Abstract
The Chinese mitten crab (Eriocheir sinensis) is a representative catadromous invertebrate of the Yangtze River and a commercial species widely cultivated in China. Both cultivated and wild crabs suffer from a variety of parasites and pathogens, which can result in catastrophic economic losses in aquaculture revenue. Polyascus gregaria, a parasitic barnacle with a highly derived morphology, is specialized in invading these crabs. This study examines the immunological mechanism in E. sinensis infected with P. gregaria. Tandem mass tags (TMT), a specialized method of mass-spectrometry, was used to analyze the infection by P. gregaria resistance at the protein level. In the hepatopancreas of infected crabs, 598 proteins differentially expressed relating to physiological change, of which, 352 were upregulated and 246 were downregulated. Based on this differential protein expression, 104 GO terms and 13 KEGG pathways were significantly enriched. Differentially expressed proteins, such as ATG, cathepsin, serpin, iron-related protein, Rab family, integrin, and lectin, are associated with the lysosome GO term and the autophagy-animal KEGG pathways, both of which likely relate to the immune response to the parasitic P. gregaria infection. These results show the benefit of taking a detailed, protein-level approach to understanding the innate immune response of aquatic invertebrates to macroparasite infection.
1. Introduction
The Chinese mitten crab (Eriocheir sinensis) is a well-known and important decapod crustacean with both ecological and economic value [1]. This migratory crustacean is native to the coastal waters of East Asia, but is now considered an invasive species throughout Europe and North America [2]. In China, more distinctive germplasm characteristics, and a high output of this native crustacean from the Yangtze River, are generally acknowledged by the public. Due to the commercial value of this species, intensive cultivation became popular after the 1950s [3] from the Yangtze River. Along with the rapid development of the large-scale aquaculture of these crabs are frequent outbreaks of viruses, bacteria, rickettsia-like organisms, and parasites, all of which have led to catastrophic economic losses for Chinese mitten crab farmers [4,5]. These infections can also cause remarkable morphological, physiological, and behavioral changes in the host [6]. Although the interactions among bacteria [7,8], funguses [9], parasites [10], and even ecological factors [6,11] in the host of the Chinese mitten crab, are represented in the literature, to date, there has been little focus on the parasitic barnacle Polyascus gregaria and its effect on the Chinese mitten crab. The hepatopancreas is an important organ in crustaceans that combines the various functions from the liver and pancreas in vertebrates [12]. It also relates the functioning of nutritional metabolism, energy storage, immune response, and other life activities in crustaceans [13,14,15]. For immune functioning, studies have demonstrated that the hepatopancreas synthesizes and excretes key immune-related molecules, such as beta-1, 3-glucan binding protein (LGBP) [16], antibacterial peptide (AMP) [17], and lectin related proteins [18], and emphasized the crucial roles in innate immune response in several crustaceans.
P. gregaria, as a member of the Rhizocephalan parasitic barnacle family, was originally classified as Sacculina polygenea [19], but was later reclassified in a new genus of Polyascus [20]. These parasites are regarded as the most specialized and divergent forms of barnacles, mainly infecting decapods, especially crab species, worldwide [21,22]. Key differences for rhizocephalans compared to other crustaceans include a high alteration of morphology, with a lack of segmentation and appendages in the adult stage. As with other barnacles, however, rhizocephalans show free-living nauplius and cypris larvae during reproduction, related to their endoparasitic lifestyles [23]. For example, Sacculinidae is one of the two larger families of Rhizocephalan, which causes host feminization [24], anecdysis, castration [18], gonadal atrophy [25], and immune response [26]. Moreover, the infection of this parasite in the Chinese mitten crab may result in the host death; the mortality of male crabs reached 13.04% and female crabs 9.09% in this research (total data not shown). For reproduction, the female host is forced to bear the parasitic barnacle eggs instead of its own, while in the male host, spermatogenesis is arrested and the gonadal tissue is degenerated, but does not disappear entirely [27]. Furthermore, changes in behavioral and external morphology, such as widening of the abdomen in parasitized males, cause the male host crabs to take on a feminine appearance [28]. For adult female barnacles, the body is made up of internal and external parts. The internal parts have hollow rootlets structures; these rootlets spread within the body of the host, including the ventral ganglionic mass, gonad, muscle, hemolymph, and hepatopancreas [27,29], where they absorb nutrients from the host [23]. The external portion of the barnacle plays a temporarily reproductive role and is critically responsible for infection in the host. The external parts generate nauplius larvae, which develop into a cypris larvae after several molts to start a new round infection.
Recently, several studies were published on Sacculinidae ecology [30,31], the histopathology and immune response in Carcinus maenas [22], the effects on the muscular system in Hemigrapsus sanguineus [5], and the molecular mechanisms of infection in Scylla olivacea [32]. Studies have specifically addressed the disease ecology of P. gregaria [33], its phylogenetic relationships [19], as well as the description of its morphological adaptation to parasitism in Hemigrapsus sanguineus [34]. As an economical crustacean, the diseases and immunity of the Chinese mitten crab are particularly important to aquaculturists. Due to the lack of an acquired immune system, the innate immune system is the first line of inducible host defense in invertebrates against bacterial, fungal, and viral pathogens [35]. This defense system includes phagocytosis, melanization, nodulation, encapsulation, and the release of relative bactericidal substances, and is essential for the survival and perpetuation of all multicellular organisms [36]. In this area, many studies have demonstrated the functional genes, crucial proteins, osmoregulation modes, antioxidant capacity, and innate immune pathways involved in various diseases in the Chinese mitten crab [14,15,37]. In addition, studies examining the protein levels of the innate immune response could provide important insights into this field of research. One such method is the use of isobaric tandem mass tag (TMT) mass-spectrometry. This technology has allowed researchers to illustrate the molecular mechanisms of inflammatory responses, infections, and temperature responses [11,38,39].
In this study, we examined the innate immune response of the male Chinese mitten crab to P. gregaria infection through TMT analysis. This proteomic analysis provides a deep insight into the relationship and interaction between the Chinese mitten crab and P. gregaria. We identified hundreds of new relevant proteins and assessed their biological importance through enrichments of gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to discriminate the immunological mechanisms involved in the response to P. gregaria infection. These results provide a novel, deep, and comprehensive view of the innate immune response in the Chinese mitten crab, especially for macroparasites, which provide an academic reference for preventing the infection by P. gregaria, to reduce the economic losses in agricultural farm. These molecular mechanisms also contribute toward pharmacological research and development, regarding new medicine against parasite infection.
