Author Contributions
Conceptualization, G.C. and S.S.; methodology, M.A. and C.D.; software, S.F. and S.S.; validation, S.S. and G.C.; formal analysis, C.D., S.F. and S.S.; investigation, M.A. and C.D.; resources, N.S., P.R. and D.G.; data curation, S.S. and S.F.; writing—original draft preparation, C.D., and S.F.; writing—review and editing, S.S. and G.C.; visualization, N.S., P.R. and D.G.; supervision, S.S. and G.C.; project administration, P.R. and D.G.; funding acquisition, N.S. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Maps of the studied area (GSA 10, southern and central Tyrrhenian Sea), Mediterranean Sea in the insert.
Figure 1.
Maps of the studied area (GSA 10, southern and central Tyrrhenian Sea), Mediterranean Sea in the insert.
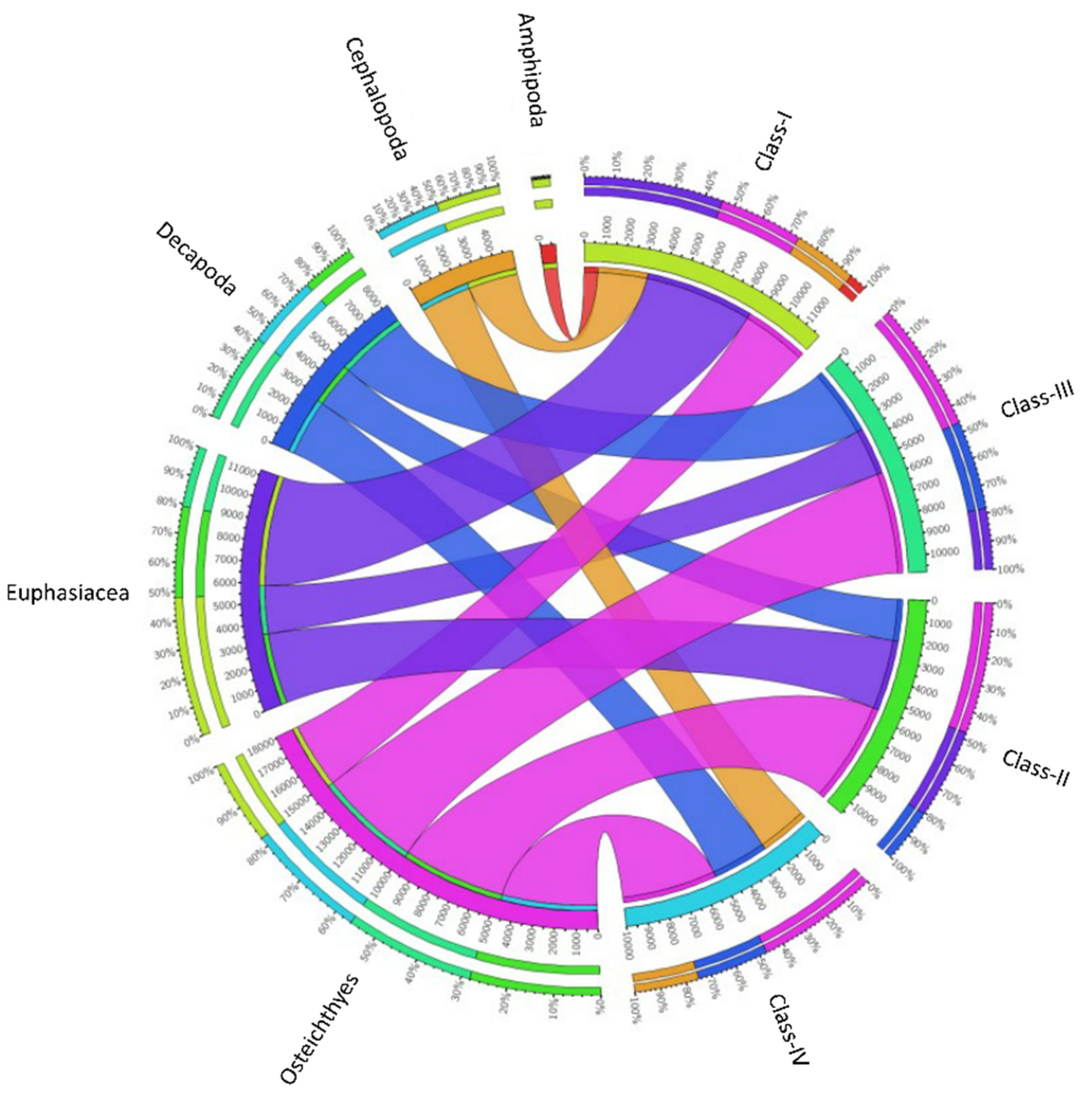
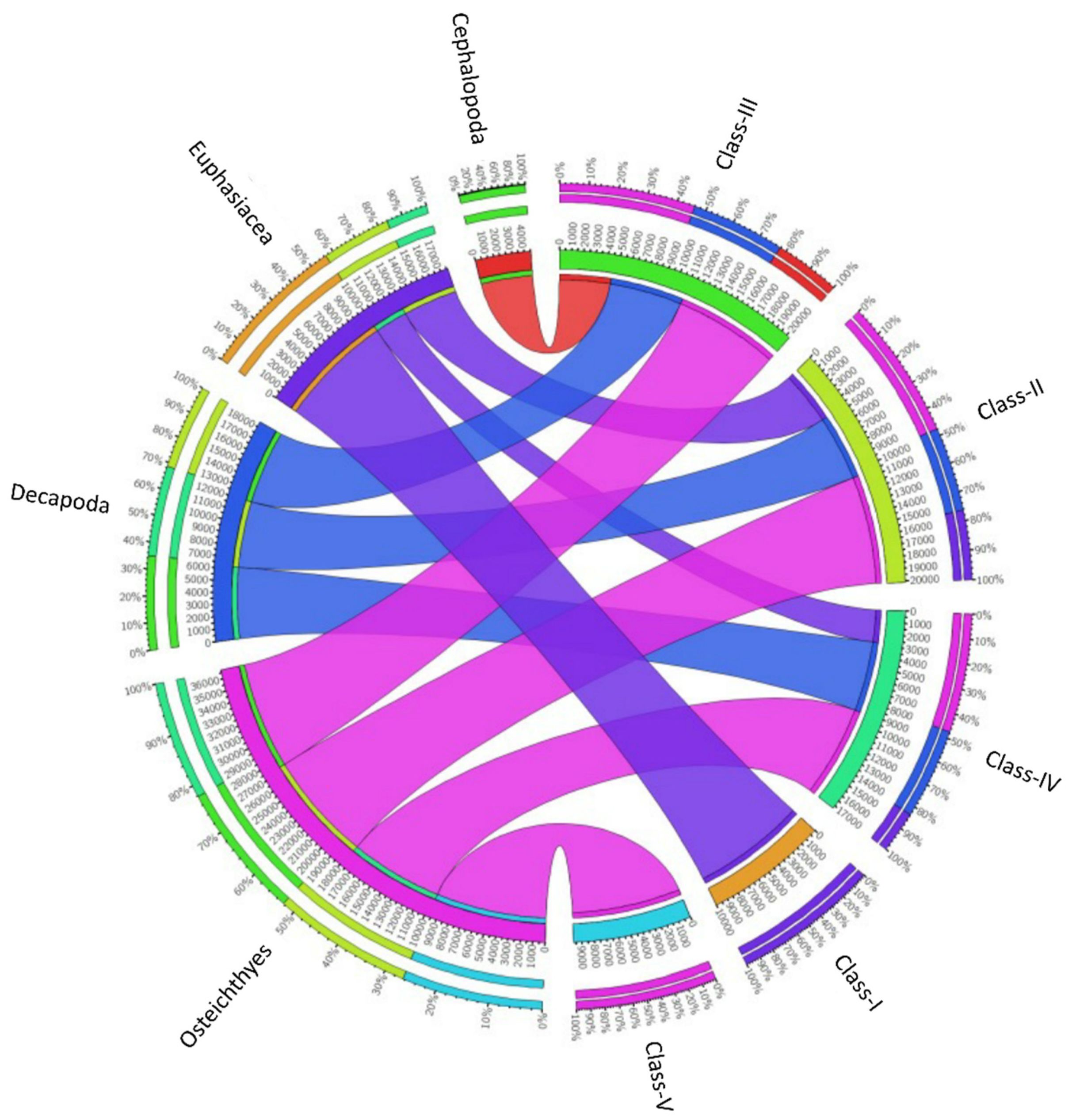
Figure 2.
