Comprehensive Genome-Wide Analysis of Wnt Gene Family and Expression Profiling during Limb Regeneration in Portunus trituberculatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the P. trituberculatus and L. vannamei Wnt Families
2.2. Phylogenetic Analysis, Gene Structure, and Motif Identification Analysis of the Wnt Family in P. trituberculatus
2.3. Chromosome Localization and Gene Duplication
2.4. Transcriptome-Based Expression Profiling and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of PtWnt Genes
2.5. Expression Patterns of the Wnt Family by qRT-PCR during Limb Regeneration in P. trituberculatus
2.6. Statistical Analysis
3. Results
3.1. Identification of Wnt Genes in P. trituberculatus and L. vannamei
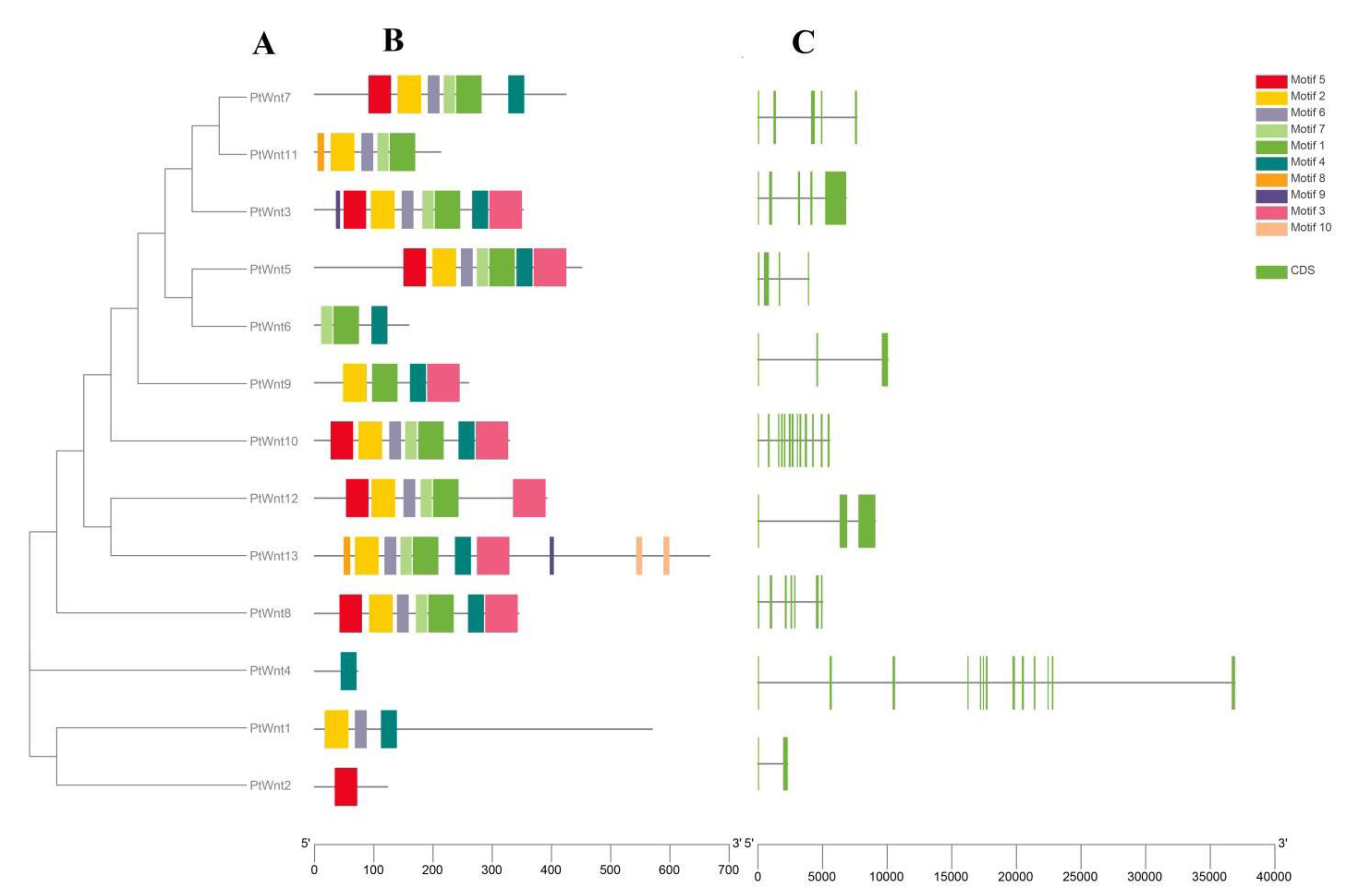
3.2. Phylogenetic Analysis
3.3. Characteristics of PtWnt Domains
3.4. Chromosomal Distribution
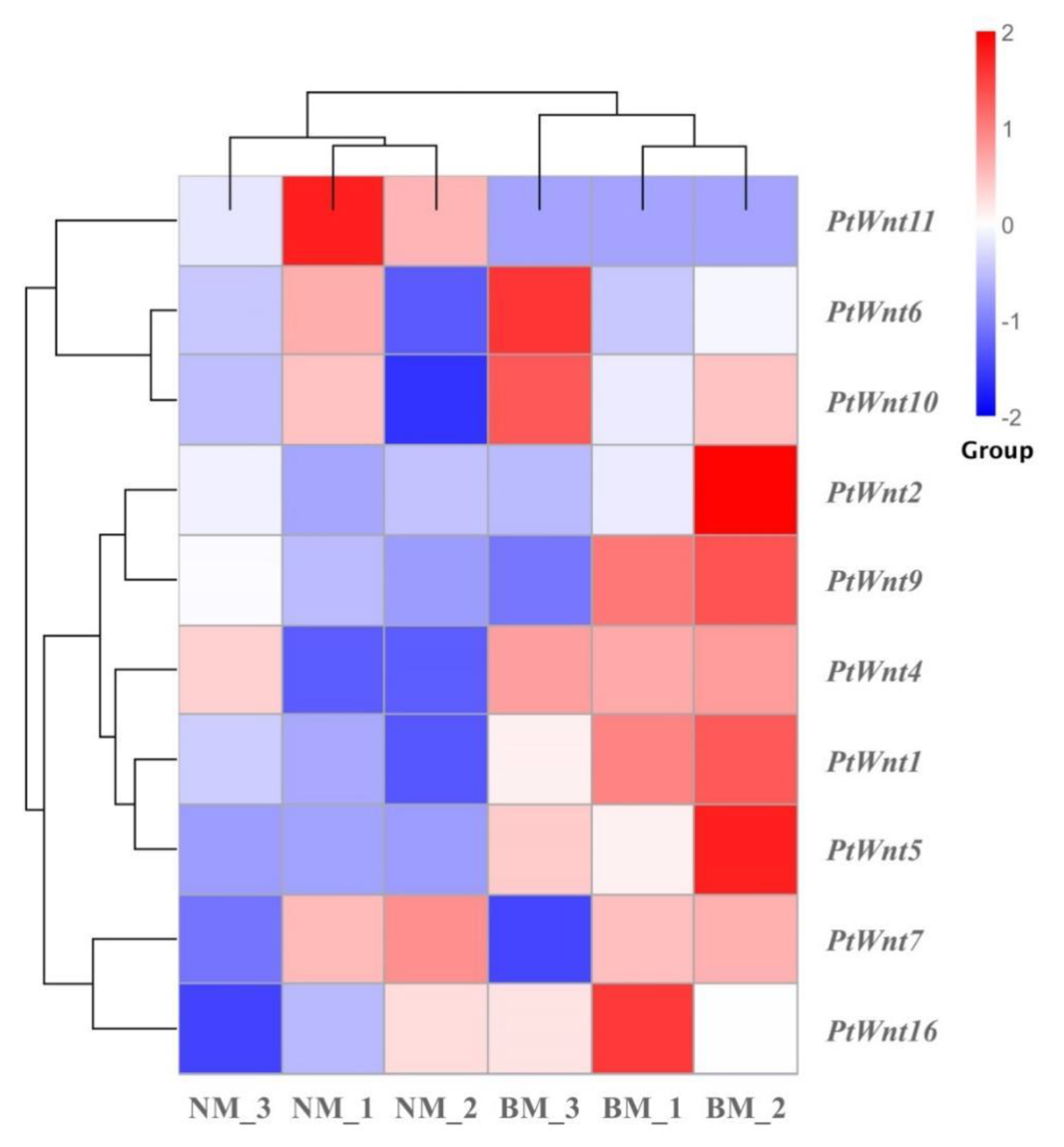
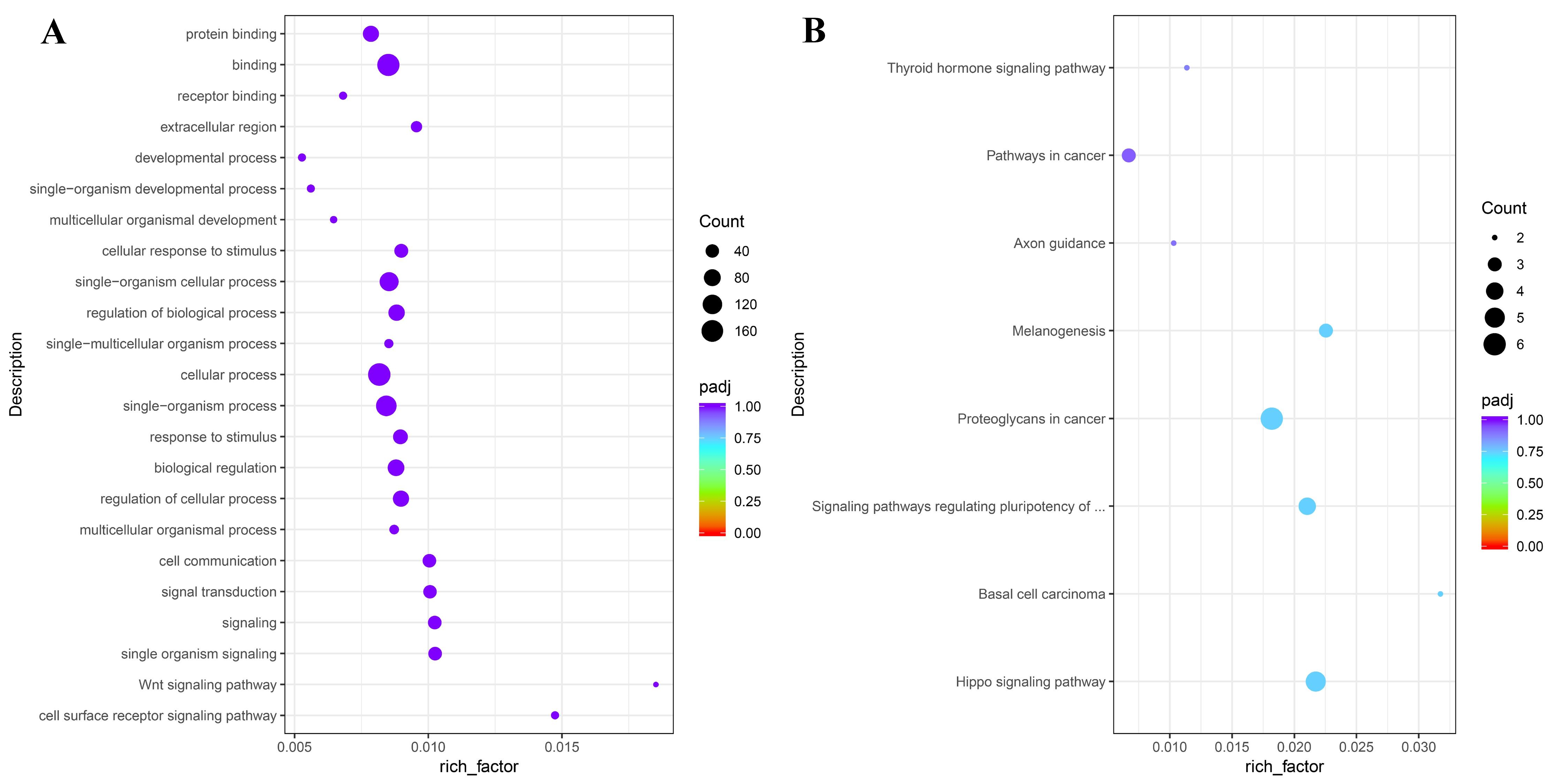
3.5. Regeneration Limb Transcriptome-Based Expression Profiling and GO and KEGG Enrichment Analysis of PtWnt Genes
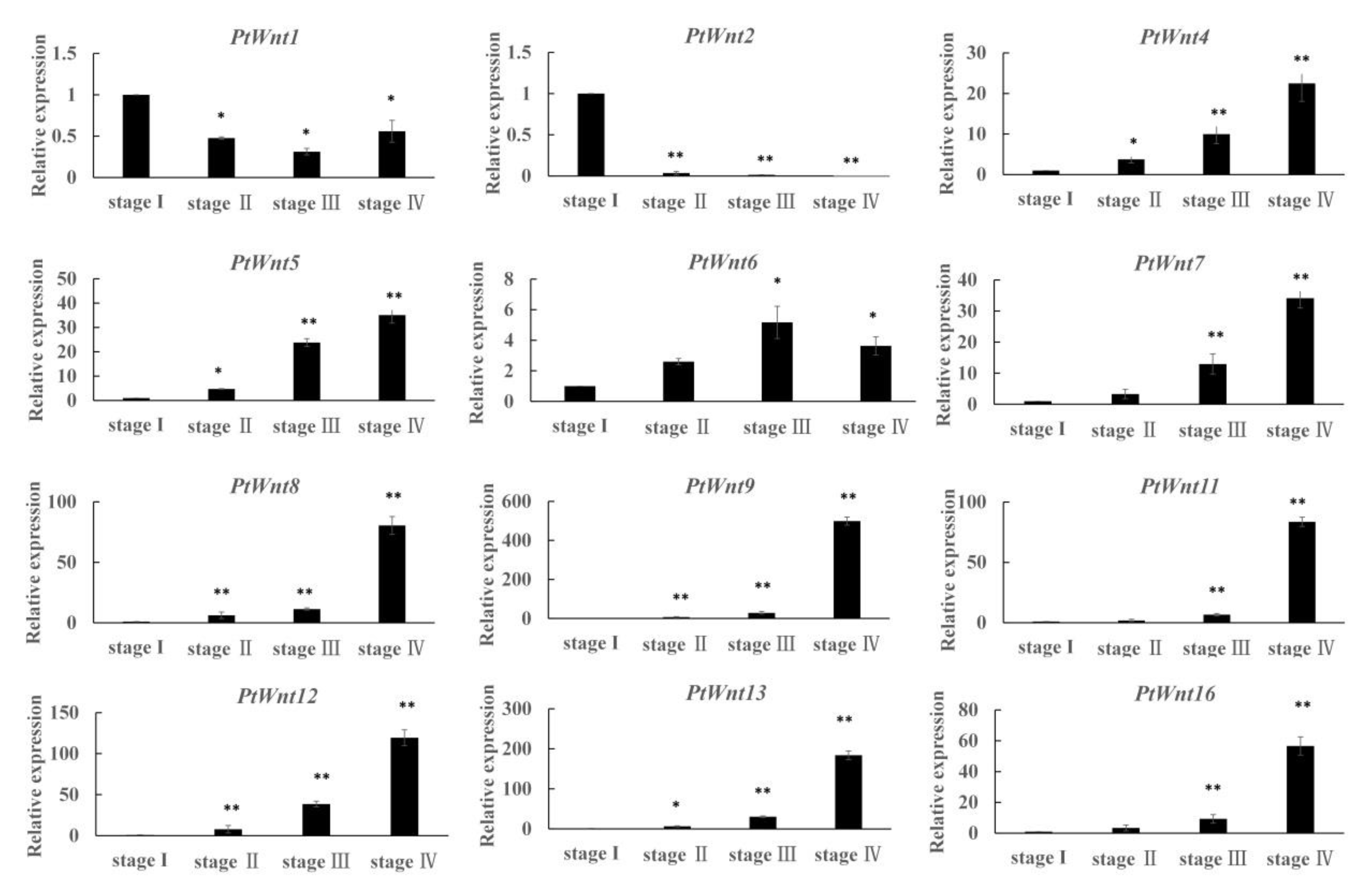
3.6. Expression Profiling of PtWnt Genes at Different Developmental Stages of Limb Regeneration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Yokoyama, H.; Ogino, H.; Stoick-Cooper, C.L.; Grainger, R.M.; Moon, R.T. Wnt/beta-catenin signaling has an essential role in the initiation of limb regeneration. Dev. Biol. 2007, 306, 170–178. [Google Scholar] [CrossRef]
- Vafopoulou, X. Mechanisms of wound repair in crayfish. ISJ 2009, 6, 125–137. [Google Scholar]
- Hopkins, P.M. Regeneration of walking legs in the fiddler crab Uca pugilator. Am. Zool. 1993, 33, 348–356. [Google Scholar] [CrossRef]
