0 | |
| 1832613275 | cytochrome c oxidase subunit 5A, mitochondrial | 16 | −6.64 | 0.000 | 181.2 | |
| 1832681058 | antithrombin-III | 51.2 | −6.64 | 0.000 | 44.5 | 163.2 |
| 1832628613 | EH domain-containing protein 4 | 62.3 | −6.64 | 0.000 | 201.9 | |
| 1832617553 | small nuclear ribonucleoprotein Sm D1 | 13.3 | −6.64 | 0.000 | 153.6 | |
| 1832651057 | uncharacterized protein si:ch1073-126c3.2 | 23.2 | −6.64 | 0.000 | 105.7 | |
| 1832661391 | complement factor I | 77.7 | −6.64 | 0.000 | 210.4 | |
| 1832670971 | LSM8 homolog, U6 small nuclear RNA associated | 10.4 | −6.64 | 0.000 | 300.0 | |
| 1832638785 | leucine-rich alpha-2-glycoprotein | 38.9 | −6.64 | 0.000 | 300.0 | |
| 1832671044 | deoxyribose-phosphate aldolase | 35 | −6.64 | 0.000 | 34.0 | 131.3 |
| 1832664977 | cytochrome c oxidase subunit 6B1 | 10.2 | −6.64 | 0.000 | 300.0 | |
| 1832613023 | transmembrane protein 258 | 9.1 | −6.64 | 0.000 | 90.0 | 105.8 |
| 1832645324 | arginine--tRNA ligase, cytoplasmic | 75.6 | −6.64 | 0.000 | 191.1 | |
| 1832612090 | heat shock 70 kDa protein 4b | 95.1 | −6.64 | 0.000 | 300.0 | |
| 1832640714 | L-threonine dehydrogenase | 41.7 | −6.64 | 0.000 | 33.9 | 128.6 |
| 1832626150 | C-reactive protein | 25.5 | −6.64 | 0.000 | 65.4 | 180.9 |
| 1832610610 | 60S ribosomal protein L39 | 6.4 | −6.64 | 0.000 | 173.9 | |
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | 104.1 | −6.64 | 0.000 | 300.0 | |
| 1832622572 | acyl-CoA thioesterase 9, tandem duplicate 1 isoform X1 | 49.1 | −6.64 | 0.000 | 79.4 | 106.4 |
| 1832631564 | E3 ubiquitin-protein ligase RBX1 | 12.3 | −6.64 | 0.000 | 102.9 | 113.6 |
| 1832700793 | TATA-binding protein-associated factor 2N isoform X1 | 43.2 | −6.64 | 0.000 | 19.6 | 133.0 |
| 1832644997 | coatomer subunit beta’ isoform X1 | 105.9 | −6.64 | 0.000 | 70.5 | 229.5 |
| 1832627111 | nesprin-2-like isoform X4 | 250.6 | −6.64 | 0.000 | 300.0 | |
| 1832670505 | coatomer subunit gamma-2 | 97.4 | −6.26 | 0.000 | 22.2 | 260.0 |
| 327239696 | complement component C6 | 11.4 | −5.36 | 0.000 | 14.4 | 266.2 |
| 1832685958 | arachidonate 15-lipoxygenase B-like isoform X1 | 76.9 | −5.04 | 0.000 | 6.9 | 247.5 |
| 1832620903 | cystatin-B | 11.9 | −4.32 | 0.000 | 17.5 | 192.7 |
| 1832656769 | vesicle-associated membrane protein 2 | 12 | −4.29 | 0.000 | 45.4 | 236.3 |
| 45269063 | immunoglobulin light-chain variable region | 11.9 | −3.91 | 0.000 | 54.4 | 202.5 |
| 1832638138 | transcription factor BTF3 | 17.7 | −3.64 | 0.000 | 79.7 | 205.7 |
| 197725770 | mitochondrial cytochrome C oxidase subunit Vb precursor | 14.9 | −3.61 | 0.000 | 74.5 | 152.0 |
| 328677247 | glyceraldehyde 3-phosphate dehydrogenase isoform, partial | 24.3 | −3.13 | 0.002 | 101.5 | 169.7 |
| 1832679851 | drebrin-like b isoform X1 | 53.6 | −3.05 | 0.003 | 31.0 | 256.8 |
| 1832650400 | prefoldin subunit 1 isoform X1 | 15.6 | −2.96 | 0.000 | 49.6 | 195.5 |