2. Materials and Methods
2.1. Sample Site and Crabs
Male Chinese mitten crabs were obtained from the Yangtze Estuary (31°10′59.06″ N–121°53′40.56″ E), Shanghai, China, in December 2020, during the spawning migration. Animals were collected from gill nets (5 mm mesh), set perpendicular to the water flow after 2–3 h in the water. Crabs were quickly stunned on ice as soon as they were caught and immediately taken to the lab. The biological information of each crab was measured and the abdomen was scrutinized to separate animals parasitized with P. gregaria from those without the parasites. Slight exfoliation was necessary to collect the parasites and to note the number. Next, 4–5 g of hepatopancreas tissue was rapidly extracted from both parasitized and non-parasitized crabs, and they were flash frozen in liquid nitrogen. All of the extracted samples were stored at −80 °C to ensure effectiveness of the tissue. The parasites collected from each host were identified based on their morphological characteristics [40]. Crabs were considered healthy (i.e., non-parasitized) if no parasite or scar was found in the abdomen, appendages, or copulatory organ, while those with at least 15 parasites evident in the abdomen were considered of the parasitized crabs (Figure 1). Six non-parasitized (mean ± SD, case length: 56.67 ± 0.98 mm) and six parasitized (mean ± SD, case length: 53.19 ± 7.02 mm; parasite count: 21 ± 4) (Table 1) were chosen to analyze the innate immune response in this present study.
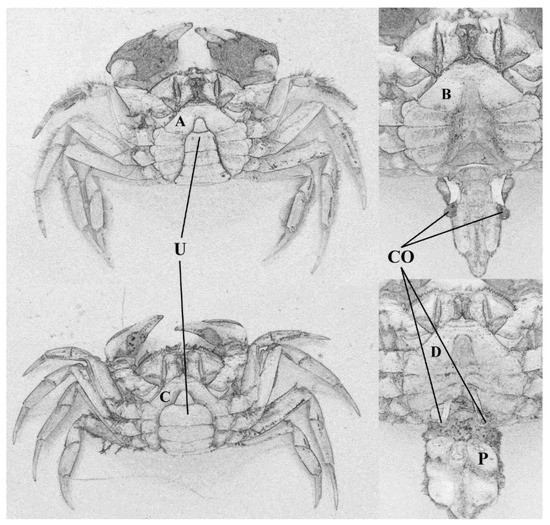
Figure 1.
Features in the male Chinese mitten crab infected with P. gregaria. (A,B): non-parasitized crab; (C,D): parasitized crab. U: umbilicus; CO: copulatory organ; P: P. gregaria.

Table 1.
The basic information of Chinese mitten crab samples.
2.2. Experimental Protein Preparation
A single-to differential (STD) buffer was added to hepatopancreas tissue and then transferred into 2 mL tubes with quartz sand (1:1). An MP Fastprep-24 Automated Homogenizer was used to homogenize the lysate in 2 cycles, 6.0 M/S for 30 s. The homogenate was sonicated and then boiled for 15 min, followed by centrifugation at 14,000 g for 40 min. The supernatant was filtered through a 0.22 µm filter and quantified with the BCA Protein Assay Kit (P0012, Beyotime) before being stored at −20 °C. To separate proteins, a 20 µg of 6X loading buffer was mixed in each sample and boiled for 5 min. The concentrations of proteins were detected (Table S1) and the proteins were separated on 12.5% SDS-PAGE gel (Figure S1) and visualized by Coomassie Blue R-250 staining.
A total of 200 μg of proteins was taken for each sample and combined with 30 μL of STD buffer (4% sodium dodecyl sulfate (SDS), 100 mM dithiothreitol (DTT), and 150 mM Tris-HCl pH 8.0). The detergent, DTT, and other low-molecular-weight components were removed using UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.5) by repeated ultrafiltration (Sartorius (Göttingen, Germany) 30 kD). Then, 100 μL of iodoacetamide (100 mM IAA in UA buffer) was added to block reduced cysteine residues and the samples were incubated for 30 min in darkness. The filters were washed three times with 100 μL of UA buffer and then twice with 100 μL of 0.1 M triethylammonium bicarbonate (TEAB) buffer. Finally, the protein suspensions were digested with 4 μg of trypsin (Promega, Madison, WI, USA) in 40 μL 0.1 M TEAB buffer overnight at 37 °C. The resulting peptides were collected as a filtrate. The peptide content (Figure S2) was estimated by UV light spectral density at 280 nm that was calculated based on the frequency of tryptophan and tyrosine in vertebrate proteins.
2.3. TMT Protein Labelling and HPLC Fractionation
The 100 μg peptide mixture of each sample was labeled using TMT reagent, according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Each TMT labeled mixture was then fractionated by RP chromatography using the Agilent 1260 infinity II HPLC. Next, the mixture was diluted with buffer A (10 mM HCOONH4, 5% ACN, pH 10.0) and loaded onto an XBridge Peptide BEH C18 Column, 130 Å, 5 µm, 4.6 mm X 100 mm column. The peptides were eluted at a flow rate of 1 mL/min with a gradient of 0–7% buffer B (10 mM HCOONH4, 85% ACN, pH 10.0) for 5 min, 7–40% buffer B for 5–40 min, 40–100% buffer B for 45–50 min, and 100% buffer B for 50–65 min. The elution was monitored at 214 nm based on the UV light trace, and fractions were collected every 1 min between 5 and 50 min.
2.4. LC-MS/MS Analysis
Each fraction was injected for nanoLC-MS/MS analysis. The peptide mixture was loaded onto the C18-reversed phase analytical column (Thermo Fisher Scientific, Acclaim PepMap RSLC 50 um × 15 cm, nano viper, P/N164943) in buffer A (0.1% Formic acid), and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% Formic acid) at a flow rate of 300 nl/min. The gradient consisted of 6% buffer B for 3 min, 6–28% buffer B for 42 min, 28–38% buffer B for 5 min, 38–100% buffer B for 5 min, and 100% buffer B for 5 min. The peptides were analyzed by a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) that was coupled to Easy nLC (Thermo Fisher Scientific) for 90 min. The mass spectrometer was operated in positive ion mode. MS data were acquired using a data-dependent top10 method, dynamically choosing the most abundant precursor ions from the survey scan (350–1800 m/z) for high-energy collisional dissociation (HCD) fragmentation. Survey scans were acquired at a resolution of 70,000 at m/z 200 with an automatic generation control (AGC) target of 3e6 and a maxIT of 50 ms. MS2 scans were acquired at a resolution of 17,500 for HCD spectra at m/z 200 with an AGC target of 2 × 105 and a maxIT of 45 ms, and isolation width was 2 m/z. Only ions with a charge state between 2 and 6, and a minimum intensity of 2 × 103 were selected for fragmentation. Dynamic exclusion for selected ions was 30 s. Normalized collision energy was 30 eV.
2.5. Data Analysis
MS/MS raw files were processed using MASCOT engine (Matrix Science, London, UK; version 2.6), analyzed in Proteome Discoverer 2.2 (Thermo Fisher Scientific), and searched against the UniProt database. The search parameters included trypsin as the enzyme used to generate peptides with a maximum of 2 missed cleavages permitted. A precursor mass tolerance of 10 ppm was specified and 0.05 Da tolerance for MS2 fragments. Except for TMT labels, carbamidomethyl(C) was set as a fixed modification. Variable modifications were oxidation (M) and acetyl (protein N-term). A peptide and protein false discovery rate (FDR) of 1% was enforced using a reverse database search strategy. Proteins with fold change >1.2 and p-value (Student’s t test) < 0.05 were considered to be differentially expressed proteins.
