Workflow for the Targeted and Untargeted Detection of Small Metabolites in Fish Skin Mucus
Abstract
1. Introduction
2. Results and Discussion
2.1. Sample Preparation for Targeted Liquid Chromatography–High-Resolution Mass Spectrometry Analysis of Fish Skin Mucus Metabolites
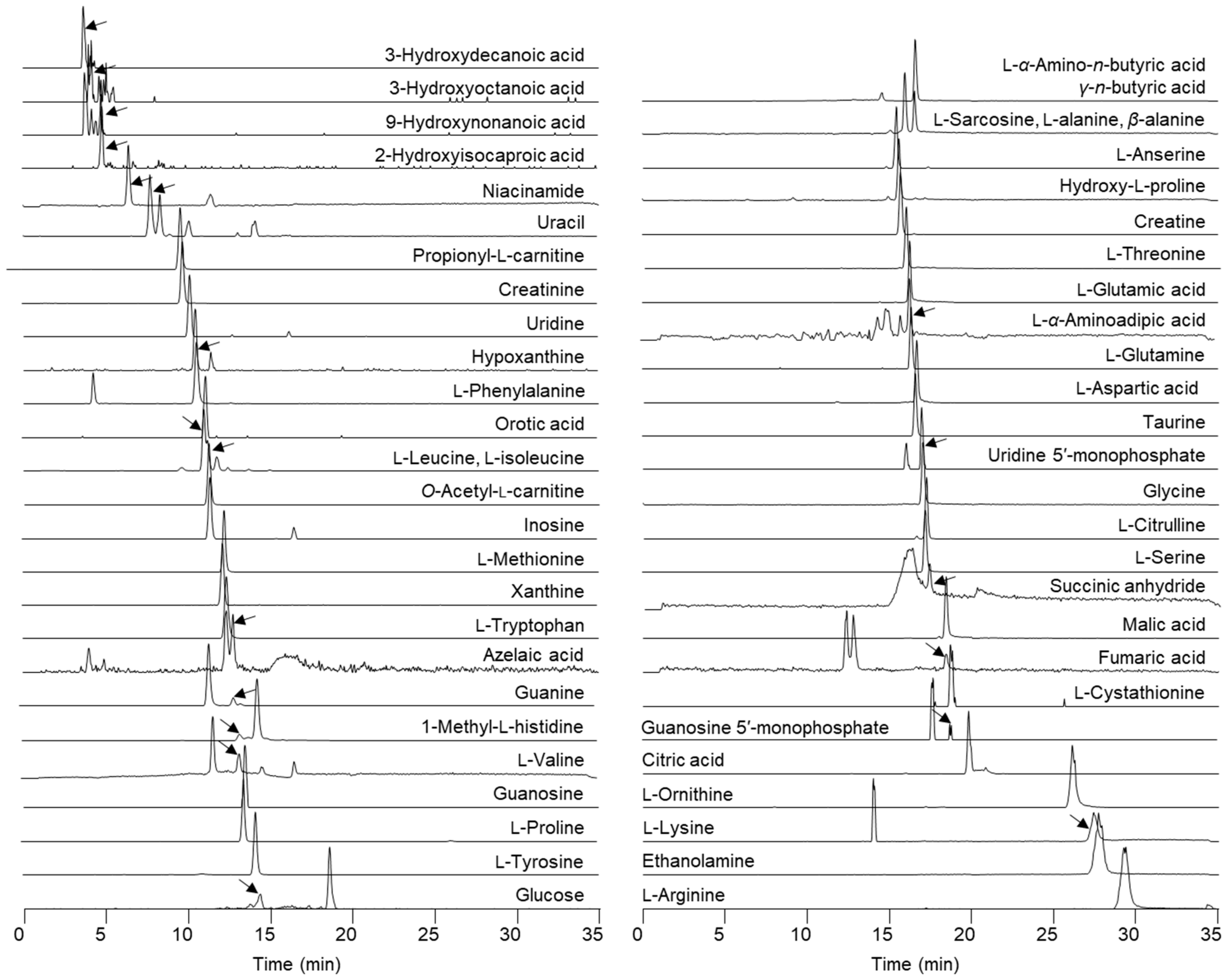
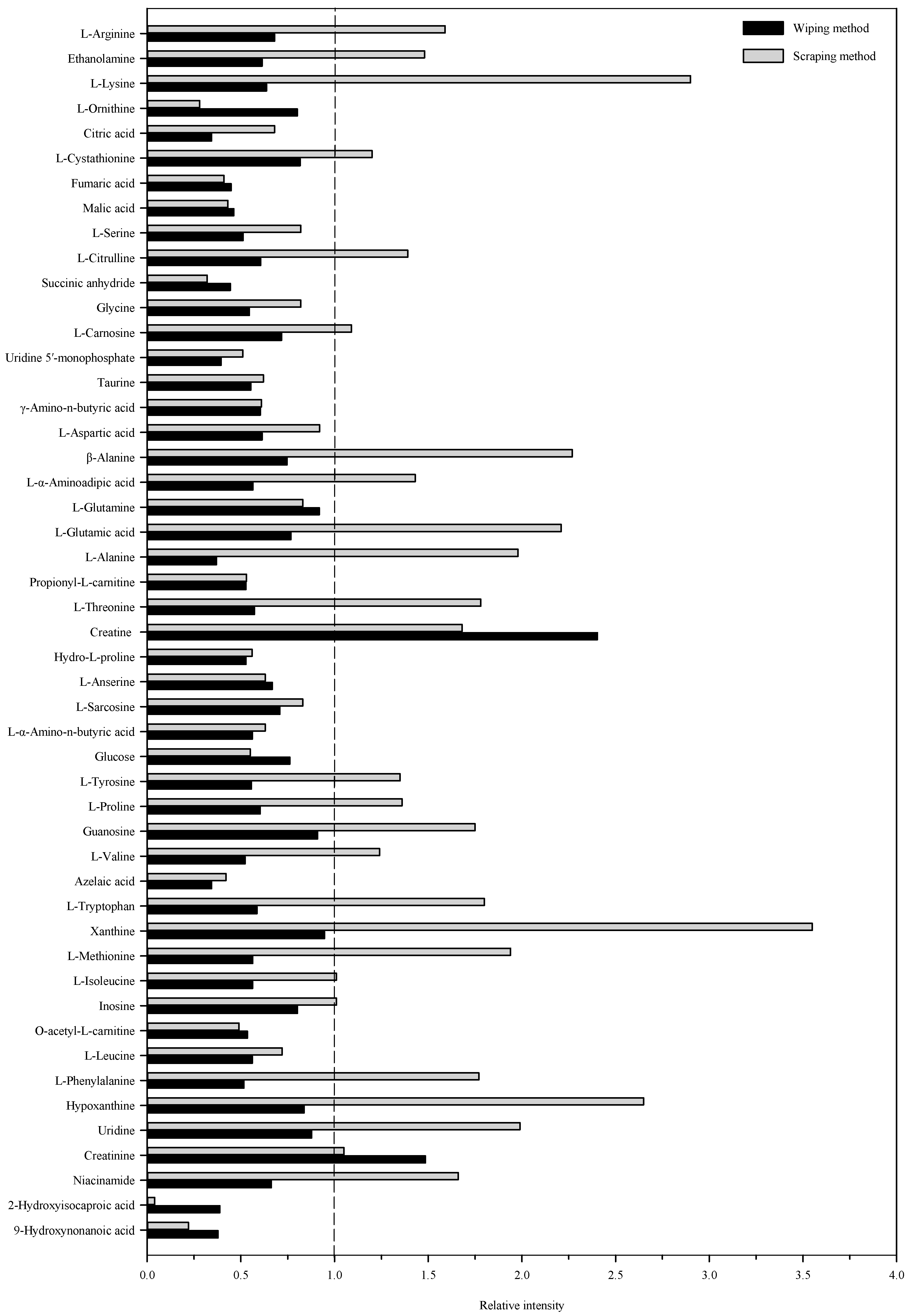
2.2. Metabolite Profiles from Targeted Liquid Chromatography–High-Resolution