The Early Immune Response of Lymphoid and Myeloid Head-Kidney Cells of Rainbow Trout (Oncorhynchus mykiss) Stimulated with Aeromonas salmonicida
Abstract
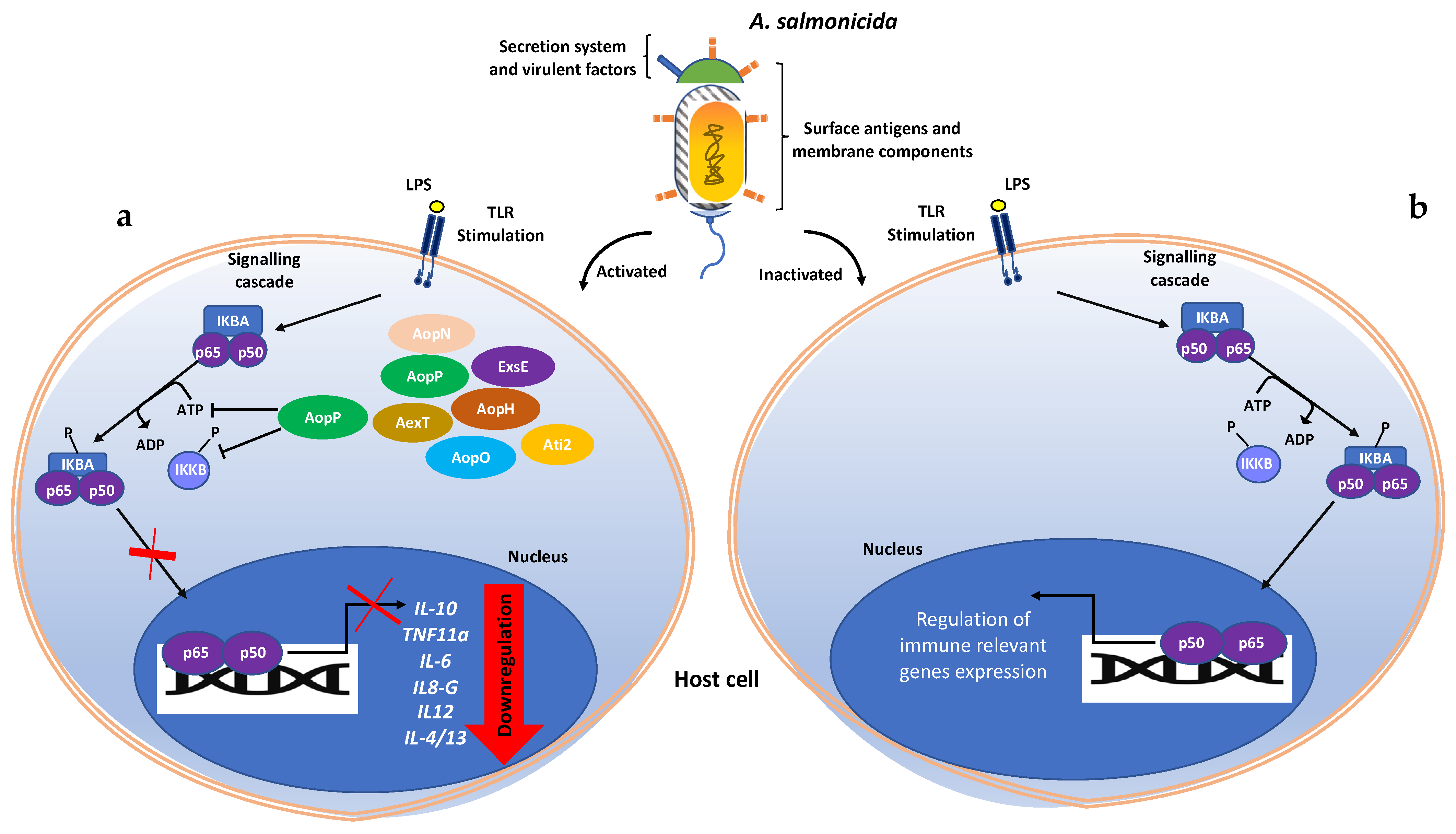
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
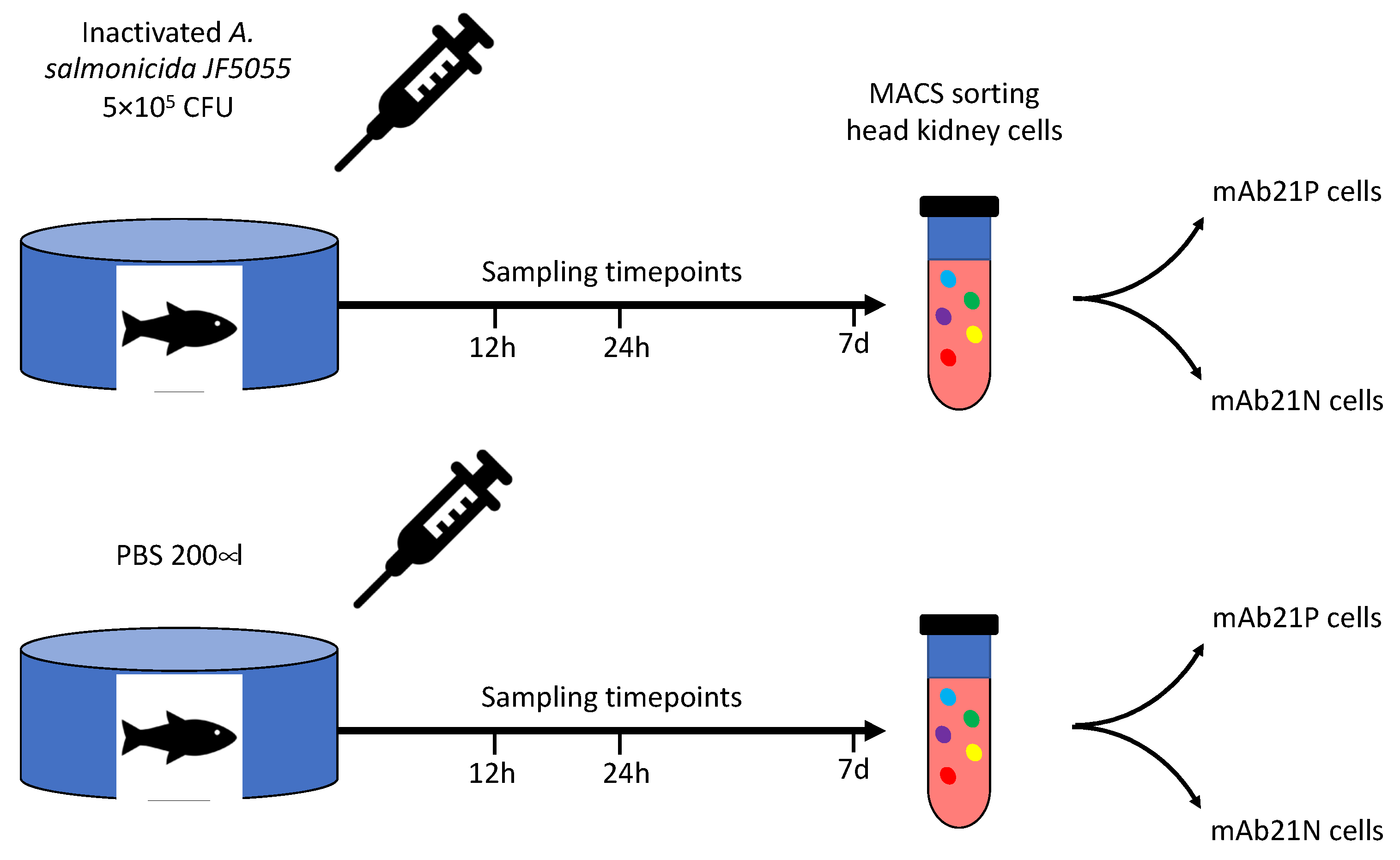
2.2. A. Salmonicida for Stimulation Experiments
2.3. Fish
2.4. Sorting of Head Kidney Cells
2.5. Primer Design and Biomark qPCR Measurements
2.6. Data Analysis
3. Results
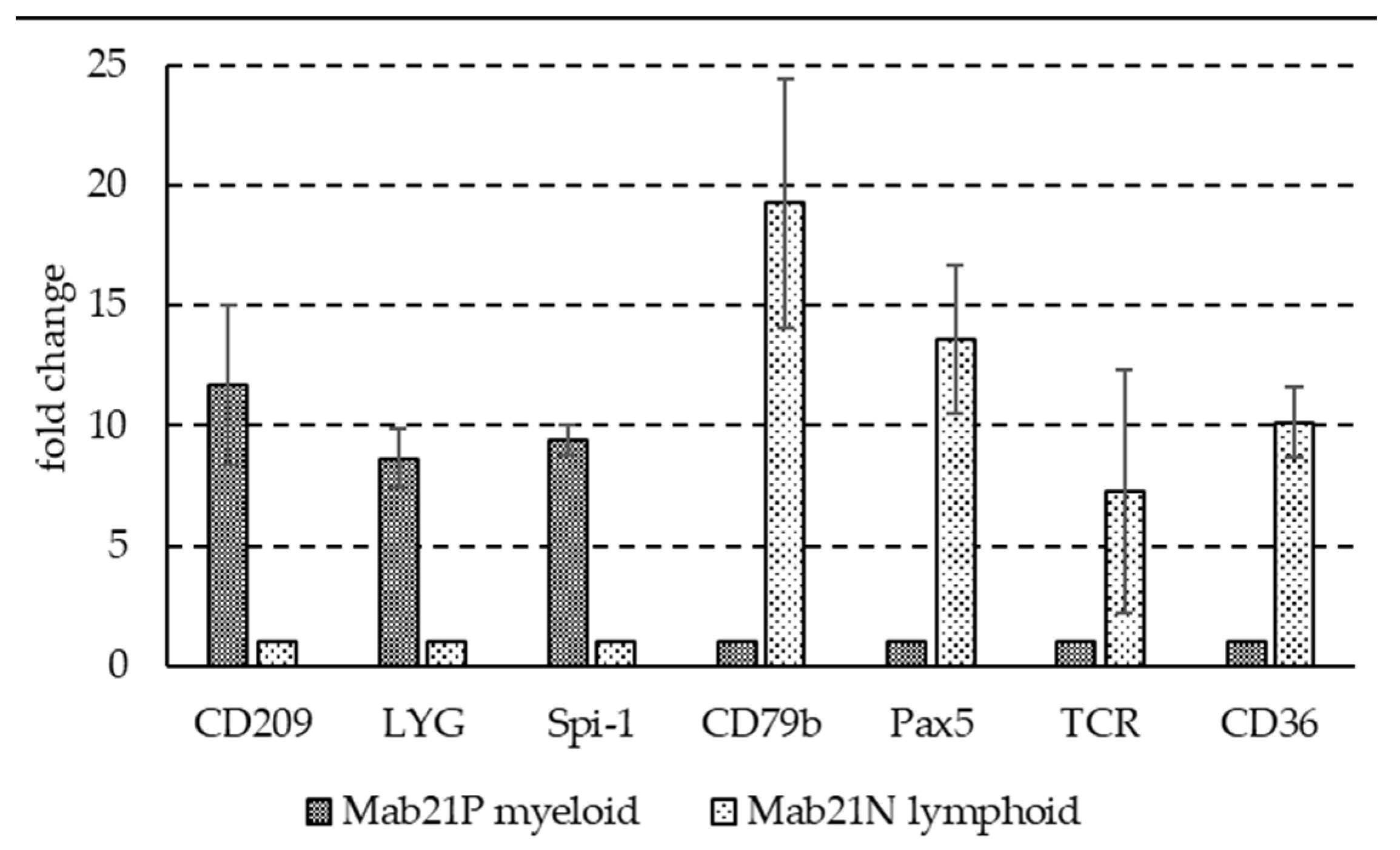
3.1. Basal Expression Profiles of Selected Immune Genes in Two Fractions Enriched in Myeloid Cells or Lymphocytes and Thrombocytes
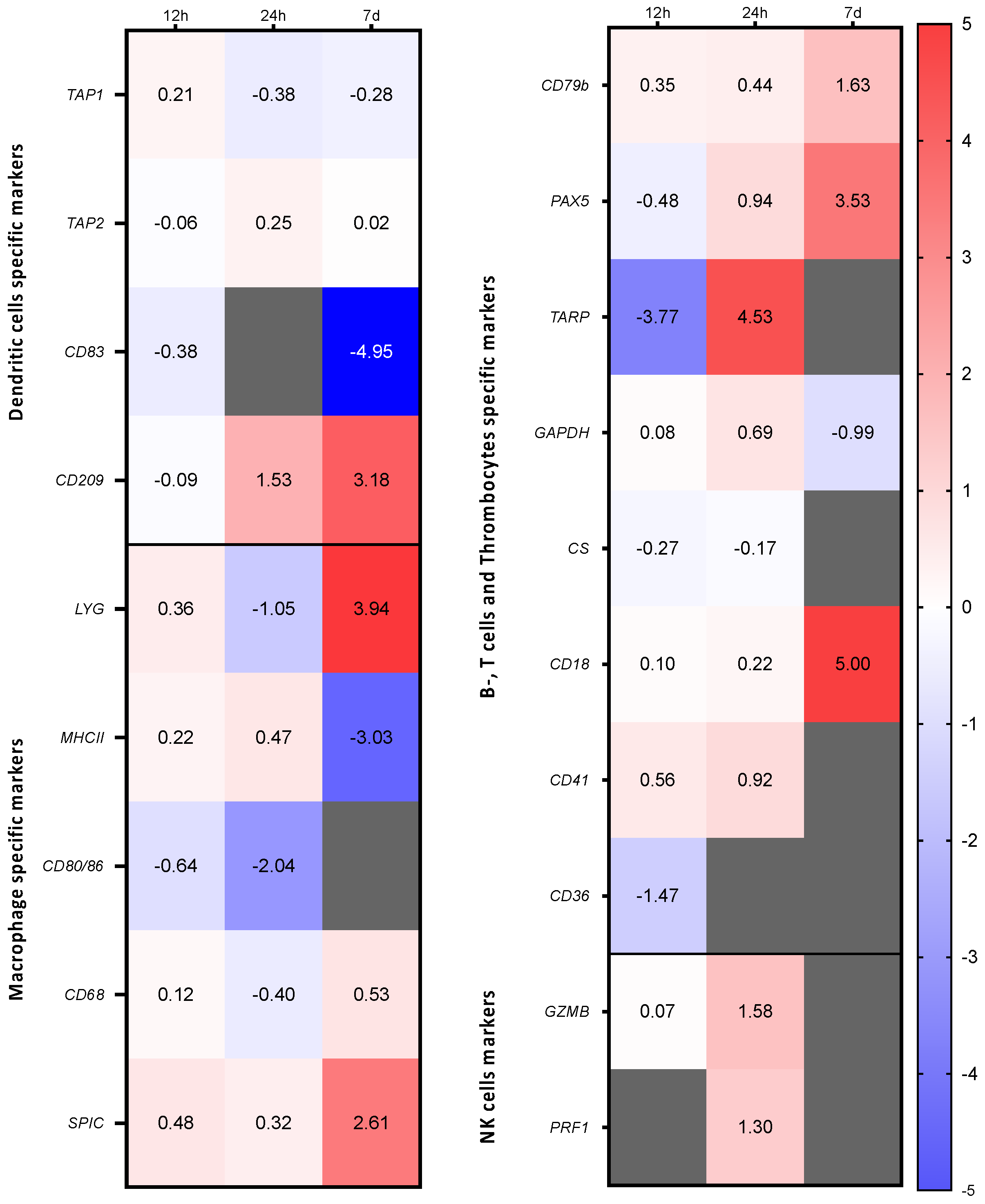
3.2. Immune Gene Expression Profiling in Two Fractions Enriched in Myeloid Cells or Lymphocytes and Thrombocytes after Stimulation with A. salmonicida
3.3. Expression Profiles of Characteristic Markers in the Cell Fraction Enriched with Lymphocytes and Thrombocytes
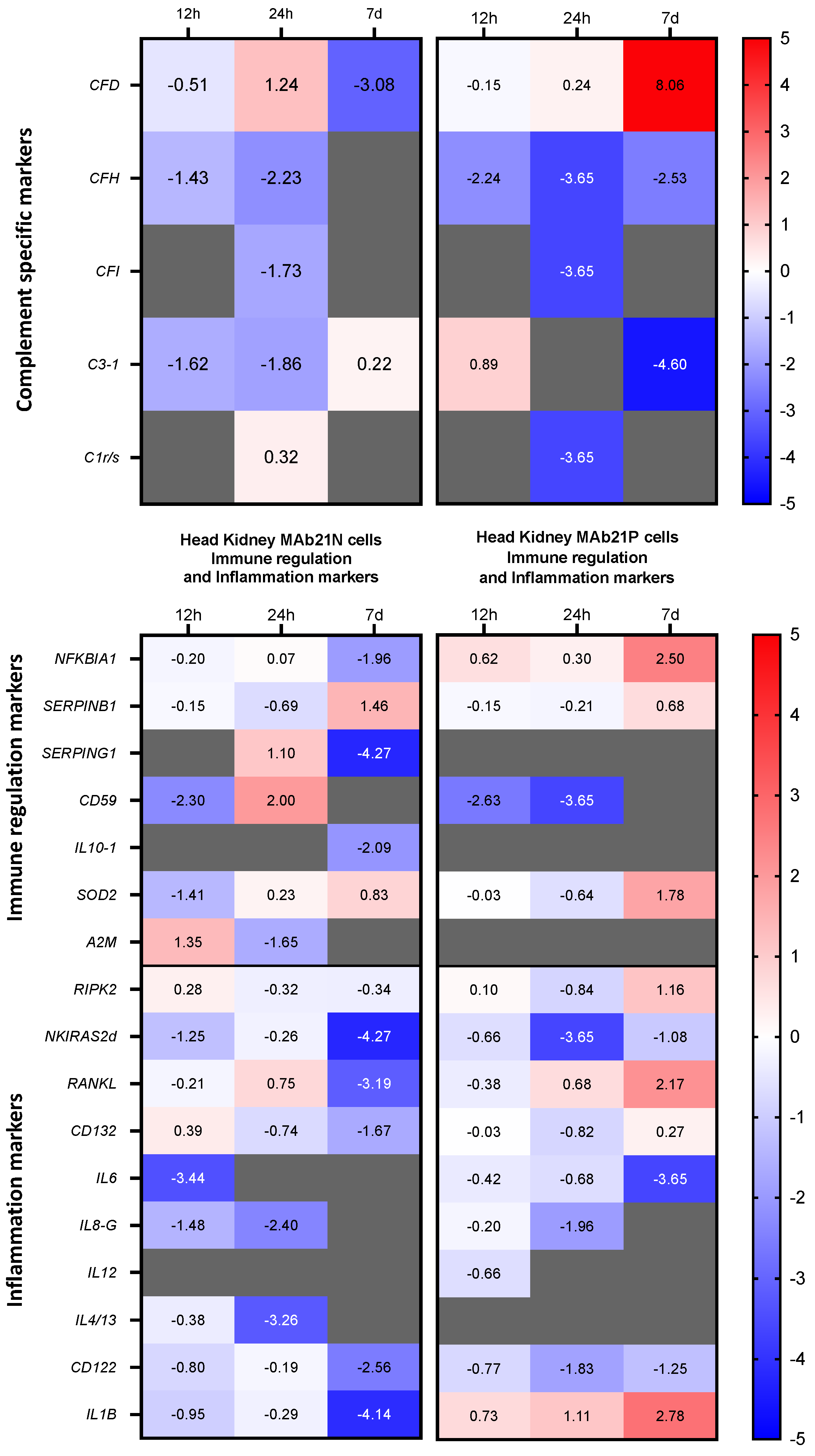
3.4. Expression Patterns of Complement- and Cytokine-encoding Genes in the Two Cell Fractions from the Head Kidney
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020; FAO: Rome, Italy, 2020. [Google Scholar]
- Scarfe, A.D.; Lee, C.-S.; O’Bryen, P.J. Aquaculture Biosecurity: Prevention, Control, and Eradication of Aquatic Animal Disease; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Flores-Kossack, C.; Montero, R.; Köllner, B.; Maisey, K. Chilean Aquaculture and the New Challenges: Pathogens, Immune Response, Vaccination and Fish Diversification. Fish Shellfish Immunol. 2020, 98, 52–67. [Google Scholar] [CrossRef]
- Magnadóttir, B. Innate Immunity of Fish (Overview). Fish Shellfish Immunol. 2006, 20, 137–151. [Google Scholar] [CrossRef]
- Uribe, C.; Folch, H.; Enriquez, R.; Moran, G. Innate and Adaptive Immunity in Teleost Fish: A Review. Veterinární Med. 2011, 56, 486–503. [Google Scholar] [CrossRef]
- Brietzke, A.; Korytář, T.; Jaros, J.; Köllner, B.; Goldammer, T.; Seyfert, H.M.; Rebl, A. Aeromonas salmonicida Infection Only Moderately Regulates Expression of Factors Contributing to Toll-like Receptor Signaling but Massively Activates the Cellular and Humoral Branches of Innate Immunity in Rainbow Trout (Oncorhynchus mykiss). J. Immunol. Res. 2015, 2015, 1–16. [Google Scholar] [CrossRef]
- Rebl, A.; Korytář, T.; Köbis, J.M.; Verleih, M.; Krasnov, A.; Jaros, J.; Kühn, C.; Köllner, B.; Goldammer, T. Transcriptome Profiling Reveals Insight into Distinct Immune Responses to Aeromonas salmonicida in Gill of Two Rainbow Trout Strains. Mar. Biotechnol. 2014, 16, 333–348. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Galán, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial Type III Secretion Systems: Specialized Nanomachines for Protein Delivery into Target Cells. Annu. Rev. Microbiol. 2014, 68, 415–438. [Google Scholar] [CrossRef]
- Vanden Bergh, P.; Frey, J. Aeromonas salmonicida Subsp. Salmonicida in the Light of Its Type-Three Secretion System. Microb. Biotechnol. 2014, 7, 381–400. [Google Scholar] [CrossRef]