- Hopkins, P.M. Limb regeneration in the fiddler crab, Uca pugilator: Hormonal and growth factor control. Am. Zool. 2001, 41, 389–398. [Google Scholar] [CrossRef]
- Cooper, R.L. Development of sensory processes during limb regeneration in adult crayfish. J. Exp. Biol. 1998, 201, 1745–1752. [Google Scholar] [CrossRef]
- Das, S.; Durica, D.S. Ecdysteroid receptor signaling disruption obstructs blastemal cell proliferation during limb regeneration in the fiddler crab, Uca pugilator. Mol. Cell. Endocrinol. 2013, 365, 249–259. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Q.; Song, X.Z.; Pang, Y.Y.; Song, Y.M.; Wang, Y.Y.; He, L.; Lv, J.H.; Cheng, Y.X.; Yang, X.Z. L-tryptophan promotes the cheliped regeneration of Chinese mitten crab (Eriocheir sinensis) through melatonin, serotonin and dopamine involvement. Aquaculture 2019, 511, 734205. [Google Scholar] [CrossRef]
- Ventura, T.; Stewart, M.J.; Chandler, J.C.; Rotgans, B.; Elizur, A.; Hewitt, A.W. Molecular aspects of eye development and regeneration in the Australian redclaw crayfish, Cherax quadricarinatus. Aquac. Fish. 2019, 4, 27–36. [Google Scholar] [CrossRef]
- Ren, Z.M.; Fu, Y.Y.; Liu, L.; Liu, X.; Wang, C.L. Expression and function of WNT4 involved in larvae development and limb regeneration in Portunus trituberculatus. J. Oceanol. Limnol. 2021, 39, 306–316. [Google Scholar] [CrossRef]
- Kawakami, Y.; Esteban, C.R.; Raya, M.; Kawakami, H.; Martí, M.; Dubova, I.; Izpisúa Belmonte, J.C. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006, 20, 3232–3237. [Google Scholar] [CrossRef]
- De Robertis, E.M. Wnt signaling in axial patterning and regeneration: Lessons from planaria. Sci. Signal. 2010, 6, pe21. [Google Scholar] [CrossRef] [PubMed]