Diet composition of the Merluccius merluccius specimens collected from the southern and central Tyrrhenian Sea during 2018. The chord diagram shows the connection among the main taxa found in the stomach contents of different hake size classes investigated. The size classes analyzed are shown on the right of the diagram. Main taxa found are shown on the left. Ribbon size in the chart codifies IRI value associated with hake size classes/prey taxa segment pair.
Figure 2.
Diet composition of the Merluccius merluccius specimens collected from the southern and central Tyrrhenian Sea during 2018. The chord diagram shows the connection among the main taxa found in the stomach contents of different hake size classes investigated. The size classes analyzed are shown on the right of the diagram. Main taxa found are shown on the left. Ribbon size in the chart codifies IRI value associated with hake size classes/prey taxa segment pair.
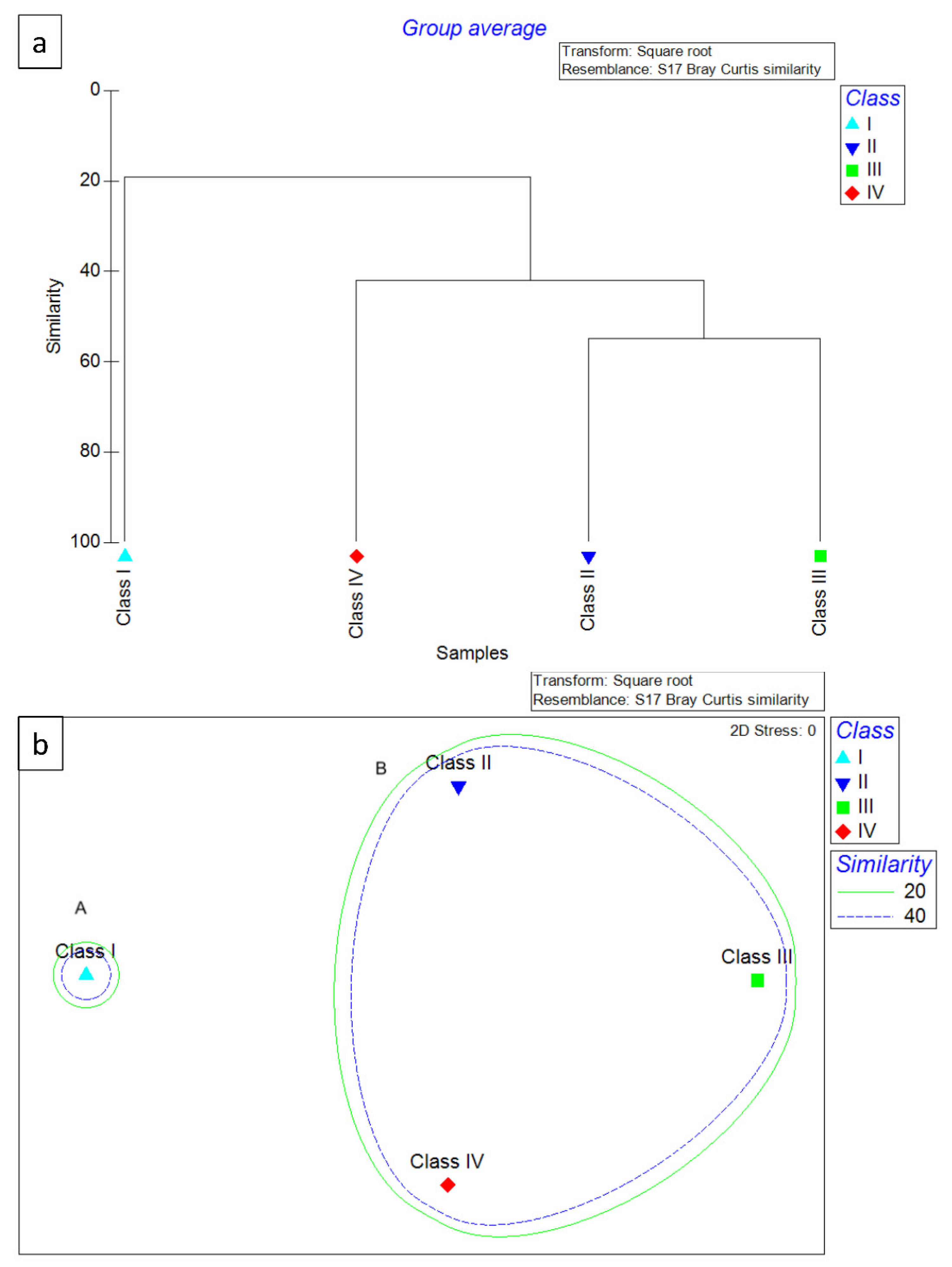
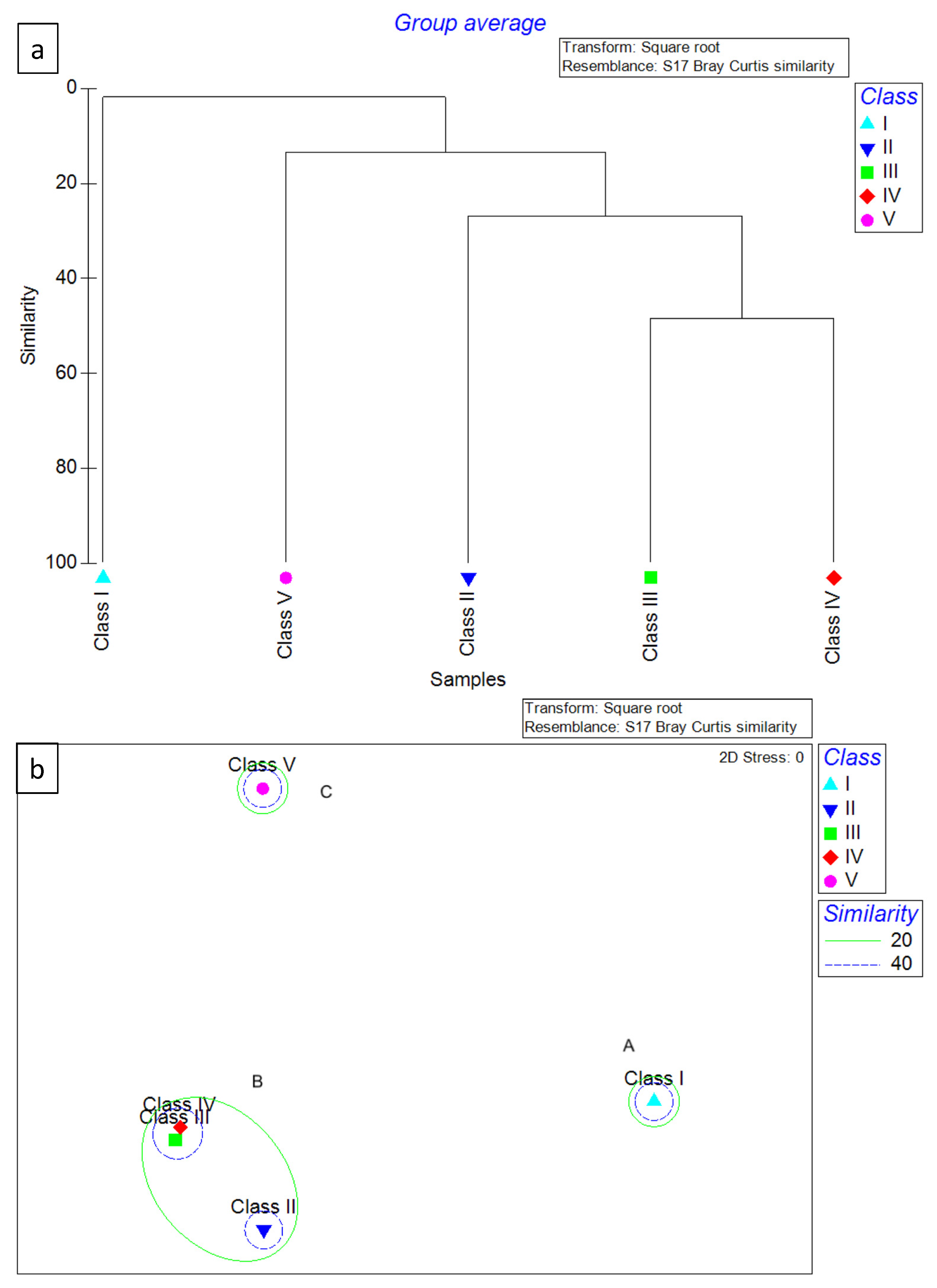
Figure 3.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the 4 hake ontogenetic stages analyzed in 2018. Cluster A included only the specimens belonging to Class I, while Cluster B included the specimens belonging to Classes II–IV.