| 221048061 | fructose-bisphosphate aldolase | 33.4 | −2.92 | 0.000 | 56.5 | 199.4 |
| 1832658469 | fibrinogen beta chain | 40.2 | −2.83 | 0.000 | 27.7 | 224.7 |
| 1832612579 | 60S ribosomal protein L27a | 16.6 | −2.65 | 0.005 | 26.6 | 166.9 |
| 222087999 | fibrinogen beta chain precursor | 16.7 | −2.61 | 0.000 | 28.6 | 197.6 |
| 1832689402 | fibrinogen gamma chain | 48.5 | −2.49 | 0.000 | 33.5 | 221.5 |
| 1832662038 | leukocyte cell-derived chemotaxin-2 | 17.1 | −2.39 | 0.000 | 39.6 | 145.9 |
| 1832604382 | histone H1 | 27.4 | −2.38 | 0.016 | 48.3 | 251.7 |
| 1832678052 | CD59B glycoprotein-like isoform X1 | 11.5 | −2.38 | 0.000 | 31.5 | 163.0 |
| 1832685624 | microfibril-associated glycoprotein 4 | 28.1 | −2.36 | 0.039 | 42.3 | 180.9 |
| 1832634516 | keratin, type I cytoskeletal 13 | 48.2 | −2.32 | 0.000 | 38.0 | 189.1 |
| 1832685241 | beta-2-glycoprotein 1 | 40.1 | −2.23 | 0.012 | 43.7 | 189.4 |
| 1832623025 | apolipoprotein Eb | 30.8 | −2.2 | 0.000 | 35.0 | 159.4 |
| 1832688344 | ADP/ATP translocase 1 | 32.9 | 3.23 | 0.000 | 28.7 | 56.1 |
| 405790938 | MHC class II antigen, partial | 18.4 | 6.64 | 0.000 | 57.5 | 137.5 |
| 1832677686 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | 19 | 6.64 | 0.000 | 89.3 | 198.3 |
| 1832616138 | testin | 62.2 | 6.64 | 0.000 | 72.2 | 227.8 |
| 295792326 | mitochondrial cytochrome c oxidase subunit 7C | 7.1 | 6.64 | 0.000 | 99.8 | 121.7 |
| Downregulated | ||||||
| 343459175 | ATP synthase, H+ transporting, mitochondrial F0 complex | 18.2 | −6.64 | 0.000 | 156.9 | 143.1 |
| 834904842 | phosphoglycerate kinase | 41 | −6.64 | 0.000 | 286.0 | 14.0 |
| 1832671502 | basic leucine zipper and W2 domain-containing protein 1-A | 48.1 | −6.64 | 0.000 | 103.3 | 80.5 |
| 1832630592 | glucosidase 2 subunit beta | 59.5 | −3.16 | 0.002 | 151.8 | 148.2 |
| 1832626146 | allograft inflammatory factor 1 | 16.6 | −3.36 | 0.000 | 194.3 | 50.7 |
| 1832685492 | 39S ribosomal protein L45, mitochondrial | 35.9 | −3.36 | 0.000 | 72.9 | 58.3 |
| 1832610590 | DNA-directed RNA polymerase I subunit RPA1 | 191 | 2.02 | 0.001 | 167.4 | 50.2 |
| 451931262 | Multidrug resistance efflux pump | 37.1 | 2.04 | 0.006 | 159.8 | 38.8 |
| 1832645146 | pigment epithelium-derived factor | 44.7 | 2.44 | 0.000 | 228.9 | 24.7 |
| 1832632244 | parvalbumin beta | 11.7 | 2.62 | 0.000 | 180.6 | 23.2 |
| 1832616865 | receptor-transporting protein 4 | 18.9 | 2.14 | 0.000 | 154.5 | 68.8 |
| 1832625915 | uncharacterized protein LOC117266159 isoform X1 | 70.4 | 2.14 | 0.000 | 145.3 | 47.5 |
| 295792242 | nucleoside diphosphate kinase B | 17 | 2.33 | 0.000 | 260.4 | 9.1 |
| 1832699882 | peptidyl-prolyl cis-trans isomerase FKBP10 | 62.4 | 3.26 | 0.000 | 241.4 | 25.8 |
| 1832702726 | serpin H1b | 45.6 | 2.32 | 0.000 | 222.4 | 39.4 |
| 1832639949 | collagen alpha-1(X) chain | 62.9 | 2.34 | 0.000 | 200.3 | 57.2 |
| 1832662114 | parvalbumin alpha | 11.5 | 4.01 | 0.000 | 269.9 | 16.8 |