- Lengfeld, T.; Watanabe, H.; Simakov, O.; Lindgens, D.; Gee, L.; Law, L.; Schmidt, H.A.; Özbek, S.; Bode, H.; Holstein, T.W. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 2009, 330, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Vallès, Y.; Giani, V.C., Jr.; Seaver, E.C.; Weisblat, D.A. Evolutionary dynamics of the Wnt gene family: A lophotrochozoan perspective. Mol. Biol. Evol. 2010, 27, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R. An ancient cluster of Wnt paralogues. Trends Genet. 2001, 17, 443. [Google Scholar] [CrossRef]
- Bolognesi, R.; Farzana, L.; Fischer, T.D.; Brown, S.J. Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr. Biol. 2008, 18, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R. Wnt signaling and stem cell control. Cell Res. 2008, 18, 523–527. [Google Scholar] [CrossRef]
- Shimizu, H.; Julius, M.A.; Giarré, M.; Zheng, Z.; Brown, A.M.; Kitajewski, J. Transformation by Wnt family proteins correlates with regulation of beta-catenin. Cell Growth Differ. 1997, 8, 1349–1358. [Google Scholar]
- Miller, J.R. The Wnts. Genome Biol. 2001, 3, REVIEWS3001. [Google Scholar] [CrossRef]
- Tada, M.; Concha, M.L.; Heisenberg, C.-P. Non-canonical Wnt signalling and regulation of gastrulation movements. Semin. Cell Dev. Biol. 2002, 13, 251–260. [Google Scholar] [CrossRef]
- Tada, M.; Smith, J.C. Xwnt11 is a target of Xenopus Brachyury: Regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development 2000, 127, 2227–2238. [Google Scholar] [CrossRef]