Figure 3.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the 4 hake ontogenetic stages analyzed in 2018. Cluster A included only the specimens belonging to Class I, while Cluster B included the specimens belonging to Classes II–IV.
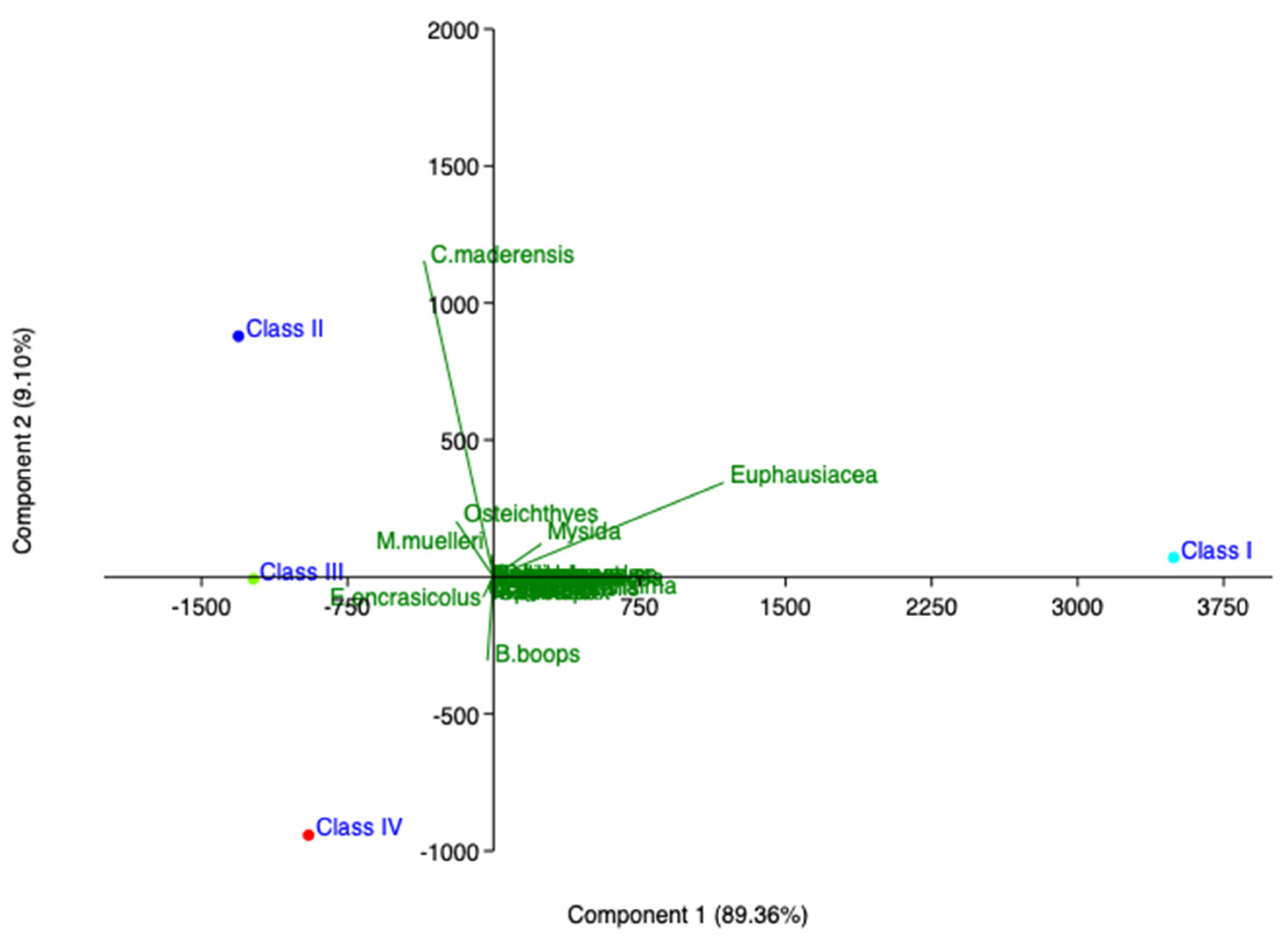
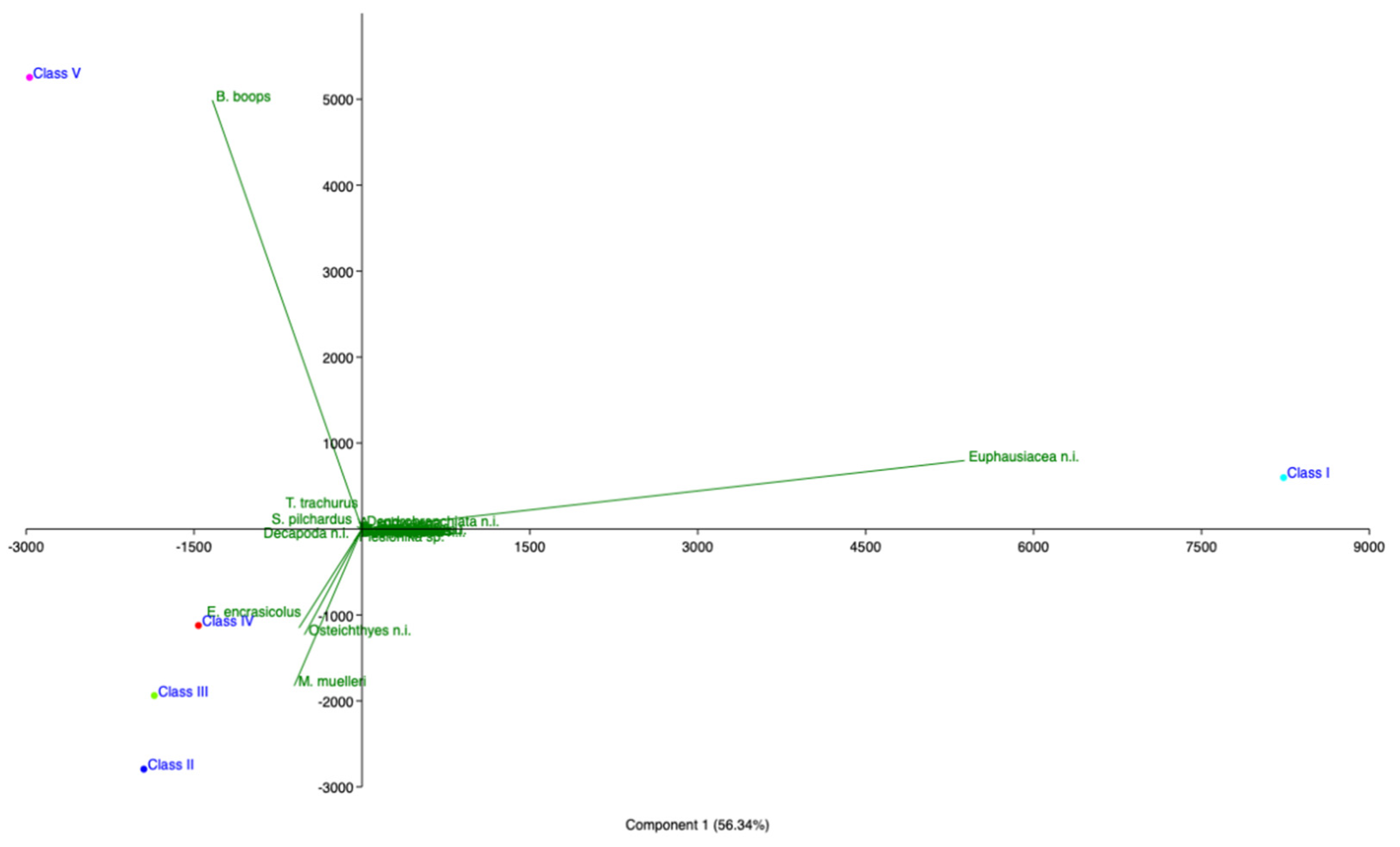
Figure 4.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2018).
Figure 4.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2018).