| 1832620905 | stefin-C | 11.7 | 3.97 | 0.000 | 171.2 | 77.6 |
| 295792244 | fructose-bisphosphate aldolase A | 39.6 | 3.61 | 0.000 | 221.9 | 48.4 |
| 1832619571 | tetranectin | 22.4 | 3.62 | 0.000 | 193.6 | 21.6 |
| 834906594 | hypothetical protein QY76_16135 | 34.4 | 2.63 | 0.000 | 175.8 | 28.5 |
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | 36 | 4.02 | 0.000 | 188.4 | 24.3 |
| 1832616029 | three prime repair exonuclease 2 | 22.1 | 3.2 | 0.000 | 179.9 | 54.9 |
| 343458999 | nattectin, partial | 18.1 | 3.76 | 0.000 | 229.8 | 20.1 |
| 834906564 | cysteine desulfhydrase | 52.8 | 6.64 | 0.000 | 300.0 | |
| 1832680021 | betaine--homocysteine S-methyltransferase 1 | 44.1 | 6.64 | 0.000 | 207.7 | 5.6 |
| 1832702196 | vasodilator-stimulated phosphoprotein isoform X1 | 44.6 | 6.64 | 0.000 | 177.5 | |
| 1832665474 | sulfotransferase 2B1 | 22.7 | 6.64 | 0.000 | 183.6 | 21.7 |
| 1832641369 | phosphotriesterase-related protein | 38.8 | 6.64 | 0.000 | 140.3 | |
| 1832645835 | ruvB-like 2 | 50.7 | 6.64 | 0.000 | 300.0 | |
| 1832635997 | proteasome subunit beta type-7 isoform X1 | 32.9 | 6.64 | 0.000 | 300.0 | |
| 1832690337 | chitinase | 50.6 | 6.64 | 0.000 | 300.0 | |
| 1832686354 | puromycin-sensitive aminopeptidase | 104.2 | 6.64 | 0.000 | 146.9 | |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | 36 | 6.64 | 0.000 | 130.0 | 104.4 |
| 1832695641 | N-alpha-acetyltransferase 10 isoform X1 | 24.8 | 6.64 | 0.000 | 109.3 | |
| 1996375967 | methionine ABC transporter substrate-binding protein | 28.8 | 6.64 | 0.000 | 125.4 | |
| 1832682447 | dedicator of cytokinesis protein 7 isoform X1 | 244.1 | 6.64 | 0.000 | 176.4 | |
| 1832697234 | L-rhamnose-binding lectin SML | 23.9 | 6.64 | 0.000 | 103.5 | |
| 1832687225 | 3-hydroxybutyrate dehydremp24ogenase type 2 | 26.4 | 6.64 | 0.000 | 121.5 | |
| 1832698896 | uncharacterized protein KIAA2013 homolog isoform X1 | 69.5 | 6.64 | 0.000 | 160.6 | |
| 1832690325 | uncharacterized protein LOC117256382 isoform X3 | 72.6 | 6.64 | 0.000 | 130.4 | |
| 1832642084 | neurogenic differentiation factor 6-B | 37.7 | 6.64 | 0.000 | 193.0 |
| Accession | Description | MW (kDa) | FC (S vs. R) | p-Value | Abundances Susceptible | Abundances Resistant |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| 1832625921 | uncharacterized protein LOC117266161 | 62.4 | 3.11 | 0.000 | 115.0 | 120.6 |
| 1832606995 | tropomyosin alpha-4 chain-like isoform X4 | 28.4 | 3.23 | 0.000 | 127.7 | 172.3 |
| 1832625753 | glyceraldehyde-3-phosphate dehydrogenase | 36 | 3.25 | 0.000 | 24.3 | 87.4 |
| 1832626146 | allograft inflammatory factor 1 | 16.6 | 4.27 | 0.000 | 50.7 | 55.0 |
| 1832632244 | parvalbumin beta | 11.7 | 6.64 | 0.000 | 23.2 | 96.3 |
| 1832691617 | ubiquinol-cytochrome-c reductase complex assembly factor 2 | 15 | 6.64 | 0.000 | 81.5 | 113.7 |
| 1832614309 | very-long-chain 3-oxoacyl-CoA reductase-A | 37.2 | 6.64 | 0.000 | 64.0 | 187.2 |