2.6. Enrichment of Pathways Analysis
All protein sequences were aligned to protein database that were assembled (Trinity, V2.4.0) and predicted (TransDecoder, V3.0.1) from the coding sequence of transcriptome, only the sequences in the top 10 and E-value ≤ 0.001 were kept. The GO term of the sequence with the top bit-score by Blast2GO was selected. Then, the annotations from GO terms to proteins was completed by the Blast2GO Command Line. After the basic annotation, InterProScan/GO (http://www.ebi.ac.uk/interpro/ (accessed on 5 October 2021)) was used to search the EBI database by motif and then add the functional information of the motif to proteins to improve annotation. Fisher’s exact test was used to enrich GO terms and KEGG pathways by comparing the number of differentially expressed proteins and total proteins correlated to GO terms. Correction for multiple hypothesis testing was carried out using standard false discovery rate (FDR) control methods.
2.7. Enrichment of Protein Domain Analysis
InterProScan (http://www.ebi.ac.uk/interpro (accessed on 1 November 2021)) was used to predict the protein families, domains, and special sites based on the protein sequence alignment method. The database was blasted and the two-tailed Fisher’s exact test was employed, aiming to test the enrichment of the differentially expressed proteins against all identified proteins. A correction for multiple hypothesis testing was performed using the FDR, and the p-values < 0.05 were considered as a significant domain.
3. Results
3.1. Identification and Quantitative Protein Profiling
The parasites were identified in six Chinese mitten crabs based on morphological characters, such as size, profile, parasitic location, color, and capture site in this study. The proteomic analysis was successfully performed on hepatopancreas tissue from six parasitized and six non-parasitized male crabs based on the TMT method, along with simultaneous identification and quantification. In total, 10,616 unique peptides and 2143 proteins were identified, of which 2046 proteins showed quantitative information. Using a 1.2-fold increase or decrease as a benchmark, 598 proteins showed differential expression between parasitized and non-parasitized crabs. Of these, 352 were upregulated and 246 were downregulated in response to P. gregaria infection. Among the upregulated proteins, several kinds of proteins related to the innate immune system were identified, such as cathepsin F and GTPase KRas, while immune system related proteins in the downregulated set included: serpin B, Autophagy-related 4 (ATG4), ATG5, ATG9, scavenger receptor class B, cathepsin D and L, and lectin (Table 2).

Table 2.
Representative differentially expressed proteins related to the innate immune system in the Chinese mitten crab, with a 1.2-fold change, with P. gregaria infection.
3.2. GO Enrichment
The analysis of GO terms that were significantly enriched in differentially expressed proteins showed a total of 104 GO terms (Table S2). Cellular component terms were especially enriched for the lysosomal lumen, plasma membrane, smooth endoplasmic reticulum, and azurophil granule membrane. Molecular function GO terms were enriched for carbohydrate binding, guanosine triphosphate (GTP) binding, GTPase activity, and others. Biological process terms were mainly enriched for antigen processing and presentation, hemocyte migration, nuclear-transcribed mRNA catabolic process, nonsense-mediated decay, and regulation of filopodium assembly (Figure 2). Of the enriched GO terms, 31 were related to immune response, including autophagosome assembly, immune effector process, astrocyte activation involved in immune response, innate immune response, and adaptive immune response (Table 3).
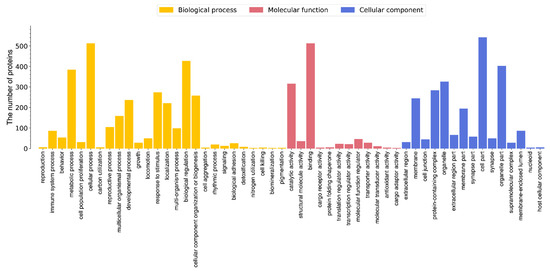
Figure 2.
Enrichment of GO terms for DEPs in the Chinese mitten crab infected with P. gregaria.

Table 3.
Enrichment of GO terms related to innate immune response in the Chinese mitten crab infected with P. gregaria.
3.3. KEGG Analysis with DEPs
In the KEGG pathway analysis, a total of 13 pathways were significantly (p < 0.05) enriched in the set of differentially expressed proteins (Figure 3, Table 4). These pathways included lysosome, protein processing in endoplasmic reticulum, extracellular matrix (ECM)_receptor interaction, other glycan degradation, proteoglycans in cancer, glycosaminoglycan degradation, focal adhesion, antigen processing and presentation, autophagy animal, endocytosis, nucleotide excision repair, and ribosome.
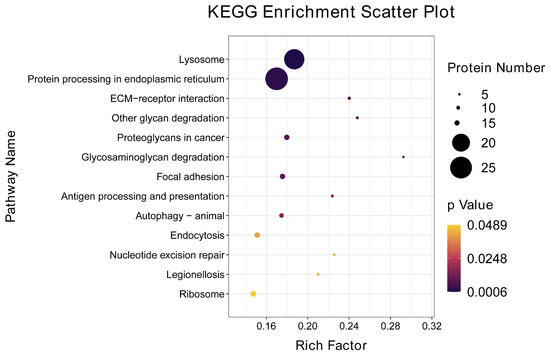
Figure 3.
Significant enrichment of KEGG pathways (p < 0.05) for DEPs in the Chinese mitten crab infected with P. gregaria.

Table 4.
Significant enrichment of KEGG pathways (p < 0.05) in the Chinese mitten crab infected with P. gregaria.
3.4. Enrichment of Protein Domain
Proteins were clustered (Figure S3) and the protein domains were further analyzed for enrichment in order to better understanding the functional aspects of the DEPs. We found significant enrichment of 32 domain categories (Table 5), including the small GTP-binding protein domain, glycoside hydrolase superfamily, glycosyl hydrolase, ferritin-like, and ferritin.

Table 5.
Significant enrichment of protein domains (p < 0.05) in the Chinese mitten crab infected with P. gregaria.
3.5. Subcellular Location of DEPs
Obtaining information about the subcellular location is an important and helpful step towards understanding the mechanism and function of proteins. In the present study, location in the cytosol was enriched with 33.9% of DEPs (n = 203) located there. The second most enriched subcellular location was the mitochondria (n = 110, 18.4% of all DEPs). The least enriched location was the peroxisome, with two DEPs (Figure 4).
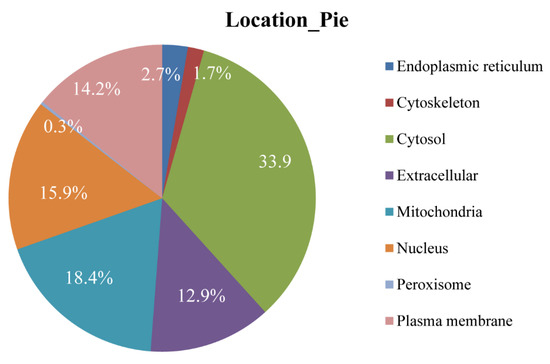
Figure 4.
The subcellular location of DEPs identified in the Chinese mitten crab infected with P. gregaria.