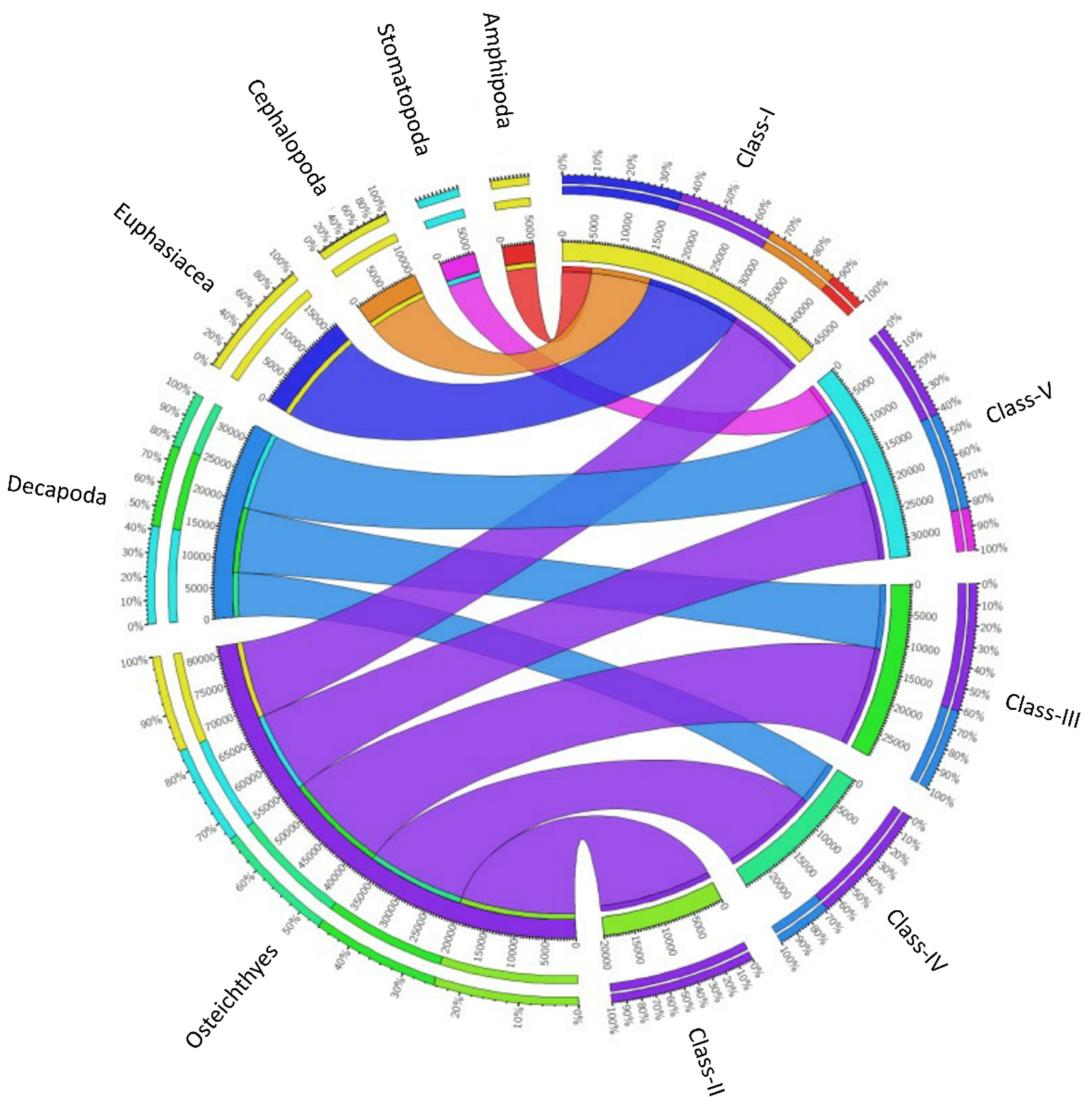
Figure 5.
Diet composition of the Merluccius merluccius specimens collected from southern and central Tyrrhenian Sea during 2019. The chord diagram shows the connection among the main taxa found in the stomach contents of different hake size classes investigated. The size classes analyzed are shown on the right of the diagram. Main taxa found are shown on the left. Ribbon size in the chart codifies IRI value associated with hake size classes/prey taxa segment pair.
Figure 5.
Diet composition of the Merluccius merluccius specimens collected from southern and central Tyrrhenian Sea during 2019. The chord diagram shows the connection among the main taxa found in the stomach contents of different hake size classes investigated. The size classes analyzed are shown on the right of the diagram. Main taxa found are shown on the left. Ribbon size in the chart codifies IRI value associated with hake size classes/prey taxa segment pair.
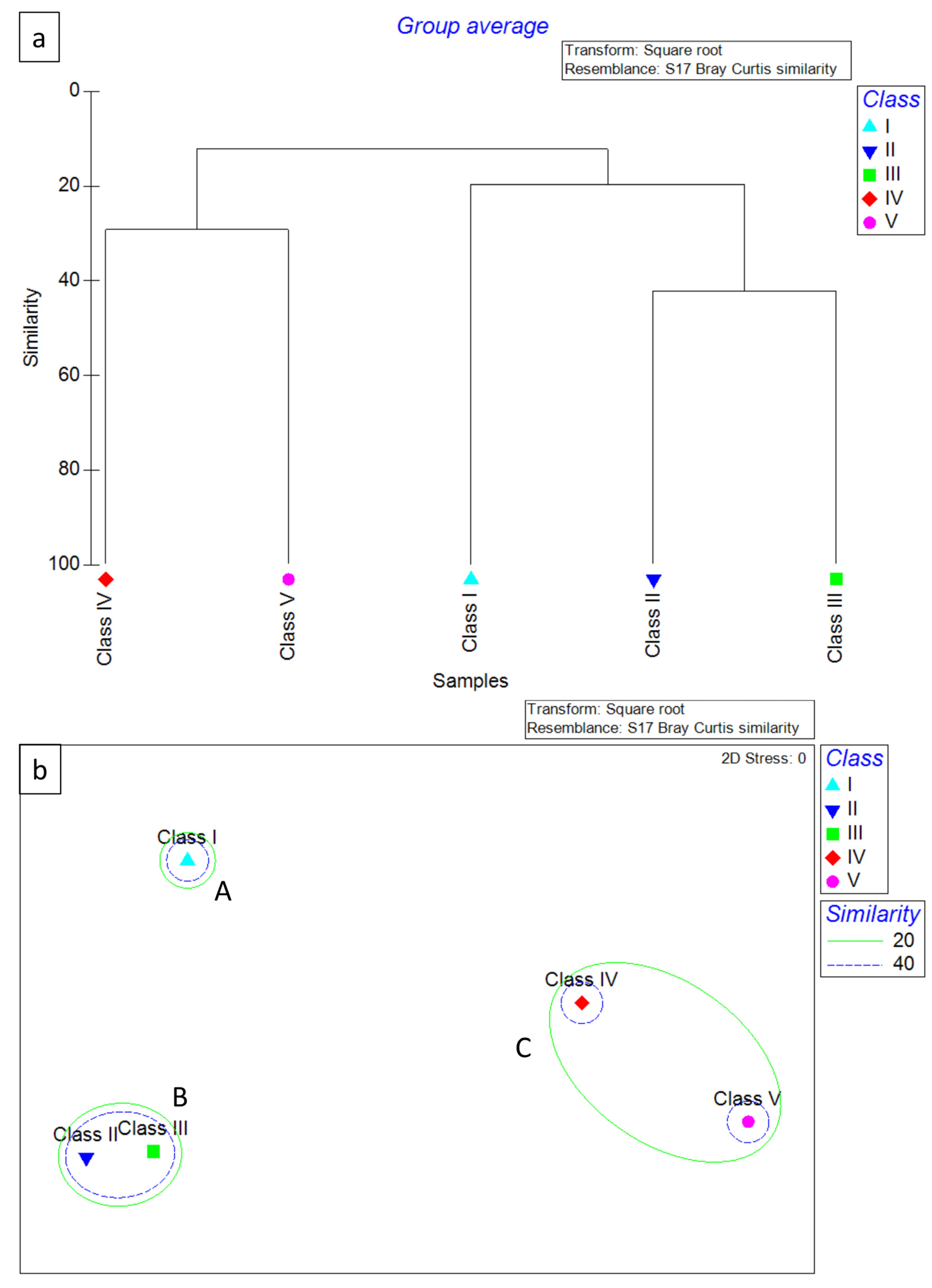
Figure 6.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the 5 hake classes analyzed in 2019. Cluster A included only the specimens belonging to Class I, cluster B included the specimens belonging to Class V, while cluster C grouped Classes II–IV.