- Burr, S.E.; Stuber, K.; Wahli, T.; Frey, J. Evidence for a Type III Secretion System in Aeromonas salmonicida Subsp. Salmonicida. J. Bacteriol. 2002, 184, 5966–5970. [Google Scholar] [CrossRef]
- Frey, J.; Origgi, F.C. Type III Secretion System of Aeromonas salmonicida Undermining the Host’s Immune Response. Front. Mar. Sci. 2016, 3, 130. [Google Scholar] [CrossRef]
- Dorrington, M.G.; Fraser, I.D.C. NF-ΚB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef]
- Abitew, A.M.; Sobti, R.C.; Sharma, V.L.; Wanchu, A. Analysis of Transporter Associated with Antigen Presentation (TAP) Genes Polymorphisms with HIV-1 Infection. Mol. Cell. Biochem. 2020, 464, 65–71. [Google Scholar] [CrossRef]
- López-Albaitero, A.; Mailliard, R.; Hackman, T.; Filho, P.A.A.; Wang, X.; Gooding, W.; Ferrone, S.; Kalinski, P.; Ferris, R.L. Maturation Pathways of Dendritic Cells Determine TAP1 and TAP2 Levels and Cross-Presenting Function. J. Immunother. 2009, 32, 465–473. [Google Scholar] [CrossRef]
- Schwenteit, J.M.; Breithaupt, A.; Teifke, J.P.; Koppang, E.O.; Bornscheuer, U.T.; Fischer, U.; Gudmundsdottir, B.K. Innate and Adaptive Immune Responses of Arctic Charr (Salvelinus alpinus L.) during Infection with Aeromonas salmonicida Subsp. Achromogenes and the Effect of the AsaP1 Toxin. Fish Shellfish Immunol. 2013, 35, 866–873. [Google Scholar] [CrossRef]
- Schetters, S.T.T.; Kruijssen, L.J.W.; Crommentuijn, M.H.W.; Kalay, H.; Ochando, J.; den Haan, J.M.M.; Garcia-Vallejo, J.J.; van Kooyk, Y. Mouse DC-SIGN/CD209a as Target for Antigen Delivery and Adaptive Immunity. Front. Immunol. 2018, 9, 1. [Google Scholar] [CrossRef]
- Buonocore, F.; Randelli, E.; Trisolino, P.; Facchiano, A.; de Pascale, D.; Scapigliati, G. Molecular Characterization, Gene Structure and Antibacterial Activity of a g-Type Lysozyme from the European Sea Bass (Dicentrarchus labrax L.). Mol. Immunol. 2014, 62, 10–18. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Kiryu, I.; Köllner, B.; Yoshiura, Y.; Ototake, M. MHC Class II Invariant Chain Homologues in Rainbow Trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2003, 15, 91–105. [Google Scholar] [CrossRef]
- Thomas, E.K.; Ostrowski, M.A.; Yu, Q.; Gu, J.X.; Kovacs, C.; Freedman, J. Cooperation of TNF Family Members CD40 Ligand, Receptor Activator of NF-kappa B Ligand, and TNF-alpha in the Activation of Dendritic Cells and the Expansion of Viral Specific CD8+ T Cell Memory Responses in HIV-1-Infected and HIV-1-Uninfected Individuals. J. Immunol. 2003, 170, 1797–1805. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/Macrosialin: Not Just a Histochemical Marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef]
- Oikawa, T.; Yamada, T.; Kihara-Negishi, F.; Yamamoto, H.; Kondoh, N.; Hitomi, Y.; Hashimoto, Y. The Role of Ets Family Transcription Factor PU.1 in Hematopoietic Cell Differentiation, Proliferation and Apoptosis. Cell Death Differ. 1999, 6, 599–608. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Zhang, X.; Wang, Y.; Zhang, W.; Wu, X.; Wang, Z. Molecular Characterization of the CD79a and CD79b and Its Role against Aeromonas hydrophila Infection in Chinese Sucker (Myxocyprinus asiaticus). Fish Physiol. Biochem. 2017, 43, 1571–1585. [Google Scholar] [CrossRef]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The Guardian of B Cell Identity and Function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef]