| 1832627296 | isovaleryl-CoA dehydrogenase, mitochondrial | 45.9 | 6.64 | 0.000 | 87.8 | 116.4 |
| 1832635569 | annexin A1 | 39.2 | 6.64 | 0.000 | 116.5 | 125.4 |
| 834904058 | malate dehydrogenase | 32.4 | 6.64 | 0.000 | 45.7 | 54.0 |
| 1832622572 | acyl-CoA thioesterase 9, tandem duplicate 1 isoform X1 | 49.1 | 6.64 | 0.000 | 106.4 | 114.2 |
| 1832617105 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | 14.1 | 6.64 | 0.000 | 67.8 | 117.4 |
| 1832667938 | sideroflexin-3 | 35.5 | −2.71 | 0.001 | 37.1 | 147.6 |
| 834906594 | hypothetical protein QY76_16135 | 34.4 | −2.26 | 0.000 | 28.5 | 95.7 |
| 1832634797 | transmembrane emp24 domain-containing protein 2 isoform X1 | 23.3 | −6.64 | 0.000 | 300.0 | |
| 1832680021 | betaine--homocysteine S-methyltransferase 1 | 44.1 | −6.64 | 0.000 | 5.6 | 86.8 |
| 1832702196 | vasodilator-stimulated phosphoprotein isoform X1 | 44.6 | −6.64 | 0.000 | 122.5 | |
| 1832665474 | sulfotransferase 2B1 | 22.7 | −6.64 | 0.000 | 21.7 | 94.7 |
| 1832641369 | phosphotriesterase-related protein | 38.8 | −6.64 | 0.000 | 159.7 | |
| 1832686354 | puromycin-sensitive aminopeptidase | 104.2 | −6.64 | 0.000 | 153.1 | |
| 1832695641 | N-alpha-acetyltransferase 10 isoform X1 | 24.8 | −6.64 | 0.000 | 190.7 | |
| 1996375967 | methionine ABC transporter substrate-binding protein | 28.8 | −6.64 | 0.000 | 174.6 | |
| 1832682447 | dedicator of cytokinesis protein 7 isoform X1 | 244.1 | −6.64 | 0.000 | 123.6 | |
| 1832697234 | L-rhamnose-binding lectin SML | 23.9 | −6.64 | 0.000 | 196.5 | |
| 1832687225 | 3-hydroxybutyrate dehydrogenase type 2 | 26.4 | −6.64 | 0.000 | 178.5 | |
| 1832698896 | uncharacterized protein KIAA2013 homolog isoform X1 | 69.5 | −6.64 | 0.000 | 139.4 | |
| 1832690325 | uncharacterized protein LOC117256382 isoform X3 | 72.6 | −6.64 | 0.000 | 169.6 | |
| 1832642084 | neurogenic differentiation factor 6-B | 37.7 | −6.64 | 0.000 | 107.0 | |
| 1832622809 | RWD domain-containing protein 1 isoform X2 | 23.2 | −6.64 | 0.000 | 300.0 | |
| Downregulated | ||||||
| 1832702676 | von Willebrand factor A domain-containing protein 5A-like isoform X1 | 69.9 | −2.56 | 0.002 | 198.0 | 102.0 |
| 1393214856 | type I glyceraldehyde-3-phosphate dehydrogenase | 36 | −3.16 | 0.000 | 104.4 | 65.7 |
| 405790938 | MHC class II antigen, partial | 18.4 | −2.82 | 0.000 | 137.5 | 105.0 |
| 1832645324 | arginine--tRNA ligase, cytoplasmic | 75.6 | −6.64 | 0.000 | 191.1 | 108.9 |
| 295792326 | mitochondrial cytochrome c oxidase subunit 7C | 7.1 | −6.64 | 0.000 | 121.7 | 78.5 |
| 1832680525 | ubiquitin-conjugating enzyme E2 L3 | 17.8 | 2.27 | 0.034 | 176.8 | 27.5 |
| 1832608362 | AP-1 complex subunit gamma-1 isoform X1 | 91.3 | 2.48 | 0.022 | 161.8 | 74.6 |
| 1832621671 | rRNA 2’-O-methyltransferase fibrillarin | 33.5 | 2.35 | 0.012 | 177.5 | 76.4 |
| 45269063 | immunoglobulin light-chain variable region, partial | 11.9 | 2.43 | 0.010 | 202.5 | 43.1 |