Figure 6.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the 5 hake classes analyzed in 2019. Cluster A included only the specimens belonging to Class I, cluster B included the specimens belonging to Class V, while cluster C grouped Classes II–IV.
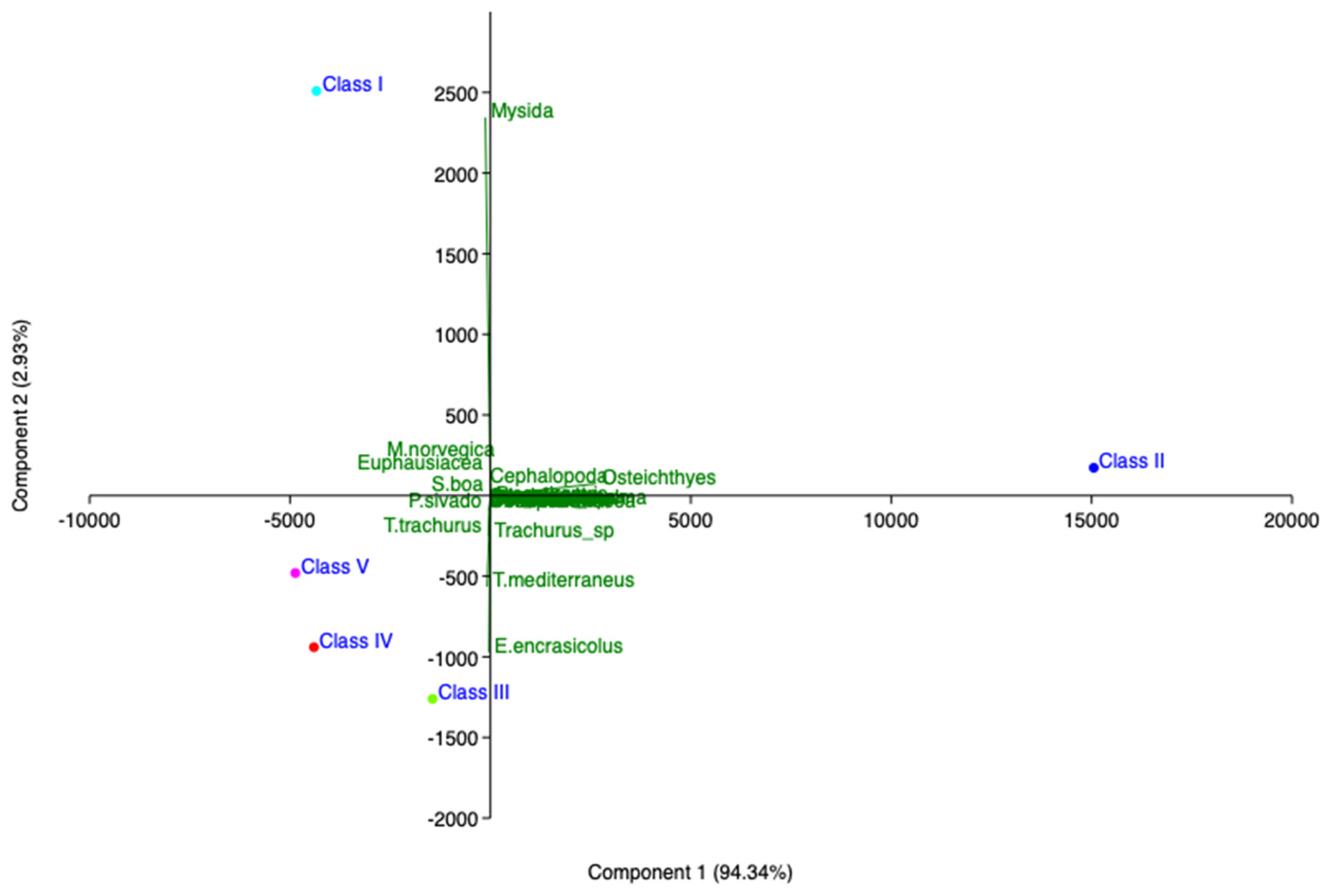
Figure 7.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2019).
Figure 7.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2019).
Figure 8.
Diet composition of the Merluccius merluccius specimens collected from southern and central Tyrrhenian Sea during 2020. The chord diagram shows the connection among the main taxa found in the stomach contents of different hake size classes investigated. The size classes analyzed are shown on the right of the diagram. Main taxa found are shown on the left. Ribbon size in the chart codifies IRI value associated with hake size classes/prey taxa segment pair.
Figure 8.
Diet composition of the Merluccius merluccius specimens collected from southern and central Tyrrhenian Sea during 2020. The chord diagram shows the connection among the main taxa found in the stomach contents of different hake size classes investigated. The size classes analyzed are shown on the right of the diagram. Main taxa found are shown on the left. Ribbon size in the chart codifies IRI value associated with hake size classes/prey taxa segment pair.
Figure 9.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the 5 hake classes analyzed in 2020. Cluster A included only the specimens belonging to Class I, cluster B included the specimens belonging to Classes II and III while the third one (cluster C) included Classes IV and V.
Figure 9.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the 5 hake classes analyzed in 2020. Cluster A included only the specimens belonging to Class I, cluster B included the specimens belonging to Classes II and III while the third one (cluster C) included Classes IV and V.
Figure 10.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2020).
Figure 10.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2020).
Figure 11.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the all hake classes analyzed in 2018–2020. Cluster A included the specimens belonging to Class I collected during 2018 and 2019. Cluster B grouped the specimens belonging to Class I collected during 2020, and Classes II–IV of all three years, except for Class IV samples collected during 2020. Cluster C included Classes IV and V from 2020. Finally, Cluster D only included specimens belonging to Class V from 2019.
Figure 11.
Dendrogram (a) and MDS ordination of Bray–Curtis similarities (b) from dietary data (square root transformation) for the all hake classes analyzed in 2018–2020. Cluster A included the specimens belonging to Class I collected during 2018 and 2019. Cluster B grouped the specimens belonging to Class I collected during 2020, and Classes II–IV of all three years, except for Class IV samples collected during 2020. Cluster C included Classes IV and V from 2020. Finally, Cluster D only included specimens belonging to Class V from 2019.
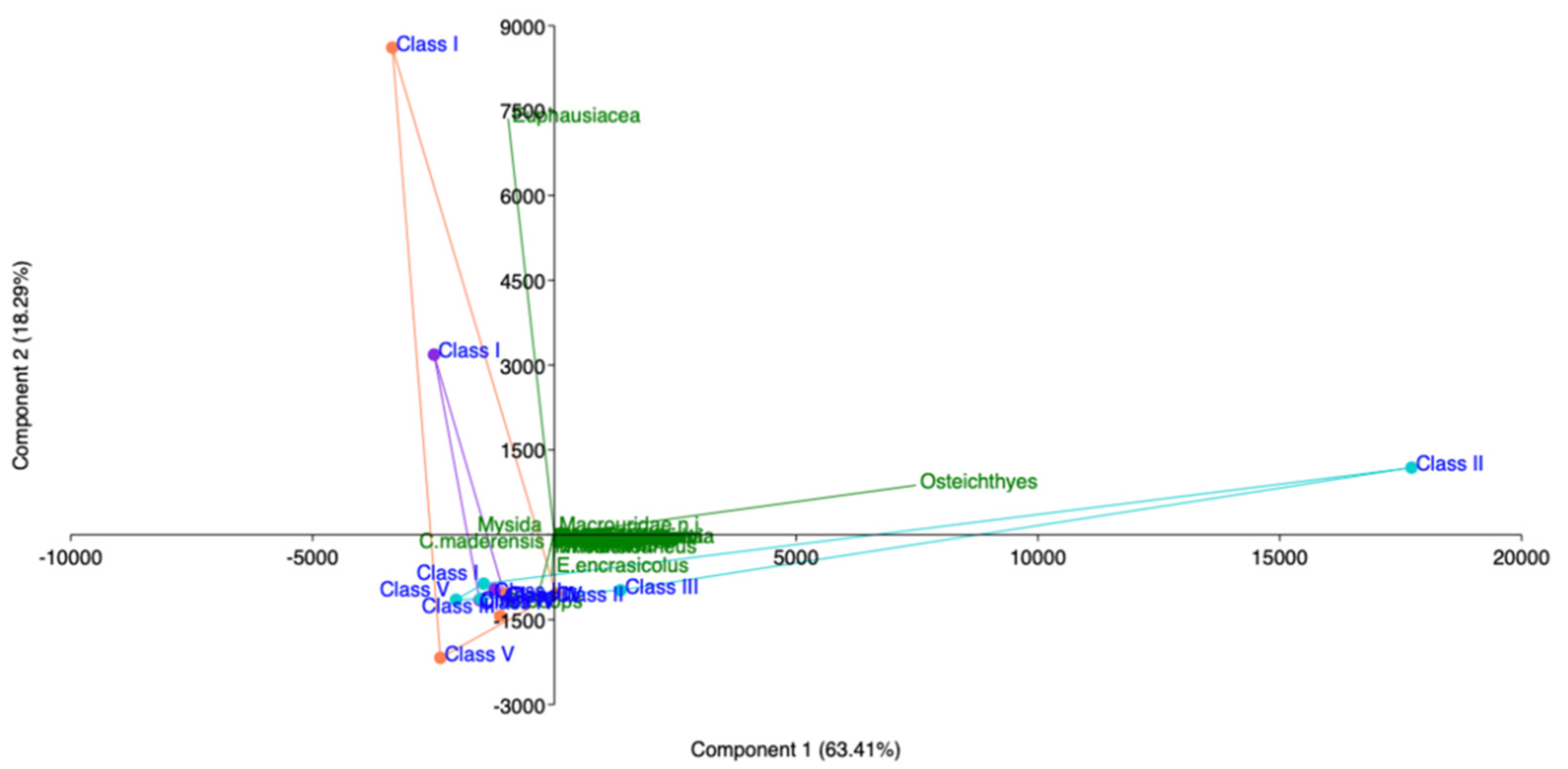
Figure 12.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2018–2020). Violet: 2018; Orange: 2019; Turquoise: 2020.