4. Discussion
The Chinese mitten crab, as an economically important crab, has become increasingly popular in the freshwater aquaculture industry; it is now widely cultured in the provinces of Jiangsu, Anhui, Hubei, and Liaoning in China [41]. Several types of hepatopancreatic diseases from bacteria, viruses, and parasites were reported on in recent years, but little attention has been devoted to the innate immune response to macroparasites in this crab species. Here, we focus on the importance of enriched GO terms, the KEGG pathway, and functional protein domains to reveal substantial insights into the innate immune response of this host–parasite system. Importantly, we found that these components, including GTPase KRas, complement 1q-binding protein (C1QBP), serpin, ATG5, lysosomal lumen term, autophagosome assembly terms, antigen processing, the presentation pathway, and the autophagy animal pathway.
Autophagy is an intracellular degradation system that plays an important role in maintaining cellular homeostasis, and is evolutionarily conserved from yeast to mammals [42]. This system is activated in response to environmental signals, from starvation, disease, and pathogen infection [43]. The targets for degradation are not only proteins, but also organelles and other cellular components. In recent years, the relationship between autophagy and disease has been explored in infections, neurodegenerative diseases, and cancers [44]. In the present study, Ras-associated binding (Rab), ATG, cathepsin, urinary gonadotropin peptide (UGP), (protein kinase D) PKD, and Psen1 were differentially expressed across parasitized and non-parasitized samples, and all have GO terms or KEGG pathways associated with autophagy. During autophagy, the autophagosomes surround the cytosolic components and then fuse with a vacuole, leading to the degradation of the target by lysosomal hydrolases. Autophagy-related (ATG) proteins play a crucial role in the regulation of this process [45]. Prior research showed that the gathering of ATG proteins to form the pre-autophagosomal structure (PAS) is the first step of autophagy [46,47], where autophagosomes are normally generated. In mammals, the overexpression of ATG4B was found to make the LC3-PE complex quickly rupture and form the stable complex of LC3, which shows it is a suppressive effector in autophagy [45]. In grouper cells, the transcriptional level of ATG5 was upregulated after infection with Singapore grouper iridovirus (SGIV) and red-spotted grouper nervous necrosis virus (RGNNV). However, the overexpression of ATG5 simultaneously decreased the expression of interferon and negatively regulated the expression of pro-inflammatory factors [48]. ATG9 has been described as a positive regulator that modulates the number of autophagosomes [49]. In a previous study on the Chinese mitten crab, the transcriptional levels of Atg12, Atg13, and Atg16L were upregulated in crabs with hepatopancreatic necrosis disease [41]. These results indicate that the upregulation of ATG family members has a positive effect on the immune response in different species. On the contrary, all ATG4 (0.78-fold), ATG5 (0.54-fold), and ATG9 (0.66-fold) proteins were significantly downregulated in crabs with P. gregaria infection in the present study. This suggests that autophagy is suppressed in the Chinese mitten crab infected by P. gregaria, and that the expression of ATG proteins is involved in this biology process, as part of the host innate immune response. Taken together, this indicates that ATG may play a suppressive role of autophagosomes generation in the autophagy process response to P. gregaria infection through diverse mechanisms in the Chinese mitten crab. These mechanisms can differ across species from different pathogens, such as the ATG family, performing differential regulations of the formation of autophagosomes and, therefore, of the autophagy process. One link that is likely very important to this process is the recognition of exogenous ligands. The scavenger receptor (SR), one of the sub-families of pattern recognition receptors (PRRs), recognizes the modified lipoproteins and danger-associated molecular patterns (DAMPs) [50]. One study showed that an increase in the expression of SRs induced by Vibrio parahaemolyticus, lipopolysaccharide (LPS), and white spot syndrome virus (WSSV) efficiently enhanced host phagocytosis to clearance bacteria [51]. Here, we found that SR protein was significantly decreased in response to P. gregaria infection in the Chinese mitten crab, likely relating its role in phagocytosis as well as the innate immune system. Interestingly, this pattern is the opposite of what was previously found in a study on Spiroplasma eriocheiris infection in the Chinese mitten crab [52]. This promotes our understanding of P. gregaria, whose infection probably silences the innate immune system through inhibition of cell recognition and autophagosome generation on the autophagy process, depending on several modulators (i.e., ATG and SR) in the Chinese mitten crab.
Lysosomes are acidic and hydrolytic organelles responsible for generating targets during endocytosis, phagocytosis, and autophagy [53]. Lysosomes receive or degrade their substrates via various pathways, including endocytosis, phagocytosis, autophagy, lysosomal proteins, soluble lysosomal hydrolases, and others [54]. Lysosome mobilization is a crucial process for phagocyte migration and bactericidal function, although the molecular mechanisms linking these processes remain unclear. Moreover, lysosomes and related organelles travel over long distances along microtubules within the cell cytoplasm during phagocytosis [55]. For lysosomes and endosomes, the active site is mostly a cysteine thiol or an aspartic acid, which functions as the key catalytic site. Some serine proteases, such as cathepsin, granzymes, and a thymus specific serine protease (TSSP), play important roles in the immune system [56]. In this study, 23 DEPs had lysosome-related lysosome GO terms and/or KEGG pathways. These included cathepsin, lysosomal alpha-glucosidase, hexosaminidase, CD63 antigen, and β-mannosidase. The best known lysosomes and cathepsins are involved in a number of important biological processes, such as intracellular protein turnover, immune response, hormone activation, remodeling of extracellular matrix (ECM), and apoptosis [57,58]. In invertebrates, the signaling pathways of MAPK and Imd are the primary components of the innate immune system, and the MAPK pathway has been shown to mediate cathepsin expression induced by all types of cells [59,60]. Moreover, JNK, ERK, p38, and Relish are regulators in these signaling pathways [61,62]. In the Chinese mitten crab, previous research has found that all of the expression key factors were decreased when cathepsin D was silenced [63]. Furthermore, using RNAi silenced cathepsin D expression caused an obvious decrease in crab immunity and resulted in a significant increase in the mortality of crabs [63]. In mice, the null expression of cathepsin D led to death shortly after birth [64]. Similarly, the expression of cathepsin L was found to distinctly increase following V. anguillarum infection in the Chinese mitten crab [65]. In addition, the over-expression of cathepsin L was homoplastically induced in black tiger shrimp (Penaeus monodon) [66] and Pacific white shrimp (Litopenaeus vannamei) [67] by lipopolysaccharide and WSSV infection, respectively. On the other hand, in this study, we found that both cathepsin D (0.68-fold) and L (0.65-fold) were dramatically inhibited in parasitized crabs, which suggests that both cathepsin D and L perform a crucial role in innate immune function in the Chinese mitten crab. Another cathepsin family member, cathepsin F, was detected as an upregulated (1.40-fold) motif in our study. This protein likely has a similar role to cathepsin S, which cleaves Ii to cross-linking and immunoprecipitation (CLIP) during major histocompatibility complex II (MHC II) Ag processing and presentation [68]. While in crabs, further analysis is needed for this protein function in lysosome-related biology.