- Szczepański, T.; Langerak, A.W.; Willemse, M.J.; Wolvers-Tettero, I.L.M.; van Wering, E.R.; van Dongen, J.J.M. T Cell Receptor Gamma (TCRG) Gene Rearrangements in T Cell Acute Lymphoblastic Leukemia Reflect “end-Stage” Recombinations: Implications for Minimal Residual Disease Monitoring. Leukemia 2000, 14, 1208–1214. [Google Scholar] [CrossRef]
- Galván-Peña, S.; Carroll, R.G.; Newman, C.; Hinchy, E.C.; Palsson-McDermott, E.; Robinson, E.K.; Covarrubias, S.; Nadin, A.; James, A.M.; Haneklaus, M.; et al. Malonylation of GAPDH Is an Inflammatory Signal in Macrophages. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Ji, D.; Yin, J.Y.; Li, D.F.; Zhu, C.T.; Ye, J.P.; Pan, Y.Q. Effects of Inflammatory and Anti-Inflammatory Environments on the Macrophage Mitochondrial Function. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Maiguel, D.; Faridi, M.H.; Wei, C.; Kuwano, Y.; Balla, K.M.; Hernandez, D.; Barth, C.J.; Lugo, G.; Donnelly, M.; Nayer, A.; et al. Small Molecule-Mediated Activation of the Integrin CD11b/CD18 Reduces Inflammatory Disease. Sci. Signal. 2011, 4, ra57. [Google Scholar] [CrossRef]
- Ali, R.A.; Wuescher, L.M.; Worth, R.G. Platelets: Essential Components of the Immune System. Curr. Trends Immunol. 2015, 16, 65. [Google Scholar]
- Fink, I.R.; Benard, E.L.; Hermsen, T.; Meijer, A.H.; Forlenza, M.; Wiegertjes, G.F. Molecular and Functional Characterization of the Scavenger Receptor CD36 in Zebrafish and Common Carp. Mol. Immunol. 2015, 63, 381–393. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and Granzymes: Function, Dysfunction and Human Pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Trapani, J.A.; Smyth, M.J. Functional Significance of the Perforin/Granzyme Cell Death Pathway. Nat. Rev. Immunol. 2002, 2, 735–747. [Google Scholar] [CrossRef]
- Gunturi, A.; Berg, R.E.; Forman, J. The Role of CD94/NKG2 in Innate and Adaptive Immunity. Immunol. Res. 2004, 30, 29–34. [Google Scholar] [CrossRef]
- Köbis, J.M.; Rebl, A.; Kühn, C.; Korytář, T.; Köllner, B.; Goldammer, T. Comprehensive and Comparative Transcription Analyses of the Complement Pathway in Rainbow Trout. Fish Shellfish Immunol. 2015, 42, 98–107. [Google Scholar] [CrossRef]
- Almitairi, J.O.M.; Girija, U.V.; Furze, C.M.; Simpson-Gray, X.; Badakshi, F.; Marshall, J.E.; Schwaeble, W.J.; Mitchell, D.A.; Moody, P.C.E.; Wallis, R. Structure of the C1r–C1s Interaction of the C1 Complex of Complement Activation. Proc. Natl. Acad. Sci. USA 2018, 115, 768–773. [Google Scholar] [CrossRef]
- Tian, B.; Nowak, D.E.; Jamaluddin, M.; Wang, S.; Brasier, A.R. Identification of Direct Genomic Targets Downstream of the NF-Kappa B Transcription Factor Mediating TNF Signaling. J. Biol. Chem. 2005, 280, 17435–17448. [Google Scholar] [CrossRef]
- Bao, J.; Pan, G.; Poncz, M.; Wei, J.; Ran, M.; Zhou, Z. Serpin Functions in Host-Pathogen Interactions. Peer J. 2018, 6, e4557. [Google Scholar] [CrossRef]