- Du, J.L.; Zhang, X.J.; Yuan, J.B.; Zhang, X.X.; Li, F.H.; Xiang, J.H. Wnt gene family members and their expression profiling in Litopenaeus vannamei. Fish Shellfish Immunol. 2018, 77, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.P.; Zhang, D.Z.; Li, H.R.; Jiang, S.H.; Zhang, H.B.; Xuan, F.J.; Ge, B.M.; Wang, Z.F.; Liu, Y.; Sha, Z.L.; et al. Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). Gigascience 2020, 9, giz161. [Google Scholar] [CrossRef]
- Lv, J.J.; Li, R.H.; Su, Z.C.; Gao, B.Q.; Ti, X.B.; Yan, D.P.; Liu, G.J.; Liu, P.; Wang, C.L.; Li, J. A chromosome-level genome of Portunus trituberculatus provides insights into its evolution, salinity adaptation and sex determination. Mol. Ecol. Resour. 2021, 22, 1606–1625. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Fu, Y.Y.; Zhu, F.; Mu, C.K.; Li, R.H.; Song, W.W.; Shi, C.; Ye, Y.F.; Wang, C.L. Transcriptomic analysis of Portunus trituberculatus reveals a critical role for WNT4 and WNT signalling in limb regeneration. Gene 2018, 658, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Y.; Liu, L.; Wang, C.L.; Zhu, F.; Liu, X. Suppression of limb regeneration by RNA interference of WNT4 in the swimming crab Portunus trituberculatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 234, 41–49. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; He, Y.H.; Xia, R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 2019, 289660. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.Z.; Yang, Q.H.; Dong, X.H.; Chi, S.Y.; Liu, H.Y.; Shi, L.L.; Tan, B.P. Molecular cloning, characterization and expression analysis of Wnt4, Wnt5, Wnt6, Wnt7, Wnt10 and Wnt16 from Litopenaeus vannamei. Fish Shellfish Immunol. 2016, 54, 445–455. [Google Scholar] [CrossRef]
- Yokoyama, H.; Maruoka, T.; Ochi, H.; Aruga, A.; Ohgo, S.; Ogino, H.; Tamura, K. Different requirement for Wnt/β-catenin signaling in limb regeneration of larval and adult Xenopus. PLoS ONE 2011, 6, e21721. [Google Scholar] [CrossRef]