| 1832611868 | transmembrane 9 superfamily member 2 isoform X1 | 76 | 2.2 | 0.004 | 152.1 | 48.3 |
| 1832685958 | arachidonate 15-lipoxygenase B-like isoform X1 | 76.9 | 2.06 | 0.004 | 247.5 | 45.6 |
| 1832671468 | coagulation factor VIIi | 50.6 | 2.29 | 0.004 | 162.7 | 32.2 |
| 1832667755 | elongation factor 1-alpha 1a | 50.5 | 2.96 | 0.003 | 146.0 | 99.7 |
| 1832702451 | LOW-QUALITY PROTEIN: apolipoprotein(a) | 122.9 | 2.34 | 0.001 | 140.0 | 27.3 |
| 1832618423 | class I histocompatibility antigen, F10 alpha chain-like isoform X1 | 40.8 | 2.5 | 0.000 | 143.6 | 71.1 |
| 62255674 | immunoglobulin mu heavy-chain | 66.3 | 2.02 | 0.000 | 162.2 | 57.2 |
| 295792244 | fructose-bisphosphate aldolase A | 39.6 | 3.38 | 0.000 | 48.4 | 29.6 |
| 157929900 | ribosomal protein L23 | 15 | 2.09 | 0.000 | 123.4 | 40.6 |
| 2022331196 | immunoglobulin M heavy-chain constant mu variant 1 | 50.8 | 2.34 | 0.000 | 196.3 | 38.7 |
| 834906005 | glyceraldehyde-3-phosphate dehydrogenase | 35.5 | 3.65 | 0.000 | 3.2 | 0.9 |
| 1832671158 | 23 kDa integral membrane protein | 26.7 | 2.98 | 0.000 | 250.1 | 49.9 |
| 1832604432 | erythroblast NAD(P)(+)-arginine ADP-ribosyltransferase | 36.7 | 3.26 | 0.000 | 198.8 | 56.8 |
| 1832697311 | nucleobindin-2a isoform X1 | 59.4 | 2.93 | 0.000 | 163.7 | 24.8 |
| 1832604652 | fibrinogen alpha chain | 13.7 | 2.28 | 0.000 | 197.6 | 66.4 |
| 1832701092 | latexin | 33 | 2.94 | 0.000 | 139.7 | 24.0 |
| 326632479 | MHC class II antigen | 26.1 | 3.14 | 0.000 | 202.4 | 32.7 |
| 1832660271 | macrosialin | 38.8 | 2.67 | 0.000 | 179.2 | 27.2 |
| 1832689402 | fibrinogen gamma chain | 48.5 | 2.28 | 0.000 | 221.5 | 45.0 |
| 1832656769 | vesicle-associated membrane protein 2 | 12 | 3.11 | 0.000 | 236.3 | 18.4 |
| 380006108 | MHC class II antigen | 10.2 | 3.19 | 0.000 | 112.0 | 100.2 |
| 1832658469 | fibrinogen beta chain | 40.2 | 2.36 | 0.000 | 224.7 | 47.6 |
| 1379027526 | ribonuclease G | 55.4 | 3.19 | 0.000 | 141.6 | 15.5 |
| 2022331198 | immunoglobulin M heavy-chain constant mu variant 2 | 50.3 | 2.52 | 0.000 | 198.7 | 48.2 |
| 158602734 | immunoglobulin light chain | 26.4 | 3.6 | 0.000 | 172.9 | 15.8 |
| 1832627647 | PTTG1-interacting protein a | 18.8 | 3.53 | 0.000 | 122.5 | 30.5 |
| 1832677686 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | 19 | 4.01 | 0.000 | 198.3 | 12.3 |
| 306008591 | heat shock protein | 61.2 | 6.64 | 0.000 | 215.7 | |
| 328677149 | hypothetical protein | 33.1 | 6.64 | 0.000 | 148.8 | 18.3 |
| 343459175 | ATP synthase, H+ transporting, mitochondrial F0 complex | 18.2 | 6.64 | 0.000 | 143.1 | |
| 1832685063 | hydroperoxide isomerase ALOXE3 | 82.7 | 6.64 | 0.000 | 300.0 | |
| 1832630592 | glucosidase 2 subunit beta | 59.5 | 6.64 | 0.000 | 148.2 | |
| 1832644368 | peroxiredoxin-6 | 24.6 | 6.64 | 0.000 | 300.0 | |
| 1832669039 | aldo-keto reductase family 1 member B1 isoform X1 | 35.7 | 6.64 | 0.000 | 194.2 | |