The activities of cathepsin, such as cystatins, stefins, tyropins, and serpins, are endogenous protein inhibitors and they tightly bind their target enzymes to prevent substrate hydrolysis [69]. Within the proteinase inhibitor superfamily, serpins are the largest and most diverse family of protease inhibitors [70], and play important roles in many immune processes, such as blood coagulation, complement activation, melanization, and phagocytosis [71]. In recent years, research on serpins in invertebrate indicated that serpins regulate the prophenoloxidase (proPO) activity in Drosophila [72], Penaeus monodon [73], and the Chinese mitten crab [74]. In invertebrates, serpins appear to be unique components of the innate immune response, and are regulated by prophenoloxidase activating enzymes (PPAEs), proteinase inhibitors, lipopolysaccharide, LGBP, and hemolin [75]. A prior study of a Chinese mitten crab infected with Vibrio anguillarum and Pichia pastoris showed that serpins were upregulated, which could be related to serine proteinase involvement in wound healing, proPO activation, phagocytosis, and other defense responses after bacterial and fungal challenges. In Hyphantria cunea, the recombinant serpin and the serine protease inhibitor aprotinin were used to investigate the relationship between serpin and PO activity, the results of which showed that the aprotinin has a stronger inhibitory activity than the recombinant protein at the same concentration, and that the increased serpin expression inhibited PO activity through competition with proPO against the target protease (PPAE) [76]. In the present study, we found a different expression pattern, in which serpin expression was significantly decreased (0.49-fold) after P. gregaria infection, which could affect the proPO system of the innate immune response in the Chinese mitten crab. It should be noted that the decrease in serpin expression does not indicate a positive effect on the proPO of the innate immune system, but the abnormal alteration showed an important role in the innate immune response to P. gregaria infection. In other words, it is possible that the differential expression of serpin mediates the autophagy process, together with ATG proteins, through the MAPK and/or IMD signaling pathways, also involved in the lysosome and autophagy-animal pathways, serving a direct role in the innate immune response to P. gregaria infection in the Chinese mitten crab.
Biology processes are inarguably complicated, polytropic, and are involved in several molecular factors. Interestingly in the present study, most DEPs relating the innate immune response were downregulated after P. gregaria infection, which suggests that P. gregaria may induce a suppressive effect on the Chinese mitten crab’s innate immune system during infection. As a hypothesis, on the one hand, this consequence may result from the long-term parasitizing [22] of P. gregaria and the newborn cyprid growing in the Chinese mitten crab. On the other hand, this may be due to the co-evolution of the host and parasite, which has resulted in reductions in resistance over time [77]. It is easily understood that the lifestyle of P. gregaria continually grows for several months [27,30], from development of the external parts to wither off, upon which, the adults release several broods of larvae [78] during parasitization. It is likely difficult to clean or kill the parasites by themselves of the host, which is beneficial for long-term parasitizing and circumventing the crab’s attention of the innate immune system [22]. Here, we identified many proteins, domains, GO terms, and KEGG pathways, which were significantly changed in response to P. gregaria infection in the Chinese mitten crab at the protein level, and screened for the crucial component relations to the innate immune system, such as ATG, cathepsin, serpins, lysosome- and autophagy-related GO terms, and KEGG pathways. Further study was performed to discuss the mechanisms of ATG, cathepsin, and serpins, which showed a response to P. gregaria infection through the autophagy process, with lysosomes participation in the innate immune response in the Chinese mitten crab. Other proteins, such as the Rab family [79], GTPase KRas [80], lectin [81], CD63 [82], and C1QBP [83] were also identified, which were shown to have immune functions in many organisms, and were significantly differentially expressed after P. gregaria infection. In this area, to better disentangle these complex signals, more research is needed on the immune response to macroparasites in crabs.
5. Conclusions
P. gregaria is a specialized crustacean parasite and draws more attention in recent years. Up to date, few researches were reported for the interactions between this parasite and its host, especially in Chinese mitten crab. Thereby, we first used TMT method to research the innate immune response against P. gregaria infection in Chinese mitten crabs. In the present study, many DEPs were identified after P. gregaria infection, and protein domain, subcellular location and Go enrichment were used to analyze protein functions. Moreover, the KEGG pathways were also analyzed to research the mechanisms responding P. gregaria infection in Chinese mitten crab. Finally, we identified DEPs such as Atgs, cathepsins, serpins, GTPase KRas, and lectin, which were mostly enriched in autophagosome assembly, innate immune response GO terms and lysosome, autophagy animal pathways to against the parasite infection through Autophagy process. The innate immune system of Chinese mitten crab was silenced after long-term parasitizing of P. gregaria. These results provided a novel understanding of the innate immune response against P. gregaria infection in crabs, as well as other crustaceans. Simultaneously, provided a basis to research the innate immune response of Chinese mitten crab and prevent the parasite infection in aquaculture industry.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/fishes6040057/s1, Figure S1: The quality control by SDS-PAGE. Figure S2: The molecular weight distribution for identified proteins. Figure S3: The heatmapimage analysis between parasitized and non-parasitized crabs. Table S1: The concentration of proteins detected in Chinese mitten crab hepatopancreas. Table S2: Significant enrichment of GO terms (p < 0.05) in Chinese mitten crab infected with P. gregaria.