- Ramachandran, R.; Zhao, X.F.; Goldman, D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc. Natl. Acad. Sci. USA 2011, 108, 15858–15863. [Google Scholar] [CrossRef]
- Meyers, J.R.; Hu, L.; Moses, A.; Kaboli, K.; Papandrea, A.; Raymond, P.A. β-catenin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Dev. 2012, 7, 30. [Google Scholar] [CrossRef]
- Smith-Bolton, R.K.; Worley, M.I.; Kanda, H.; Hariharan, I.K. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell 2009, 16, 797–809. [Google Scholar] [CrossRef]
- Shah, M.V.; Namigai, E.K.O.; Suzuki, Y. The role of canonical Wnt signaling in leg regeneration and metamorphosis in the red flour beetle Tribolium castaneum. Mech. Dev. 2011, 128, 342–358. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.T.; Nie, H.T.; Wang, Z.X.; Yan, X.W. Genome-wide identification and transcriptome-based expression profiling of Wnt gene family in Ruditapes philippinarum. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100709. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.D.S.; Choudhury, M.; Majlish, A.-N.K.; Islam, T.; Ghosh, A. Comprehensive genome-wide analysis of Glutathione S-transferase gene family in potato (Solanum tuberosum L.) and their expression profiling in various anatomical tissues and perturbation conditions. Gene 2018, 639, 149–162. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, C.X.; Zhang, Y.L.; Wang, Q.; Li, W.X.; Tian, Y.; Cong, P.H. Bioinformatics and expression analysis of the LysM gene family in apple. Sci. Agric. Sin. 2014, 47, 2602–2612. [Google Scholar] [CrossRef]