- Morgan, P. CD59. In The Complement Facts Book: Second Edition; Elsevier: Amsterdam, The Netherlands, 2018; pp. 361–367. ISBN 9780128104200. [Google Scholar]
- Carla Piazzon, M.; Lutfall, G.; Forlenzaa, M. IL10, a Tale of an Evolutionarily Conserved Cytokine across Vertebrates. Crit. Rev. Immunol. 2016, 36, 99–129. [Google Scholar] [CrossRef]
- Peterman, E.M.; Sullivan, C.; Goody, M.F.; Rodriguez-Nunez, I.; Yoder, J.A.; Kim, C.H. Neutralization of Mitochondrial Superoxide by Superoxide Dismutase 2 Promotes Bacterial Clearance and Regulates Phagocyte Numbers in Zebrafish. Infect. Immun. 2015, 83, 430–440. [Google Scholar] [CrossRef]
- Ellis, A.E. Inhibition of the Aeromonas salmonicida Extracellular Protease by A2-Macroglobulin in the Serum of Rainbow Trout. Microb. Pathog. 1987, 3, 167–177. [Google Scholar] [CrossRef]
- Wu, X.M.; Chen, W.Q.; Hu, Y.W.; Cao, L.; Nie, P.; Chang, M.X. RIP2 Is a Critical Regulator for NLRs Signaling and MHC Antigen Presentation but Not for MAPK and PI3K/Akt Pathways. Front. Immunol. 2018, 9, 726. [Google Scholar] [CrossRef]
- Sarais, F.; Rebl, H.; Verleih, M.; Ostermann, S.; Krasnov, A.; Köllner, B.; Goldammer, T.; Rebl, A. Characterisation of the Teleostean ΚB-Ras Family: The Two Members NKIRAS1 and NKIRAS2 from Rainbow Trout Influence the Activity of NF-ΚB in Opposite Ways. Fish Shellfish Immunol. 2020, 106, 1004–1013. [Google Scholar] [CrossRef]
- Glenney, G.W.; Wiens, G.D. Early Diversification of the TNF Superfamily in Teleosts: Genomic Characterization and Expression Analysis. J. Immunol. 2007, 178, 7955–7973. [Google Scholar] [CrossRef] [PubMed]
- Ratthé, C.; Girard, D. Interleukin-15 Enhances Human Neutrophil Phagocytosis by a Syk-Dependent Mechanism: Importance of the IL-15Rα Chain. J. Leukoc. Biol. 2004, 76, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Secombes, C.J. Identification and Expression Analysis of Two Fish-Specific IL-6 Cytokine Family Members, the Ciliary Neurotrophic Factor (CNTF)-like and M17 Genes, in Rainbow Trout Oncorhynchus mykiss. Mol. Immunol. 2009, 46, 2290–2298. [Google Scholar] [CrossRef]
- Rebl, A.; Rebl, H.; Korytář, T.; Goldammer, T.; Seyfert, H.M. The Proximal Promoter of a Novel Interleukin-8-Encoding Gene in Rainbow Trout (Oncorhynchus mykiss) Is Strongly Induced by CEBPA, but not NF-κB p65. Dev. Comp. Immunol. 2014, 46, 155–164. [Google Scholar] [CrossRef]
- Vignali, D.A.A.; Kuchroo, V.K. IL-12 Family Cytokines: Immunological Playmakers. Nat. Immunol. 2012, 13, 722–728. [Google Scholar] [CrossRef]
- Li, J.H.; Shao, J.Z.; Xiang, L.X.; Wen, Y. Cloning, Characterization and Expression Analysis of Pufferfish Interleukin-4 CDNA: The First Evidence of Th2-Type Cytokine in Fish. Mol. Immunol. 2007, 44, 2078–2086. [Google Scholar] [CrossRef]
- Wang, T.; Hu, Y.; Wangkahart, E.; Liu, F.; Wang, A.; Zahran, E.; Maisey, K.R.; Liu, M.; Xu, Q.; Imarai, M.; et al. Interleukin (IL)-2 Is a Key Regulator of T Helper 1 and T Helper 2 Cytokine Expression in Fish: Functional Characterization of Two Divergent IL2 Paralogs in Salmonids. Front. Immunol. 2018, 9, 1683. [Google Scholar] [CrossRef]