Author Contributions
Conceptualization, Y.Y., K.L. and F.Z.; methodology, Y.Y., F.M., Y.Z., K.L. and F.Z.; software, Y.Y. and P.R.; validation, K.L. and F.Z.; formal analysis, Y.Y.; investigation, Y.Y., J.D., L.L. and P.R.; resources, Y.Y., F.M., J.D., L.L. and Y.W. (Yatao Wu); data curation, Y.Y., F.M. and Y.Z.; writing—original draft preparation, Y.Y.; writing—review and editing, Y.Y.; visualization, Y.Y.; supervision, K.L. and F.Z.; project administration, Y.Y., Y.W. (Yinping Wang), K.L.; funding acquisition, K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2019YFD0901203); investigation of fishery resources and environment in the lower Yangtze River, the Ministry of Agriculture and Rural Affairs of China (CJDC-2017-22); investigation of fishery resources of the Yangtze Estuary, Special Financial Funds of Shanghai Government (JC202004).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Animal Care and Use Committee of the Freshwater Fisheries Research Center at the Chinese Academy of Fishery Sciences. The analysis was carried out following the Guidelines for the Care and Use of Laboratory Animals set by the Animal Care and Use Committee of the Freshwater Fisheries Research Center (2003WXEP61). All operations were carried out with field permit no. 20181AC1128.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, M.; Ge, J.; Yu, J.; Su, S.; Li, J.; Tang, Y. Molecular insights into information processing and developmental and immune regulation of Eriocheir sinensis megalopa under hyposaline stress. Genomics 2020, 112, 4647–4656. [Google Scholar] [CrossRef] [PubMed]
- Gillard, M.; Thiébaut, G.; Deleu, C.; Leroy, B. Present and future distribution of three aquatic plants taxa across the world: Decrease in native and increase in invasive ranges. Biol. Invasions 2017, 19, 2159–2170. [Google Scholar] [CrossRef]
- Qiu, G.; Xiong, L.; Liu, Z.; Yan, Y.; Shen, H. A first generation microsatellite-based linkage map of the Chinese mitten crab Eriocheir sinensis and its application in quantitative trait loci (QTL) detection. Aquaculture 2016, 451, 223–231. [Google Scholar] [CrossRef]
- Bonami, J.; Zhang, S. Viral diseases in commercially exploited crabs: A review. J. Invertebr. Pathol. 2011, 106, 6–17. [Google Scholar] [CrossRef]
- Miroliubov, A.; Borisenko, I.; Nesterenko, M.; Korn, O.; Lianguzova, A.; Ilyutkin, S.; Lapshin, N.; Dobrovolskij, A.A. Muscular system in the interna of Polyascus polygenea and Sacculina pilosella (Cirripedia: Rhizocephala: Sacculinidae). Invertebr. Zool. 2019, 16, 48–56. [Google Scholar] [CrossRef]
- Yuan, Q.; Wang, Q.; Zhang, T.; Li, Z.; Liu, J. Effects of water temperature on growth, feeding and molting of juvenile Chinese mitten crab Eriocheir sinensis. Aquaculture 2017, 468, 169–174. [Google Scholar] [CrossRef]
- Veillet, A. Recherches Sur le Parasitisme des Crabes et des Galathées par les Khizocéphales et les Épicarides; Annales de l’Institut Oceanographique Monaco: Monte-Carlo, Monaco, 1945; Volume 22, pp. 193–341. [Google Scholar]
- Wang, J.; Yang, B.; Wang, W.; Song, X.; Jiang, Q.; Qiu, L.; Wang, L.; Song, L. The enhanced immune protection in Chinese mitten crab Eriocheir sinensis against the second exposure to bacteria Aeromonas hydrophila. Front. Immunol. 2019, 10, 2041. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Bao, J.; Xing, Y.; Feng, C.; Li, X.; Chen, Q. Proteomic analysis of the hemolymph after Metschnikowia bicuspidata infection in the Chinese mitten crab Eriocheir sinensis. Front. Immunol. 2021, 12, 659–723. [Google Scholar]
- Wang, W.; Chen, J. Ultrastructural study on a novel microsporidian, Endoreticulatus eriocheir sp. nov. (Microsporidia, Encephalitozoonidae), parasite of Chinese mitten crab, Eriocheir sinensis (Crustacea, Decapoda). J. Invertebr. Pathol. 2007, 94, 77–83. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Zhou, J.; Zou, J.; Fan, L. New insights into the immune regulation and tissue repair of Litopenaeus vannamei during temperature fluctuation using TMT-based proteomics. Fish Shellfish Immunol. 2020, 106, 975–981. [Google Scholar] [CrossRef]
- Sousa, L.G.; Cuartas, E.I.; Petriella, A.M. Fine structural analysis of the epithelial cells in the hepatopancreas of Palaemonetes argentinus (Crustacea, Decapoda, Caridea) in intermoult. Biocell 2005, 29, 25. [Google Scholar] [CrossRef]
- Gao, T.; Xu, Y.; Wang, K.; Deng, Y.; Yang, Y.; Lu, Q.; Pan, J.; Xu, Z. Comparative LC-MS based non-targeted metabolite profiling of the Chinese mitten crab Eriocheir sinensis suffering from hepatopancreatic necrosis disease (HPND). Aquaculture 2018, 491, 338–345. [Google Scholar] [CrossRef]
- Li, X.; Cui, Z.; Liu, Y.; Song, C.; Shi, G. Transcriptome analysis and discovery of genes involved in immune pathways from hepatopancreas of microbial challenged mitten crab Eriocheir sinensis. PLoS ONE 2013, 8, e68233. [Google Scholar] [CrossRef]
- Li, S.; Jia, Z.; Li, X.; Geng, X.; Sun, J. Calmodulin is a stress and immune response gene in Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2014, 40, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Pain, A.; Klimpel, K.; Dhar, A. The lipopolysaccharide and β-1, 3-glucan binding protein gene is upregulated in white spot virus-infected shrimp (Penaeus stylirostris). J. Virol. 2002, 76, 7140–7149. [Google Scholar] [CrossRef]
- Ried, C.; Wahl, C.; Miethke, T.; Wellnhofer, G.; Landgraf, C.; Schneider-Mergener, J.; Adolf, H. High affinity endotoxin-binding and neutralizing peptides based on the crystal structure of recombinant Limulus anti-lipopolysaccharide factor. J. Biol. Chem. 1996, 271, 28120–28127. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.; Bartlett, T.; Browdy, C.; Chapman, R.; Warr, G. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific White Shrimp, Litopenaeus vannamei, and the Atlantic White Shrimp, L. setiferus. Dev. Comp. Immunol. 2001, 25, 565–577. [Google Scholar] [CrossRef]
- Glenner, H.; Lützen, J.; Takahashi, T. Molecular and morphological evidence for a monophyletic clade of asexually reproducing Rhizocephala: Polyascus, new genus (Cirripedia). J. Crustacean Biol. 2003, 23, 548–557. [Google Scholar] [CrossRef]
- Jorgen, L.; Tohru, T. Sacculina polygenea, a new species of rhizocephalan (Cirripedia: Rhizocephala) from Japan, parasitic on the intertidal crab Hemigrapsus sanguineus (De Haan, 1835) (Decapoda: Brachyura: Grapsidae). Crustacean Res. 1997, 26, 103–108. [Google Scholar]