| 1832638111 | hyaluronan and proteoglycan link protein 1 | 40 | 6.64 | 0.000 | 28.3 | |
| 1832616138 | testin | 62.2 | 6.64 | 0.000 | 227.8 | |
| 1832670971 | LSM8 homolog, U6 small nuclear RNA associated | 10.4 | 6.64 | 0.000 | 300.0 | |
| 1832638785 | leucine-rich alpha-2-glycoprotein | 38.9 | 6.64 | 0.000 | 300.0 | |
| 1832604382 | histone H1 | 27.4 | 6.64 | 0.000 | 251.7 | |
| 327239696 | complement component C6 | 11.4 | 6.64 | 0.000 | 266.2 | 19.4 |
| 1832664977 | cytochrome c oxidase subunit 6B1 | 10.2 | 6.64 | 0.000 | 300.0 | |
| 1832620903 | cystatin B | 11.9 | 6.64 | 0.000 | 192.7 | 89.8 |
| 1832692739 | protein SSUH2 homolog | 39.7 | 6.64 | 0.000 | 234.0 | |
| 834904842 | phosphoglycerate kinase | 41 | 6.64 | 0.000 | 14.0 | |
| 1832656308 | neural cell adhesion molecule 1a isoform X4 | 109.2 | 6.64 | 0.000 | 130.5 | 14.2 |
| 1832670505 | coatomer subunit gamma-2 | 97.4 | 6.64 | 0.000 | 260.0 | 17.8 |
| 1832612090 | heat shock 70 kDa protein 4b | 95.1 | 6.64 | 0.000 | 300.0 | |
| 1832640826 | papilin b, proteoglycan-like sulfated glycoprotein | 63 | 6.64 | 0.000 | 83.8 | 54.5 |
| 1832641526 | uroporphyrinogen decarboxylase | 41.3 | 6.64 | 0.000 | 142.3 | |
| 1042045193 | glycine dehydrogenase (aminomethyl-transferring) | 104.1 | 6.64 | 0.000 | 300.0 | |
| 1832673253 | 60S ribosomal protein L8 | 28 | 6.64 | 0.000 | 198.2 | |
| 834904288 | TDP-fucosamine acetyltransferase | 25.4 | 6.64 | 0.000 | 94.0 | 6.5 |
| 1832638138 | transcription factor BTF3 | 17.7 | 6.64 | 0.000 | 205.7 | 14.6 |
| 1832643512 | rho GTPase-activating protein 12-like isoform X1 | 100.9 | 6.64 | 0.000 | 170.3 | |
| 1832644997 | coatomer subunit beta’ isoform X1 | 105.9 | 6.64 | 0.000 | 229.5 | |
| 1832627111 | nesprin-2-like isoform X4 | 250.6 | 6.64 | 0.000 | 300.0 |
| Pathway ID | Description | Protein Count | p-Value |
|---|---|---|---|
| 1100 | Metabolic pathways | 30 | 0.000 |
| 190 | Oxidative phosphorylation | 9 | 0.000 |
| 10 | Glycolysis/Gluconeogenesis | 7 | 0.000 |
| 1200 | Carbon metabolism | 8 | 0.000 |
| 1230 | Biosynthesis of amino acids | 6 | 0.000 |
| 4260 | Cardiac muscle contraction | 6 | 0.000 |
| 3010 | Ribosome | 5 | 0.001 |
| 30 | Pentose phosphate pathway | 3 | 0.001 |
| 4672 | Intestinal immune network for IgA production | 3 | 0.001 |
| 51 | Fructose and mannose metabolism | 3 | 0.001 |
| 260 | Glycine, serine and threonine metabolism | 3 | 0.002 |
| 4514 | Cell adhesion molecules (CAMs) | 4 | 0.005 |
| 4145 | Phagosome | 4 | 0.007 |
| 5168 | Herpes simplex virus 1 infection | 4 | 0.014 |
| 630 | Glyoxylate and dicarboxylate metabolism | 2 | 0.015 |
| 270 | Cysteine and methionine metabolism | 2 | 0.029 |
| 72 | Synthesis and degradation of ketone bodies | 1 | 0.047 |
| Accession | Description | MW (kDa) | FC (S vs. R) | p-Value (S vs. R) | FC (C vs. R) | p-Value (C vs. R) | Abundances Susceptible | Abundances Resistant | Abundance Control |
|---|---|---|---|---|---|---|---|---|---|