- Sigh, J.; Lindenstrøm, T.; Buchmann, K. Expression of Pro-Inflammatory Cytokines in Rainbow Trout (Oncorhynchus mykiss) during an Infection with Ichthyophthirius Multifiliis. Fish Shellfish Immunol. 2004, 17, 75–86. [Google Scholar] [CrossRef]
- Ellis, A.E. Innate Host Defense Mechanisms of Fish against Viruses and Bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.H.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.F.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a Novel Dendritic Cell–Specific ICAM-3 Receptor That Supports Primary Immune Responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Jiménez-Cantizano, R.M.; Infante, C.; Martin-Antonio, B.; Ponce, M.; Hachero, I.; Navas, J.I.; Manchado, M. Molecular Characterization, Phylogeny, and Expression of c-Type and g-Type Lysozymes in Brill (Scophthalmus rhombus). Fish Shellfish Immunol. 2008, 25, 57–65. [Google Scholar] [CrossRef]
- Gomard, T.; Michaud, H.-A.; Tempé, D.; Thiolon, K.; Pelegrin, M.; Piechaczyk, M. An NF-ΚB–Dependent Role for JunB in the Induction of Proinflammatory Cytokines in LPS-Activated Bone Marrow–Derived Dendritic Cells. PLoS ONE 2010, 5, e9585. [Google Scholar] [CrossRef]
- Aerts-Toegaert, C.; Heirman, C.; Tuyaerts, S.; Corthals, J.; Aerts, J.L.; Bonehill, A.; Thielemans, K.; Breckpot, K.; Breckpot, K. CD83 Expression on Dendritic Cells and T Cells: Correlation with Effective Immune Responses. Eur. J. Immunol. 2007, 37, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, N.C.; Rao, K.V.S.; Natarajan, K. CD80 Expression Is Induced on Activated B Cells Following Stimulation by CD86. Scand. J. Immunol. 2002, 55, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Lane, P. Regulation of T and B Cell Responses by Modulating Interactions between CD28/CTLA4 and Their Ligands, CD80 and CD86. Ann. N. Y. Acad. Sci. 1997, 815, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Lüder, C.G.K.; Lang, T.; Beuerle, B.; Gross, U. Down-Regulation of MHC Class II Molecules and Inability to up-Regulate Class I Molecules in Murine Macrophages after Infection with Toxoplasma Gondii. Clin. Exp. Immunol. 1998, 112, 308. [Google Scholar] [CrossRef]
- Korytář, T.; Jaros, J.; Verleih, M.; Rebl, A.; Kotterba, G.; Kühn, C.; Goldammer, T.; Köllner, B. Novel Insights into the Peritoneal Inflammation of Rainbow Trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2013, 35, 1192–1199. [Google Scholar] [CrossRef]
- Trendel, N.; Kruger, P.; Gaglione, S.; Nguyen, J.; Pettmann, J.; Sontag, E.D.; Dushek, O. Perfect Adaptation of CD8+ T Cell Responses to Constant Antigen Input over a Wide Range of Affinity Is Overcome by Costimulation. Sci. Signal. 2021, 14, 666. [Google Scholar] [CrossRef]
- Hu, Q.; Ao, Q.; Zhu, J. Response of Chemokine Receptors CXCR2 and Integrin Β2 after Streptococcus agalactiae and Aeromonas hydrophila Challenge in GIFT Strain of Nile Tilapia Oreochromis Niloticus. Dev. Comp. Immunol. 2021, 115, 103897. [Google Scholar] [CrossRef]
- Silverstein, R.L. Type 2 Scavenger Receptor CD36 in Platelet Activation: The Role of Hyperlipemia and Oxidative Stress. Clin. Lipidol. 2009, 4, 767. [Google Scholar] [CrossRef][Green Version]
- Ferdous, F.; Scott, T.R. A Comparative Examination of Thrombocyte/Platelet Immunity. Immunol. Lett. 2015, 163, 32–39. [Google Scholar] [CrossRef]