- Høeg, J.; Lützen, J. Crustacea Rhizocephala; Christiansen, M.E., Ed.; Norwegian University Press: Oslo, Norway, 1985; Volume 6, pp. 1–92. [Google Scholar]
- Rowley, A.; Davies, C.; Malkin, S.; Bryan, C.; Thomas, J.; Batista, F.; Coates, C. Prevalence and histopathology of the parasitic barnacle, Sacculina carcini in shore crabs, Carcinus maenas. J. Invertebr. Pathol. 2020, 171, 107338. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.; Wu, Y.; Tung, T.; Wang, G.; Toullec, J.; Liu, S.; Huang, W.; Lee, C. Metabolic effects of parasitisation by the barnacle Polyascus plana (Cirripedia: Rhizocephala: Sacculinidae) on a grapsid host, Metopograpsus thukuhar. Dis. Aquat. Org. 2016, 119, 199–206. [Google Scholar] [CrossRef]
- Waiho, K.; Fazhan, H.; Glenner, H.; Ikhwanuddin, M. Infestation of parasitic rhizocephalan barnacles Sacculina beauforti (Cirripedia, Rhizocephala) in edible mud crab Scylla Olivacea. PeerJ 2017, 5, e3419. [Google Scholar] [CrossRef]
- Tsuchida, K.; Lützen, J.; Nishida, M. Sympatric three-species infection by Sacculina parasites (Cirripedia: Rhizocephala: Sacculinidae) of an intertidal grapsoid crab. J. Crustacean Biol. 2006, 26, 474–479. [Google Scholar] [CrossRef]
- Mouritsen, K.N.; Jensen, T. The effect of Sacculina carcini infections on the fouling, burying behaviour and condition of the shore crab, Carcinus maenas. Ophelia 2006, 2, 270–275. [Google Scholar]
- Heg, J.T. The biology and life cycle of the Rhizocephala (Cirripedia). J. Mar. Biol. Assoc. 1995, 75, 517–550. [Google Scholar] [CrossRef]
- Kristensen, T.; Nielsen, A.; Jørgensen, A.; Mouritsen, K.; Glenner, H.; Christensen, J.; Lützen, J.; Høeg, J. The selective advantage of host feminization: A case study of the green crab Carcinus maenas and the parasitic barnacle Sacculina carcini. Mar. Biol. 2012, 159, 2015–2023. [Google Scholar] [CrossRef]
- Adam, P.; Rowley, A.F. Tissue changes in the shore crab Carcinus maenas as a result of infection by the parasitic barnacle Sacculina carcini. Dis. Aquat. Org. 2008, 80, 75–79. [Google Scholar]
- Lützen, J. Growth, reproduction, and life span in Sacculina carcini Thompson (Cirripedia: Rhizocephala) in the Isefjord, Denmark. Sarsia 1984, 69, 91–105. [Google Scholar] [CrossRef]
- Trédez, F.; Rabet, N.; Bellec, L.; Audebert, F. Synchronism of naupliar development of Sacculina carcini Thompson, 1836 (Pancrustacea, Rhizocephala) revealed by precise monitoring. Helgol. Mar. Res. 2016, 70, 26. [Google Scholar] [CrossRef]
- Waiho, K.; Fazhan, H.; Zhang, Y.; Afiqah-Aleng, N.; Moh, J.; Ikhwanuddin, M.; Hassan, M.; Norainy, M.; Ma, H. Gonadal transcriptomic analysis of the mud crab Scylla olivacea infected with rhizocephalan parasite Sacculina beauforti. Genomics 2020, 112, 2959–2969. [Google Scholar] [CrossRef]
- Tang, B.; Wang, Q.; Chen, L.; Yang, S. Features of an intersex Chinese mitten crab, Eriocheir Japonica sinensis (decapoda, brachyura). Freshw. Fish 2005, 78, 371–377. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Aratake, H. Morphological modifications caused by Sacculina polygenea in Hemigrapsus sanguineus (De Haan) (Brachyura: Grapsidae). Crustacean Res. 1997, 26, 125–145. [Google Scholar] [CrossRef]
- Iwanaga, S.; Lee, B.-L. Recent advances in the innate immunity of invertebrate animals. BMB Rep. 2005, 38, 128–150. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Lee, B.L.; Söderhäll, K. Cell-mediated immunity in arthropods: Hematopoiesis, coagulation, melanization and opsonization. Immunobiology 2006, 211, 213–236. [Google Scholar] [CrossRef]
- Kong, P.; Wang, L.; Zhang, H.; Zhou, Z.; Qiu, L.; Gai, Y.; Song, L. Two novel secreted ferritins involved in immune defense of Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2010, 28, 604–612. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, A.; Ao, W.; Wang, Z.; Yuan, J.; Song, Q.; Wei, D.; Ye, H. Proteomic analysis of serum proteins from HIV/AIDS patients with Talaromyces marneffei infection by TMT labeling-based quantitative proteomics. Clin. Proteom. 2018, 15, 40. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Jiang, Z. Immune induction identified by TMT proteomics analysis in Fusobacterium nucleatum autoinducer-2 treated macrophages. Expert Rev. Proteom. 2020, 17, 175–185. [Google Scholar] [CrossRef]
- Du, N.S. Crustaceology; Scence Press: Beijing, China, 1987; Volume 1. [Google Scholar]
- Yan, B.; Liu, X.; Zhou, Y.; Zhang, M.; Fang, P.; Jiang, M.; Yuan, R.; Hu, X.; Gao, G.; Xue, R.; et al. Transcriptomic analysis reveals that hepatopancreatic necrosis disease in Eriocheir sinensis (Chinese mitten crabs) may be the result of autophagy and apoptosis. Aquaculture 2020, 515, 734579. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Sun, M.X.; Huang, L.; Wang, R.; Yu, Y.L.; Li, C.; Li, P.P.; Hu, X.C.; Hao, H.P.; Ishag, H.A.; Mao, X. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 2012, 8, 1434–1447. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pedro, J.; Levine, B.; Green, D.; Kroemer, G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017, 16, 487–511. [Google Scholar] [CrossRef]
- Maruyama, T.; Noda, N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2018, 71, 72–78. [Google Scholar] [CrossRef]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Clarendon Press 2007, 12, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, J.; Zhang, X.; Wei, S.; Huang, X.; Huang, Y.; Wei, J.; Qin, Q. Fish Autophagy Protein 5 Exerts Negative Regulation on Antiviral Immune Response against Iridovirus and Nodavirus. Front. Immunol. 2019, 10, 517. [Google Scholar] [CrossRef]
- Jin, M.; Klionsky, D.J. Transcriptional regulation of ATG9 by the Pho23-Rpd3 complex modulates the frequency of autophagosome formation. Autophagy 2014, 10, 1681–1682. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef]
- Luzio, J.P.; Pryor, P.R.; Bright, N.A. Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 2007, 8, 622–632. [Google Scholar] [CrossRef]
- Hou, L.; Zhou, H.; Wan, H.; Liu, Z.; Wang, L.; Cheng, Y.; Wu, X.; Gu, W.; Wang, W.; Meng, Q. TMT-based quantitative proteomic analysis of Eriocheir sinensis hemocytes and thoracic ganglion during Spiroplasma eriocheiris infection. Fish Shellfish Immunol. 2020, 96, 126–137. [Google Scholar] [CrossRef]
- Hipolito, V.E.B.; Ospina-Escobar, E.; Botelho, R.J. Lysosome remodelling and adaptation during phagocyte activation. Cell Microbiol. 2018, 20, e12824. [Google Scholar] [CrossRef]