| 1832634797 | transmembrane emp24 domain-containing protein 2 isoform X1 | 23.3 | −6.64 | 0.00 | −6.64 | 0.00 | 300.0 | ||
| 1832702196 | vasodilator-stimulated phosphoprotein isoform X1 | 44.6 | −6.64 | 0.00 | 3.87 | 0.00 | 122.5 | 177.5 | |
| 1832641369 | phosphotriesterase-related protein | 38.8 | −6.64 | 0.00 | 0.89 | 0.65 | 159.7 | 140.3 | |
| 1832686354 | puromycin-sensitive aminopeptidase | 104.2 | −6.64 | 0.00 | −1.25 | 0.38 | 153.1 | 146.9 | |
| 1832695641 | N-alpha-acetyltransferase 10 isoform X1 | 24.8 | −6.64 | 0.00 | −0.80 | 0.65 | 190.7 | 109.3 | |
| 1996375967 | methionine ABC transporter substrate-binding protein | 28.8 | −6.64 | 0.00 | 1.70 | 0.13 | 174.6 | 125.4 | |
| 1832682447 | dedicator of cytokinesis protein 7 isoform X1 | 244.1 | −6.64 | 0.00 | 0.81 | 0.65 | 123.6 | 176.4 | |
| 1832697234 | L-rhamnose-binding lectin SML | 23.9 | −6.64 | 0.00 | −0.93 | 0.60 | 196.5 | 103.5 | |
| 1832687225 | 3-hydroxybutyrate dehydrogenase type 2 | 26.4 | −6.64 | 0.00 | −6.64 | 0.00 | 178.5 | 121.5 | |
| 1832698896 | uncharacterized protein KIAA2013 homolog isoform X1 | 69.5 | −6.64 | 0.00 | 0.47 | 0.78 | 139.4 | 160.6 | |
| 1832690325 | uncharacterized protein LOC117256382 isoform X3 | 72.6 | −6.64 | 0.00 | −6.64 | 0.00 | 169.6 | 130.4 | |
| 1832642084 | neurogenic differentiation factor 6-B | 37.7 | −6.64 | 0.00 | 6.64 | 0.00 | 107.0 | 193 | |
| 1832622809 | RWD domain-containing protein 1 isoform X2 | 23.2 | −6.64 | 0.00 | −6.64 | 0.00 | 300.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nurhikmah; Christianus, A.; Wan Solahudin, W.M.S.; Lau, B.Y.C.; Ismail, I.S.; Fei, L.C. Skin Mucus Proteome Analysis Reveals Disease-Resistant Biomarker Signatures in Hybrid Grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) against Vibrio alginolyticus. Fishes 2022, 7, 278. https://doi.org/10.3390/fishes7050278
Nurhikmah, Christianus A, Wan Solahudin WMS, Lau BYC, Ismail IS, Fei LC. Skin Mucus Proteome Analysis Reveals Disease-Resistant Biomarker Signatures in Hybrid Grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) against Vibrio alginolyticus. Fishes. 2022; 7(5):278. https://doi.org/10.3390/fishes7050278
Chicago/Turabian StyleNurhikmah, Annie Christianus, Wan Mohd Syazwan Wan Solahudin, Benjamin Yii Chung Lau, Intan Safinar Ismail, and Low Chen Fei. 2022. "Skin Mucus Proteome Analysis Reveals Disease-Resistant Biomarker Signatures in Hybrid Grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) against Vibrio alginolyticus" Fishes 7, no. 5: 278. https://doi.org/10.3390/fishes7050278
APA StyleNurhikmah, Christianus, A., Wan Solahudin, W. M. S., Lau, B. Y. C., Ismail, I. S., & Fei, L. C. (2022). Skin Mucus Proteome Analysis Reveals Disease-Resistant Biomarker Signatures in Hybrid Grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂) against Vibrio alginolyticus. Fishes, 7(5), 278. https://doi.org/10.3390/fishes7050278