- Köllner, B.; Fischer, U.; Rombout, J.H.W.M.; Taverne-Thiele, J.J.; Hansen, J.D. Potential Involvement of Rainbow Trout Thrombocytes in Immune Functions: A Study Using a Panel of Monoclonal Antibodies and RT-PCR. Dev. Comp. Immunol. 2004, 28, 1049–1062. [Google Scholar] [CrossRef]
- Zamora, C.; Cantó, E.; Nieto, J.C.; Ortiz, M.A.; Juarez, C.; Vidal, S. Functional Consequences of CD36 Downregulation by TLR Signals. Cytokine 2012, 60, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tian, M.; Zhang, L.; Fu, Q.; Song, L.; Yang, N. Expression Profiling and Functional Characterization of CD36 in Turbot (Scophthalmus maximus L.). Fish Shellfish Immunol. 2018, 81, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Van der Sar, A.M.; Spaink, H.P.; Zakrzewska, A.; Bitter, W.; Meijer, A.H. Specificity of the Zebrafish Host Transcriptome Response to Acute and Chronic Mycobacterial Infection and the Role of Innate and Adaptive Immune Components. Mol. Immunol. 2009, 46, 2317–2332. [Google Scholar] [CrossRef]
- Merselis, L.C.; Rivas, Z.P.; Munson, G.P. Breaching the Bacterial Envelope: The Pivotal Role of Perforin-2 (MPEG1) within Phagocytes. Front. Immunol. 2021, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Heesterbeek, D.A.C.; Angelier, M.L.; Harrison, R.A.; Rooijakkers, S.H.M. Complement and Bacterial Infections: From Molecular Mechanisms to Therapeutic Applications. J. Innate Immun. 2018, 10, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Lambris, J.D.; Ricklin, D.; Geisbrecht, B.V. Complement Evasion by Human Pathogens. Nat. Rev. Microbiol. 2008, 6, 132. [Google Scholar] [CrossRef]
- Merino, S.; Alberti, S.; Tomasl, J.M. Aeromonas salmonicida Resistance to Complement-Mediated Killing. Infect. Immun. 1994, 62, 5483–5490. [Google Scholar] [CrossRef]
- Menanteau-Ledouble, S.; Kumar, G.; Saleh, M.; El-Matbouli, M. Aeromonas salmonicida: Updates on an Old Acquaintance. Dis. Aquat. Org. 2016, 120, 49–68. [Google Scholar] [CrossRef]
- Volanakis, J.E. Transcriptional Regulation of Complement Genes. Annu. Rev. Immunol. 1995, 13, 277–305. [Google Scholar] [CrossRef] [PubMed]

| Cell Population | Gene Symbol | Primer 5′–3′ | Length (bp) | Accession No. | Function | Ref. |
|---|---|---|---|---|---|---|
| Dendritic cells | TAP1 | CACTCCTGGCAGGGGCTACTT | 176 | XM_021559784 | Antigen presentation | [14] |
| CCTTATTTCATACGCTTTGGAGC | ||||||
| TAP2 | CATCTGTGAGACGTTTATCCCTT | 99 | XM_024386707 | Antigen presentation | [15] | |
| TCATGTACGCCATTGGAGGCAT | ||||||
| CD83 | GTCTGCATTCTAGCTGCCTACT | 128 | XM_021593617 | Immune cell interactions | [16] | |
| ACGTAAGCCTGGGGTCCAGTA | ||||||
| CD209 | ATCTCTCAGGTACCGGAAGAGT | 127 | HG428763 | Intercellular adhesion, antigen uptake | [17] | |
| GACTGTCTGGAGAGAGGAGCA | ||||||
| Macrophages | LYG | GCAGGTTGACAAGCGCTACCA | 118 | BT073825 | Hydrolyzation of the bacterial cell wall | [18] |
| AAAGGGGGAATTTCAGCCTACAA | ||||||
| DAA (MHCII) | CAGTGATTCAGATGGAGTGAATAT | 131 | FR688130 | Cell surface proteins with a key role in adaptive immunity | [19] | |
| AGATTTCCTTCCCTGGATATTATG | ||||||