- Raposo, G.; Fevrier, B.; Stoorvogel, W.; Marks, M. Lysosomes related organelles: A view from immunity and pigmentation. Cell Struct. Funct. 2002, 27, 443–456. [Google Scholar] [CrossRef][Green Version]
- Labrousse, A.M.; Meunier, E.; Record, J.; Labernadie, A.; Beduer, A.; Vieu, C.; Safta, T.; Maridonneau-Parini, I. Frustrated phagocytosis on micro-patterned immune complexes to characterize lysosome movements in live macrophages. Front. Immunol. 2011, 2, 51. [Google Scholar] [CrossRef]
- Colbert, J.; Matthews, S.; Miller, G.; Watts, C. Diverse regulatory roles for lysosomal proteases in the immune response. Eur. J. Immunol. 2009, 39, 2955–2965. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Turk, V. Lysosomal cysteine proteases: More than scavengers. Biochim. Biophys. Acta 2000, 1477, 98–111. [Google Scholar] [CrossRef]
- Turk, B.; Stoka, V.; Rozman-Pungercar, J.; Cirman, T.; Droga-Mazovec, G.; Oreic, K.; Turk, V. Apoptotic Pathways: Involvement of lysosomal proteases. Biol. Chem. 2002, 383, 1035–1044. [Google Scholar] [CrossRef]
- Keegan, P.M.; Wilder, C.L.; Platt, M.O. Tumor necrosis factor alpha stimulates cathepsin K and V activity via juxtacrine monocyte-endothelial cell signaling and JNK activation. Mol. Cell Biochem. 2012, 367, 65–72. [Google Scholar] [CrossRef]
- Tsukuba, T.; Yanagawa, M.; Kadowaki, T.; Takii, R.; Okamoto, Y.; Sakai, E.; Okamoto, K.; Yamaoto, K. Cathepsin E deficiency impairs autophagic proteolysis in macrophages. PLoS ONE 2013, 8, e82415. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, Y.X.; Liu, J.; Yang, C.; Feng, Q.; Xu, W. A regulatory pathway, Ecdysone-transcription factor Relish-Cathepsin L, is involved in insect fat body dissociation. PLoS Genet. 2013, 9, e1003273. [Google Scholar] [CrossRef]
- Ning, M.; Yuan, M.; Liu, M.; Gao, Q.; Wei, P.; Gu, W.; Wang, W.; Meng, Q. Characterization of cathepsin D from Eriocheir sinensis involved in Spiroplasma eriocheiris infection. Dev. Comp. Immunol. 2018, 86, 1–8. [Google Scholar] [CrossRef]
- Tulone, C.; Uchiyama, Y.; Novelli, M.; Grosvenor, N.; Softig, P.; Chain, B. Haematopoietic development and immunological function in the absence of cathepsin D. BMC Immunol. 2007, 8, 22. [Google Scholar] [CrossRef]
- Li, W.; Jin, X.; He, L.; Jiang, H.; Gong, Y.; Xie, Y.; Wang, J. Molecular cloning, characterization, expression and activity analysis of cathepsin L in Chinese mitten crab, Eriocheir sinensis. Fish Shellfish Immunol. 2010, 29, 1010–1018. [Google Scholar] [CrossRef]
- Qiu, L.; Jiang, S.; Huang, J.; Wang, W.; Zhang, D.; Wu, Q.; Yang, K. Molecular cloning and mRNA expression of cathepsin C gene in black tiger shrimp (Penaeus monodon). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2008, 150, 320–325. [Google Scholar] [CrossRef]
- Zhao, Z.; Yin, Z.; Weng, S.; Guan, H.; Li, S.; Xing, K.; Chan, S.; He, J. Profiling of differentially expressed genes in hepatopancreas of white spot syndrome virus-resistant shrimp (Litopenaeus vannamei) by suppression subtractive hybridisation. Fish Shellfish Immunol. 2007, 22, 520–534. [Google Scholar] [CrossRef]
- Shi, G.P.; Bryant, R.A.R.; Riese, R.; Verhelst, S.; Driessen, C.; Li, Z.; Bromme, D.; Ploegh, H.; Chapman, H. Role for Cathepsin F in invariant chain processing and Major Histocompatibility Complex Class II peptide loading by Macrophages. J. Exp. Med. 2000, 191, 1177–1186. [Google Scholar] [CrossRef]
- Turk, B.; Turk, D.; Salvesen, G. Regulating cysteine protease activity: Essential role of protease inhibitors as guardians and regulators. Curr. Pharm. Des. 2002, 8, 1623–1637. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, Y.; Jiang, H.; Xu, S.; Zhao, X.; Wang, J. Antibacterial activity of serine protease inhibitor 1 from kuruma shrimp Marsupenaeus japonicus. Dev. Comp. Immunol. 2014, 44, 261–269. [Google Scholar] [CrossRef]
- Rimphanitchayakit, V.; Tassanakajon, A. Structure and function of invertebrate Kazal-type serine proteinase inhibitors. Dev. Comp. Immunol. 2009, 34, 377–386. [Google Scholar] [CrossRef]
- Tang, H.; Kambris, Z.; Lemaitre, B.; Hashimoto, C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev. Cell 2008, 15, 617–626. [Google Scholar] [CrossRef]
- Wetsaphan, N.; Rimphanitchayakit, V.; Tassanakajon, A.; Somboonwiwat, K. PmSERPIN3 from black tiger shrimp Penaeus monodon is capable of controlling the proPO system. Dev. Comp. Immunol. 2013, 41, 110–119. [Google Scholar] [CrossRef]
- Wang, L.; Ma, Z.; Yang, J.; Gai, Y.; Zhou, Z.; Wang, L.; Yue, F.; Song, L. Identification and characterization of a serine protease inhibitor Esserpin from the Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2013, 34, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Söderhäll, K.; Cerenius, L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 1998, 10, 23–28. [Google Scholar] [CrossRef]
- Park, D.; Shin, S.; Hong, S.; Park, H. Immunological detection of Serpin in the Fall webworm, Hyphantria cunea and its inhibitory activity on the prophenoloxidase system. Mol. Cells 2000, 10, 186–192. [Google Scholar] [CrossRef]
- Koskella, B. Resistance gained, resistance lost: An explanation for host-parasite coexistence. PLoS Biol. 2018, 16, e3000013. [Google Scholar] [CrossRef]
- Jensen, A.; Schneider, M.; Høeg, J.; Glenner, H.; Lützen, J. Variation in juvenile stages and success of male acquisition in Danish and French populations of the parasitic barnacle Sacculina carcini (Cirripedia: Rhizocephala) parasitizing the shore crab Carcinus maenas. Mar. Biol. Res. 2019, 15, 191–203. [Google Scholar] [CrossRef]
- Han, F.; Zhang, X. Characterization of a ras-related nuclear protein (Ran protein) up-regulated in shrimp antiviral immunity. Fish Shellfish Immunol. 2007, 23, 937–944. [Google Scholar] [CrossRef]
- Scheele, J.S.; Marks, R.E.; Boss, G.R. Signaling by small GTPases in the immune system. Immunol. Rev. 2007, 218, 92–101. [Google Scholar] [CrossRef]
- Patin, E.C.; Orr, S.J.; Schaible, U.E. Macrophage Inducible C-Type Lectin as a Multifunctional Player in Immunity. Front. Immunol. 2017, 8, 861. [Google Scholar] [CrossRef]
- Liyanage, D.; Omeka, W.; Yang, H.; Lim, C.; Kwon, H.; Choi, C.; Lee, J. Expression profiling, immune functions, and molecular characteristics of the tetraspanin molecule CD63 from Amphiprion clarkii. Dev. Comp. Immunol. 2021, 123, 104168. [Google Scholar] [CrossRef]
- Wang, Y.; Su, J.; Yuan, B.; Fu, D.; Niu, Y.; Yue, D. The role of C1QBP in CSF-1-dependent PKCζ activation and macrophage migration. Exp. Cell Res. 2018, 362, 11–16. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).