Figure 12.
Principal component analysis (PCA) of hake diet in southern and central Tyrrhenian Sea (2018–2020). Violet: 2018; Orange: 2019; Turquoise: 2020.
Table 1.
Numbers of sampled and analyzed stomachs (AS), with Vacuity Index (VI%) and numbers of Empty (E), Full < 50% (F < 50), Full > 50% (F > 50) and bursting stomachs (B) by ontogenetic classes in the different years.
Table 1.
Numbers of sampled and analyzed stomachs (AS), with Vacuity Index (VI%) and numbers of Empty (E), Full < 50% (F < 50), Full > 50% (F > 50) and bursting stomachs (B) by ontogenetic classes in the different years.
Size
Classes | 2018 | 2019 | 2020 |
|---|
| | E | F < 50 | F > 50 | B | AS | VI % | E | F < 50 | F > 50 | B | AS | VI % | E | F < 50 | F > 50 | B | AS | VI % |
| I | 2 | 5 | 4 | 11 | 22 | 9.09 | 5 | 2 | 0 | 0 | 7 | 71.42 | 0 | 5 | 2 | 7 | 14 | 0 |
| II | 18 | 4 | 16 | 31 | 69 | 26.08 | 6 | 1 | 2 | 5 | 14 | 42.85 | 0 | 0 | 0 | 3 | 3 | 0 |
| III | 6 | 11 | 8 | 22 | 47 | 12.76 | 11 | 3 | 6 | 8 | 28 | 39.28 | 0 | 3 | 2 | 8 | 13 | 0 |
| IV | 8 | 9 | 12 | 14 | 43 | 18.60 | 40 | 25 | 8 | 21 | 94 | 42.55 | 8 | 5 | 0 | 6 | 19 | 42.10 |
| V | 2 | 2 | 0 | 0 | 4 | 50 | 0 | 1 | 5 | 8 | 14 | 0 | 6 | 4 | 4 | 6 | 20 | 30 |
| ∑ | 36 | 29 | 40 | 78 | 185 | 19.45 | 62 | 32 | 21 | 42 | 157 | 39.49 | 14 | 17 | 8 | 30 | 69 | 20.28 |
Table 2.
Diet composition of the whole hake’s series from southern and central Tyrrhenian Sea sampled in 2018. In the columns are reported the diet index values (%F, %W, %N, IRI and %IRI) for each prey category.
Table 2.
Diet composition of the whole hake’s series from southern and central Tyrrhenian Sea sampled in 2018. In the columns are reported the diet index values (%F, %W, %N, IRI and %IRI) for each prey category.
| TAXON | %F | %W | %N | IRI | %IRI |
|---|
| MOLLUSCA | | | | | |
| Cephalopoda n.i. | 3.82 | 1.07 | 1.97 | 11.62 | 0.64 |
| CRUSTACEA | | | | | |
| Amphipoda | | | | | |
| Amphipoda n.i. | 0.64 | 0.01 | 0.33 | 0.21 | 0.01 |
| Decapoda | | | | | |
| Parapenaeus longirostris | 1.27 | 0.99 | 0.66 | 2.10 | 0.12 |
| Solenocera membranacea | 0.64 | 0.46 | 0.33 | 0.50 | 0.03 |
| Alpheus glaber | 1.27 | 0.11 | 0.66 | 0.98 | 0.05 |
| Chlorotocus crassicornis | 1.27 | 0.28 | 0.66 | 1.20 | 0.07 |
| Processa acutirostris | 0.64 | 0.43 | 0.99 | 0.90 | 0.05 |
| Decapoda n.i. | 1.91 | 0.56 | 0.99 | 2.95 | 0.16 |
| Dendrobranchiata n.i. | 5.10 | 0.98 | 2.63 | 18.43 | 1.02 |
| Euphausiacea | | | | | |
| Meganyctiphanes norvegica | 0.64 | 0.13 | 4.93 | 3.22 | 0.18 |
| Stylocheiron longicorne | 0.64 | 0.01 | 0.33 | 0.21 | 0.01 |
| Euphausiacea n.i. | 10.83 | 1.81 | 31.91 | 365.10 | 20.16 |
| Mysida n.i. | 7.01 | 0.29 | 18.75 | 133.42 | 7.37 |
| OSTEICHTHYES | | | | | |
| Ceratoscopelus maderensis | 20.38 | 13.04 | 11.84 | 507.17 | 28.00 |
| Diaphus holti | 1.27 | 0.00 | 0.66 | 0.84 | 0.05 |
| Boops boops | 1.27 | 30.34 | 0.66 | 39.49 | 2.18 |
| Callionymus sp. | 1.27 | 0.50 | 0.66 | 1.47 | 0.08 |
| Macroramphosus scolopax | 1.27 | 1.94 | 0.66 | 3.30 | 0.18 |
| Maurolicus muelleri | 2.55 | 1.51 | 1.32 | 7.21 | 0.40 |
| Engraulis encrasicolus | 3.82 | 16.70 | 1.97 | 71.36 | 3.94 |
| Peristedion cataphractum | 1.27 | 0.63 | 0.66 | 1.64 | 0.09 |
| Cepola macrophthalma | 1.27 | 0.00 | 0.66 | 0.84 | 0.05 |
| Chlorophthalmus agassizi | 5.10 | 5.49 | 1.97 | 38.04 | 2.10 |
| Notoscopelus elongatus | 1.27 | 0.16 | 0.66 | 1.05 | 0.06 |
| Gobiidae n.i. | 1.27 | 0.50 | 0.66 | 1.48 | 0.08 |
| Sparidae n.i. | 1.27 | 5.92 | 0.66 | 8.38 | 0.46 |
| Osteichthyes n.i. | 21.02 | 16.13 | 11.84 | 588.00 | 32.47 |
Table 3.
Diet composition in the five hake size classes sampled in timeframe 2018/2020. In the column are reported the IRI% values for each prey category.
Table 3.