| CD80/86 | GCGTCGGCTGCTTCGAAGGT | 152 | NM_001160477 | Co-stimulation of T-cells | [20] | |
| AGACTCCCAAACCACCTGTATG | ||||||
| CD68 | GACACTGGAAAGACAGGAGTATT | 115 | XM_021578316 | Scavenger receptor and antigen processor | [21] | |
| TTCAAGGAGGGCTTCATCACCT | ||||||
| SPIC | CACCTGGTCCTGCATCAGAAG | 127 | NM_001124513 | Immune gene expression | [22] | |
| CTGGGACTATCACGCCACTCA | ||||||
| B- and T-cells | CD79B | TGAACCTCTCAGTGGCTTTAAAC | 111 | XM_021565350 | Signalling through B-cell receptors | [23] |
| TCTGTGTGGTGTCGGACCGAA | ||||||
| PAX5 | AAGTATCCGTCCCGGGGTGAT | 144 | NM_001124682 | Development of lymphoid progenitors | [24] | |
| GACTATTGGCTGAGAGAGTGTG | ||||||
| TARP | GAGAAGTGGAGTGGGACCAGA | 105 | XM_021619909 | Antigen presentation | [25] | |
| AGGTGAGTGACGGGGGACAC | ||||||
| Pan-T-cells and Thrombocytes | GAPDH | TCAACGGATTTGGCCGTATTGG | 134 | NM_001124246 | Influence cytokine production | [26] |
| GTTCAAGTATGACTCCACCCAC | ||||||
| CS | CACCTTCAACGAGGTTTACCCC | 131 | XM_021610150 | Metabolic regulator | [27] | |
| GAGATGTGCTTTTGGATCTTGTC | ||||||
| CD18 | AGTGAGTTGAGGGTTACATAGGA | 148 | XM_021579244 | Leukocyte migration and adhesion | [28] | |
| GTATGTTCCAAAACAGTGATCAAC | ||||||
| CD41 | CAGTTTAGCAACAAAACCATCAGT | 161 | XM_021624569 | Leukocyte migration and adhesion | [29] | |
| TGACACAAAAGGTGATGAGGTTTA | ||||||
| CD36 | GACTGTTACAAAGGAATCGGTCAT | 112 | XM_021577070 | Ligand binding | [30] | |
| ACAGTTCTGGCTCTTTGACGTG | ||||||
| Natural Killer cells | GZMB | TCCTTTCCTCTGCTGGAGCCT | 94 | XM_021598076 | Destruction of infected and/or transformed cells | [31] |
| TATATGGTCTCTCTGCAACACAG | ||||||
| PRF1 | GCGGGTATTACAGCTATCGAGTA | 161 | XM_021558434 | Destruction of infected and/or transformed cells | [32] | |
| ATTACATTACCAAGGTGAGCCTG | ||||||
| KLRD1 (CD94) | TGGGGCAACGATCGGCTCAAA | 114 | XM_021559366 | “Missing-self” discrimination | [33] | |
| GAATCCTGCCACAGCAGTGGA | ||||||
| Complement system | CFD | GACAAGTCATGAGCCCCAAG | 151 | NM_001246346 | Serine protease activity of the alternative complement pathway | [34] |
| GTGCCGAAAGTGGGTATTGT | ||||||
| CFH | GCTGGACCAAGACACTTGGC | 166 | NM_001124410 | Complement regulation | [34] | |
| CCTCTACCGGGGGTTGGTG | ||||||
| CFI | ACCCAGTGTTTGCAAGAGAACC | 167 | XM_021593383 | Inactivation of C3b/C4b factors | [34] | |
| CAGTTGGCGATCAGAGAGACG | ||||||
| C3-1 | AGCCTCTGACCAGGGAGATATT | 164 | L24433 | Opsonisation, elimination of pathogens | [34] | |
| GACGATGTCAGGGAGTTTGAAC | ||||||
| C1r/s | AACCAGAGGGGACTCTGTCCA | 182 | NM_001124380 | Initiation of classical complement pathway | [35] | |
| TGGACAGAGTCCCCTCTGGTT | ||||||
| Immune regulation | NFKBIA | AACCCTGGAGGAAAACAGTGAC | 153 | NM_001124368 | Inhibition of NF-κB pathways | [36] |
| GAACAATCAGAGACAGACGGCG | ||||||
| SERPINB1 | TACCAGTTCGTTGAGACGTTCC | 116 | NM_001124515 | Reduction of tissue damage; cell differentiation; immune activation | [37] | |
| ATCAAGAACCTATTGGCGGAGG | ||||||
| SERPING1 | AAGGAATGACGAACGGCAAACG | 169 | NM_001124379 | Activation of the C1 complex | [37] | |
| TCAGCTGTCTCACAGTAGTACAT | ||||||
| CD59 | GATTGAGTGGGCAAAGTATTGTAT | 167 | XM_021606996 | Inhibition of the membrane-attack complex | [38] | |