- Yuan, G.P.; He, S.S.; Bian, S.X.; Han, X.L.; Liu, K.; Cong, P.H.; Zhang, C.X. Genome-wide identification and expression analysis of major latex protein (MLP) family genes in the apple (Malus domestica Borkh.) genome. Gene 2020, 733, 144275. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Y.; Chai, F.M.; Li, S.H.; Xin, H.P.; Liang, Z.C. Genome-wide identification and characterization of the 14-3-3 family in Vitis vinifera L. during berry development and cold- and heat-stress response. BMC Genom. 2018, 19, 579. [Google Scholar] [CrossRef] [PubMed]
- Takata, H.; Terada, K.; Oka, H.; Sunada, Y.; Moriguchi, T.; Nohno, T. Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Dev. Dyn. 2007, 236, 2800–2807. [Google Scholar] [CrossRef] [PubMed]
- von Maltzahn, J.; Chang, N.C.; Bentzinger, C.F.; Rudnicki, M.A. Wnt signaling in myogenesis. Trends Cell Biol. 2012, 22, 602–609. [Google Scholar] [CrossRef]
- Ozhan, G.; Weidinger, G. Wnt/β-catenin signaling in heart regeneration. Cell Regen. 2015, 4, 3. [Google Scholar] [CrossRef]
- Tanaka, E.M.; Reddien, P.W. The cellular basis for animal regeneration. Dev. Cell 2011, 21, 172–185. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Genomic Position | Chr | Chromosome Length | ORF | Gene Stand | Protein Length (aa) | Theoretical pI | Molecular Weight (kDa) |
|---|---|---|---|---|---|---|---|---|
| PtWnt1 | 5,505,983–5,542,897 | 3 | 10,158,951 | 1710 | reverse | 569 | 5.39 | 58.91 |
| PtWnt2 | 5,544,234–5,546,536 | 3 | 10,158,951 | 369 | reverse | 122 | 10.04 | 13.65 |
| PtWnt3 | 28,210,259–28,217,084 | 40 | 29,382,946 | 1056 | reverse | 351 | 9.09 | 39.62 |
| PtWnt4 | 6,906,935–6,911,930 | 28 | 12,784,577 | 219 | forward | 72 | 10.54 | 8.03 |