Diet composition in the five hake size classes sampled in timeframe 2018/2020. In the column are reported the IRI% values for each prey category.
| | 2018 | 2019 | 2020 |
|---|
| TAXON/CLASS | I | II | III | IV | V | I | II | III | IV | V | I | II | III | IV | V |
|---|
| MOLLUSCA | | | | | | | | | | | | | | | |
| Cephalopoda n.i. | 0.88 | | | 2.77 | | | | 1.11 | | | 3.92 | | | | |
| CRUSTACEA | | | | | | | | | | | | | | | |
| Amphipoda | | | | | | | | | | | | | | | |
| Amphipoda n.i. | 0.06 | | | | | | | | | | 0.28 | | | | |
| Decapoda | | | | | | | | | | | | | | | |
| Eusergestes arcticus | | | | | | | | | 0.54 | | | | | | 0.44 |
| Parapenaeus longirostris | | | 1.13 | | | | | | 0.63 | | | | | | |
| Aristaeomorpha foliacea | | | | | | | | | 0.15 | | | | | | |
| Solenocera membranacea | | | 0.27 | | | | | | | | | | 0.98 | | |
| Alpheus glaber | | 0.37 | | | | | | | 0.55 | | | | | | |
| Pasiphaea multidentata | | | | | | | | | | | | | | | 2.12 |
| Pasiphaea sivado | | | | | | | | | 0.17 | | | | | | 39.50 |
| Chlorotocus crassicornis | | | | 2.26 | | | | | 0.27 | | | | | | |
| Processa acutirostris | | | 0.58 | | | | | | | | | | 2.36 | | |
| Sergestes sp. | | | | | | | | | | | | | | | 0.45 |
| Pasiphaea sp. | | | | | | | | | | | | | | | 1.82 |
| Dardanus sp. | | | | | | | | | | | | | | 1.07 | |
| Plesionika sp. | | | | | | | 2.94 | | 0.15 | | | | | | |
| Decapoda n.i. | | 0.39 | 5.02 | 2.28 | | | | 6.41 | 11.88 | | | | 0.85 | | 0.88 |
| Dendrobranchiata n.i. | | | | | | | | | | | | | | | |
| Stomatopoda | | | | | | | | | | | | | | | |
| Parasquilla ferussaci | | | | | | | | | | | | | | | 1.05 |
| Euphausiacea | | | | | | | | | | | | | | | |
| Meganyctiphanes norvegica | 0.93 | | | | | | | | 0.66 | | 4.31 | | | | |
| Stylocheiron longicorne | 0.06 | | | | | | | | | | 0.28 | | | | |
| Euphausiacea n.i. | 78.24 | 4.48 | 1.30 | | | 100.00 | 1.13 | | | | 4.11 | | | | |
| Mysida n.i. | 16.72 | 2.96 | | | | | | | | | 71.11 | | | | |
| OSTEICHTHYES | | | | | | | | | | | | | | | |
| Ceratoscopelus maderensis | | 60.08 | 36.62 | 18.94 | | | | | 2.94 | 0.74 | | | | | 3.21 |
| Diaphus holti | | 0.29 | | | | | | | | | | | | | 0.56 |
| Lampanyctus crocodilus | | | | | | | | | | | | | | | 0.55 |
| Stomias boa | | | | | | | | | | | | | | | 0.83 |
| Argentina sphyraena | | | | | | | | | | 1.62 | | | | | |
| Boops boops | | | | 22.26 | | | | | | 92.51 | | | | | 0.70 |
| Callionymus sp. | | 0.67 | | | | | | | | | | | | | |
| Macroramphosus scolopax | | | | 3.36 | | | | | | | | | | | 0.47 |
| Echiodon dentatus | | | | | | | | | 0.62 | | | | | | |
| Gaidropsarus biscayensis | | | | | | | | | 0.27 | | | | | | |
| Maurolicus muelleri | | 3.50 | | | | | 65.76 | 0.94 | | | | | | | 0.49 |
| Engraulis encrasicolus | | | 14.68 | 5.17 | | | | 73.71 | 13.67 | 0.85 | | | 41.17 | | |
| Peristedion cataphractum | | 0.77 | | | | | | | | | | | | | 0.42 |
| Cepola macrophthalma | | | | 2.07 | | | | | | | | | | | 0.91 |
| Physiculus dalwigki | | | | | | | | | 0.12 | | | | | | |
| Sardina pilchardus | | | | | | | | | 13.25 | 1.31 | | | | | |
| Chlorophthalmus agassizi | | 0.00 | 7.06 | 2.74 | | | | | 0.26 | | | | | | 0.83 |
| Nettastoma melanura | | | | | | | | | | | | | | 1.07 | |
| Trachurus trachurus | | | | | | | | | | 2.21 | | | | 13.55 | 38.03 |
| Trachurus mediterraneus | | | | | | | | | | | | | | 48.33 | 3.30 |
| Trachurus sp. | | | | | | | | | | | | | | 21.90 | |
| Notoscopelus elongatus | | | | 2.18 | | | | | | | | | | | |
| Myctophidae n.i. | | | | | | | | | 0.17 | | | | | | |
| Sparidae n.i. | | | | 6.01 | | | | | 0.84 | | | | | | |
| Carapidae n.i. | | | | | | | | 0.67 | 0.12 | | | | | | |
| Gobiidae n.i. | | 0.68 | | | | | | | 0.12 | | | | | | |
| Macrouridae n.i. | | | | | | | | | 0.13 | | | | | | |
| Osteichthyes n.i. | 3.10 | 25.80 | 33.33 | 29.96 | | | 30.17 | 17.16 | 52.49 | 0.77 | 15.99 | 100 | 56.64 | 14.08 | 3.45 |
Table 4.
Results of the SIMPER analysis conducted between European hake trophic groups from 2018. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table shows results obtained for the trophic cluster B, which includes Classes II–IV. Trophic cluster A = less than 2 size groups.
Table 4.
Results of the SIMPER analysis conducted between European hake trophic groups from 2018. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table shows results obtained for the trophic cluster B, which includes Classes II–IV. Trophic cluster A = less than 2 size groups.
| Group B | Species | Av.IRI val | Av.Sim | Contrib% | Cum.% |
|---|
| Average similarity: | Osteichthyes n.i. | 28.98 | 19.2 | 41.42 | 41.42 |
| 46.36 | C.maderensis | 33.16 | 17.87 | 38.55 | 79.97 |
| | Decapoda n.i. | 7.7 | 3.47 | 7.48 | 87.45 |
| | E.encrasicolus | 10.6 | 2.45 | 5.28 | 92.74 |
Table 5.
Diet composition of the whole hake series from southern and central Tyrrhenian Sea sampled in 2018. In the columns are reported the diet index values (%F, %W, %N, IRI and %IRI) for each prey category.
Table 5.
Diet composition of the whole hake series from southern and central Tyrrhenian Sea sampled in 2018. In the columns are reported the diet index values (%F, %W, %N, IRI and %IRI) for each prey category.
| TAXON | %F | %W | %N | IRI | %IRI |
|---|
| MOLLUSCA | | | | | |
| Cephalopoda n.i. | 1.00 | 0.35 | 0.77 | 1.12 | 0.07 |
| CRUSTACEA | | | | | |
| Decapoda | | | | | |
| Parapenaeus longirostris | 1.00 | 0.57 | 3.08 | 3.65 | 0.22 |
| Aristaeomorpha foliacea | 1.00 | 0.13 | 0.77 | 0.90 | 0.05 |
| Alpheus glaber | 2.00 | 0.13 | 1.54 | 3.33 | 0.20 |
| Chlorotocus crassicornis | 1.00 | 0.10 | 1.54 | 1.64 | 0.10 |
| Eusergestes arcticus | 2.00 | 0.11 | 1.54 | 3.29 | 0.20 |
| Pasiphaea sivado | 1.00 | 0.22 | 0.77 | 0.99 | 0.06 |
| Plesionika sp. | 2.00 | 0.39 | 1.54 | 3.86 | 0.23 |
| Decapoda n.i. | 6.00 | 0.70 | 4.62 | 31.87 | 1.91 |
| Dendrobranchiata n.i. | 6.00 | 0.33 | 4.62 | 29.67 | 1.78 |
| Euphausiacea | | | | | |
| Meganyctiphanes norvegica | 1.00 | 0.21 | 3.85 | 4.05 | 0.24 |
| Euphausiacea n.i. | 5.00 | 0.02 | 13.08 | 65.48 | 3.93 |
| OSTEICHTHYES | | | | | |
| Ceratoscopelus maderensis | 5.00 | 0.54 | 4.62 | 25.76 | 1.55 |
| Argentina sphyraena | 1.00 | 0.72 | 1.54 | 2.26 | 0.14 |
| Boops boops | 7.00 | 42.05 | 5.38 | 332.06 | 19.92 |
| Echiodon dentatus | 2.00 | 0.27 | 1.54 | 3.61 | 0.22 |
| Gaidropsarus biscayensis | 1.00 | 0.12 | 1.54 | 1.66 | 0.10 |
| Maurolicus muelleri | 5.00 | 1.60 | 6.15 | 38.79 | 2.33 |
| Engraulis encrasicolus | 12.00 | 14.83 | 10.77 | 307.17 | 18.43 |
| Physiculus dalwigki | 1.00 | 0.00 | 0.77 | 0.77 | 0.05 |
| Sardina pilchardus | 6.00 | 15.51 | 4.62 | 120.78 | 7.25 |
| Chlorophthalmus agassizi | 1.00 | 0.58 | 0.77 | 1.35 | 0.08 |
| Trachurus trachurus | 1.00 | 7.67 | 0.77 | 8.44 | 0.51 |
| Myctophidae n.i. | 1.00 | 0.21 | 0.77 | 0.98 | 0.06 |
| Sparidae n.i. | 1.00 | 2.94 | 0.77 | 3.71 | 0.22 |
| Carapidae n.i. | 2.00 | 0.00 | 1.54 | 3.08 | 0.18 |
| Gobiidae n.i. | 1.00 | 0.00 | 0.77 | 0.77 | 0.05 |
| Macrouridae n.i. | 1.00 | 0.03 | 0.77 | 0.80 | 0.05 |
| Osteichthyes n.i. | 23.00 | 9.68 | 19.23 | 664.90 | 39.89 |
Table 6.