| CATACCCTGTTACATAACATTGCT | ||||||
| IL10 | TGCCCAGTGCAGACGTGTACC | 137 | NM_001245099 | Anti-inflammatory function | [39] | |
| TACACCACTTGAAGAGCCCCG | ||||||
| SOD2 | TCCCTGACCTGACCTACGAC | 201 | XR_00247449 | Oxidative stress | [40] | |
| GAGGTTTAATGGAGGAGGCC | ||||||
| A2M | GGGAGGAAGGATGAGATGAGTA | 184 | XM_021582312 | Inhibition of cytokine-induced inflammation | [41] | |
| CTAACAGTGGAGCTTCAGGACC | ||||||
| Inflammation | RIPK2 | TGTTGGCGAAAGGGAGAGGAAT | 105 | KJ184523 | Modulation of innate and adaptive immune responses | [42] |
| GTACATGAGCAATGGCTCTCTG | ||||||
| NKIRAS2a | TGCATGTCTGCCTGTCTCTTTTT | 201 | XM_021557705 | Regulation of NF-κB signalling | [43] | |
| TGAGCCCGCAATATGATTGGCA | ||||||
| RANKL | GAGAGCATCGACTGGGAAAATGT | 125 | XM_021620403 | Regulation of interactions between T-cells and dendritic cells | [44] | |
| TGTTCTGGGTACTCTGACACCA | ||||||
| IL2RG (CD132) | ACCCCCAATGTAAACTGCCTGA | 112 | NM_001124356 | Cytokine signalling involved in the stimulation of phagocytosis | [45] | |
| TTTCAGCAGCAGGTTCATCAAAG | ||||||
| IL6 | GTGTTAGTTAAGGGGAATCCAGT | 128 | NM_001124657 | Proinflammatory cytokine and anti-inflammatory myokine | [46] | |
| CCTTGCGGAACCAACAGTTTGT | ||||||
| CXCL8 | ATATAACACTTGTTACCAGCGAGA | 106 | HG917307 | Chemoattraction | [47] | |
| ATTACTGAGGAGATGAGTCTGAG | ||||||
| IL12 | ACATTCAGTGAGAGTGCGTGTC | 118 | HE798148 | Differentiation of naïve T-cells | [48] | |
| ACAAGGGGATCCTTCCTCACAA | ||||||
| IL4/13 | CTGTCAGAGGAACTTCTGGAAAC | 131 | NM_001246341 | Regulation of inflammatory processes | [49] | |
| GTGAAAAATGACGCGTTTGGTGA | ||||||
| IL2RB (CD122) | AGAGGACAGTGGCGGTAATGAT | 94 | XM_021622445 | Cytokine signalling involved in T-cell-mediated immune responses | [50] | |
| CTCACAACCTCCAAGGACTGTT | ||||||
| IL1B | GAGAGTGCTGTGGAAGAACATAT | 157 | NM_001124347 | Inflammation | [51] | |
| ATGAATGAGGCTATGGAGCTGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarais, F.; Montero, R.; Ostermann, S.; Rebl, A.; Köllner, B.; Goldammer, T. The Early Immune Response of Lymphoid and Myeloid Head-Kidney Cells of Rainbow Trout (Oncorhynchus mykiss) Stimulated with Aeromonas salmonicida. Fishes 2022, 7, 12. https://doi.org/10.3390/fishes7010012
Sarais F, Montero R, Ostermann S, Rebl A, Köllner B, Goldammer T. The Early Immune Response of Lymphoid and Myeloid Head-Kidney Cells of Rainbow Trout (Oncorhynchus mykiss) Stimulated with Aeromonas salmonicida. Fishes. 2022; 7(1):12. https://doi.org/10.3390/fishes7010012
Chicago/Turabian StyleSarais, Fabio, Ruth Montero, Sven Ostermann, Alexander Rebl, Bernd Köllner, and Tom Goldammer. 2022. "The Early Immune Response of Lymphoid and Myeloid Head-Kidney Cells of Rainbow Trout (Oncorhynchus mykiss) Stimulated with Aeromonas salmonicida" Fishes 7, no. 1: 12. https://doi.org/10.3390/fishes7010012
APA StyleSarais, F., Montero, R., Ostermann, S., Rebl, A., Köllner, B., & Goldammer, T. (2022). The Early Immune Response of Lymphoid and Myeloid Head-Kidney Cells of Rainbow Trout (Oncorhynchus mykiss) Stimulated with Aeromonas salmonicida. Fishes, 7(1), 12. https://doi.org/10.3390/fishes7010012