| PtWnt5 | 4,822,813–4,826,764 | 30 | 12,291,475 | 1353 | reverse | 450 | 9.34 | 50.85 |
| PtWnt6 | 4,925,695–4,935,753 | 30 | 12,291,475 | 477 | forward | 158 | 9.67 | 17.95 |
| PtWnt7 | 4,941,929–4,949,590 | 30 | 12,291,475 | 1272 | forward | 423 | 9.87 | 47.80 |
| PtWnt8 | 18,612,647–18,621,734 | 24 | 20,371,438 | 1035 | forward | 344 | 9.37 | 38.59 |
| PtWnt9 | 18,843,485–18,849,022 | 24 | 20,371,438 | 780 | reverse | 259 | 8.97 | 28.59 |
| PtWnt10 | 987 | 328 | 9.86 | 36.29 | ||||
| PtWnt11 | 639 | 212 | 9.68 | 24.57 | ||||
| PtWnt12 | 1176 | 391 | 9.85 | 43.25 | ||||
| PtWnt13 | 2001 | 666 | 9.49 | 73.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; He, J.; Liu, L.; Huang, X.; Xu, Y.; Wang, C. Comprehensive Genome-Wide Analysis of Wnt Gene Family and Expression Profiling during Limb Regeneration in Portunus trituberculatus. Fishes 2022, 7, 258. https://doi.org/10.3390/fishes7050258
Fu Y, He J, Liu L, Huang X, Xu Y, Wang C. Comprehensive Genome-Wide Analysis of Wnt Gene Family and Expression Profiling during Limb Regeneration in Portunus trituberculatus. Fishes. 2022; 7(5):258. https://doi.org/10.3390/fishes7050258
Chicago/Turabian StyleFu, Yuanyuan, Jie He, Lei Liu, Xinlian Huang, Yuankai Xu, and Chunlin Wang. 2022. "Comprehensive Genome-Wide Analysis of Wnt Gene Family and Expression Profiling during Limb Regeneration in Portunus trituberculatus" Fishes 7, no. 5: 258. https://doi.org/10.3390/fishes7050258
APA StyleFu, Y., He, J., Liu, L., Huang, X., Xu, Y., & Wang, C. (2022). Comprehensive Genome-Wide Analysis of Wnt Gene Family and Expression Profiling during Limb Regeneration in Portunus trituberculatus. Fishes, 7(5), 258. https://doi.org/10.3390/fishes7050258