Results of the SIMPER analysis conducted between European hake trophic groups from 2019. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table shows results obtained for the trophic cluster B, which includes Classes II–IV. Trophic cluster A and C = less than 2 size groups.
Table 6.
Results of the SIMPER analysis conducted between European hake trophic groups from 2019. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table shows results obtained for the trophic cluster B, which includes Classes II–IV. Trophic cluster A and C = less than 2 size groups.
| Group | Species | Av.IRI val | Av.Sim | Contrib% | Cum.% |
|---|
| Average similarity: | Osteichthyes n.i. | 37.15 | 23.51 | 68.76 | 68.76 |
| 34.19 | E. encrasicolus | 27.54 | 4.16 | 12.18 | 80.94 |
| | Decapoda n.i. | 11.75 | 3.88 | 11.36 | 92.3 |
Table 7.
Diet composition of the whole hake series from southern and central Tyrrhenian Sea sampled in 2020. In the columns are reported the diet index values (%F, %W, %N, IRI and %IRI) for each prey category.
Table 7.
Diet composition of the whole hake series from southern and central Tyrrhenian Sea sampled in 2020. In the columns are reported the diet index values (%F, %W, %N, IRI and %IRI) for each prey category.
| TAXON | %F | %W | %N | IRI | %IRI |
|---|
| MOLLUSCA | | | | | |
| Cephalopoda n.i. | 4.60 | 0.02 | 2.68 | 12.43 | 0.84 |
| CRUSTACEA | | | | | |
| Amphipoda | | | | | |
| Amphipoda n.i. | 1.15 | 0.01 | 0.67 | 0.78 | 0.05 |
| Decapoda | | | | | |
| Eusergestes arcticus | 1.15 | 0.09 | 0.67 | 0.87 | 0.06 |
| Solenocera membranacea | 1.15 | 0.33 | 0.67 | 1.16 | 0.08 |
| Pasiphaea multidentata | 2.30 | 0.66 | 1.34 | 4.61 | 0.31 |
| Pasiphaea sivado | 5.75 | 3.73 | 10.74 | 83.15 | 5.65 |
| Processa acutirostris | 1.15 | 0.32 | 2.01 | 2.68 | 0.18 |
| Sergestes sp. | 1.15 | 0.11 | 0.67 | 0.90 | 0.06 |
| Pasiphaea sp. | 1.15 | 0.51 | 2.68 | 3.67 | 0.25 |
| Dardanus sp. | 1.15 | 0.01 | 0.67 | 0.79 | 0.05 |
| Decapoda n.i. | 3.45 | 0.27 | 1.34 | 5.57 | 0.38 |
| Stomatopoda | | | | | |
| Parasquilla ferussaci | 1.15 | 0.64 | 1.34 | 2.28 | 0.15 |
| Euphausiacea | | | | | |
| Meganyctiphanes norvegica | 1.15 | 0.09 | 10.07 | 11.68 | 0.79 |
| Stylocheiron longicorne | 1.15 | 0.01 | 0.67 | 0.78 | 0.05 |
| Euphausiacea n.i. | 3.45 | 0.04 | 2.01 | 7.08 | 0.48 |
| Mysida n.i. | 10.34 | 0.12 | 24.83 | 258.12 | 17.54 |
| OSTEICHTHYES | | | | | |
| Ceratoscopelus maderensis | 2.30 | 1.03 | 2.01 | 7.00 | 0.48 |
| Diaphus holti | 1.15 | 0.40 | 0.67 | 1.23 | 0.08 |
| Boops boops | 1.15 | 0.78 | 0.67 | 1.67 | 0.11 |
| Engraulis encrasicolus | 4.60 | 8.86 | 2.68 | 53.07 | 3.61 |
| Macroramphosus scolopax | 1.15 | 0.17 | 0.67 | 0.97 | 0.07 |
| Maurolicus muelleri | 1.15 | 0.23 | 0.67 | 1.04 | 0.07 |
| Lampanyctus crocodilus | 1.15 | 0.39 | 0.67 | 1.23 | 0.08 |
| Peristedion cataphractum | 1.15 | 0.04 | 0.67 | 0.82 | 0.06 |
| Nettastoma melanura | 1.15 | 0.01 | 0.67 | 0.79 | 0.05 |
| Chlorophthalmus agassizi | 1.15 | 1.14 | 0.67 | 2.08 | 0.14 |
| Cepola macrophthalma | 1.15 | 1.35 | 0.67 | 2.32 | 0.16 |
| Stomias boa | 1.15 | 1.14 | 0.67 | 2.08 | 0.14 |
| Trachurus mediterraneus | 5.75 | 22.12 | 4.03 | 150.29 | 10.21 |
| Trachurus trachurus | 5.75 | 40.55 | 3.36 | 252.32 | 17.14 |
| Trachurus sp. | 3.45 | 8.74 | 2.01 | 37.08 | 2.52 |
| Osteichthyes n.i. | 25.29 | 6.10 | 16.11 | 561.45 | 38.14 |
Table 8.
Results of the SIMPER analysis conducted between European hake trophic groups from 2020. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table shows results obtained for the trophic clusters B (including size Classes II and III) and C (including size Classes IV and V). Trophic cluster A = less than 2 size groups.
Table 8.
Results of the SIMPER analysis conducted between European hake trophic groups from 2020. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table shows results obtained for the trophic clusters B (including size Classes II and III) and C (including size Classes IV and V). Trophic cluster A = less than 2 size groups.
| Group B | Species | Av.IRI val | Av.Sim | Contrib% | Cum.% |
|---|
| Average similarity: | Osteichthyes n.i. | 100.3 | 42.33 | 100 | 100 |
| 42.33 | | | | | |
| Group C | | | | | |
| Average similarity: | T. trachurus | 24.79 | 17.86 | 60.81 | 60.81 |
| 29.36 | Osteichthyes n.i. | 16.02 | 5.8 | 19.76 | 80.57 |
| | T. mediterraneus | 26.31 | 5.71 | 19.43 | 100 |
Table 9.
Results of the SIMPER analysis conducted between European hake trophic groups collected between 2018 and 2020. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table show results obtained for the trophic clusters A–C. Trophic cluster D = less than 2 size groups.
Table 9.
Results of the SIMPER analysis conducted between European hake trophic groups collected between 2018 and 2020. The average similarity between cluster groups is reported. The percentage and cumulative contribution of typifying species within-group similarity of the identified hake trophic groups is shown. The table show results obtained for the trophic clusters A–C. Trophic cluster D = less than 2 size groups.
| Group A | Species | Av.IRI val | Av.Sim | Contrib% | Cum.% |
|---|
| Average similarity: | Euphausiacea | 83.2 | 58.22 | 100 | 100 |
| 58.22 | |
| Group B | | | | | |
| Average similarity: | Osteichthyes n.i. | 47.31 | 22.39 | 66.92 | 66.92 |
| 33.46 | E. encrasicolus | 18.42 | 3.77 | 11.28 | 78.2 |
| | Decapoda | 7.31 | 2.22 | 6.62 | 84.83 |
| | C. maderensis | 11.93 | 1.98 | 5.92 | 90.75 |
| Group C | | | | | |
| Average similarity: | T. trachurus | 24.79 | 17.86 | 60.81 | 60.81 |
| 29.36 | Osteichthyes n.i. | 16.02 | 5.8 | 19.76 | 80.57 |
| | T. mediterraneus | 26.31 | 5.71 | 19.43 | 100 |