Mass Spectrometry of Skin Mucus
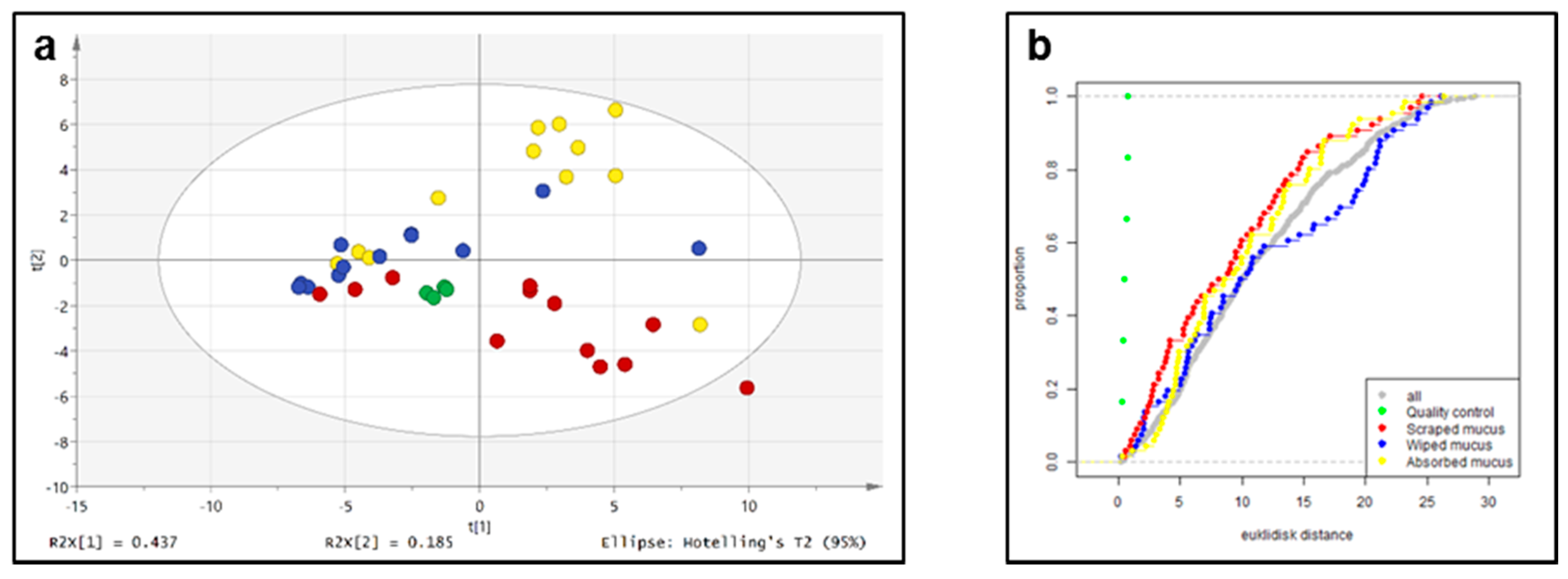
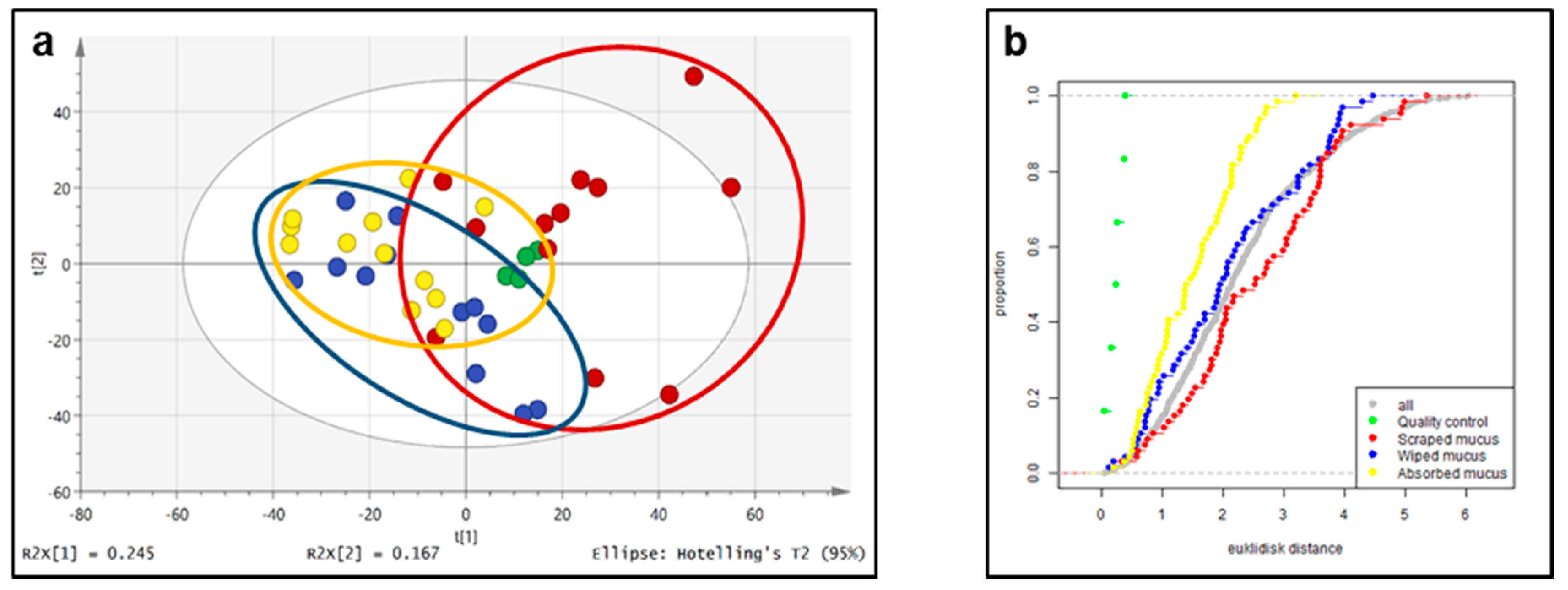
2.3. Untargeted Liquid Chromatography–High-Resolution Mass Spectrometryof Fish Skin Mucus
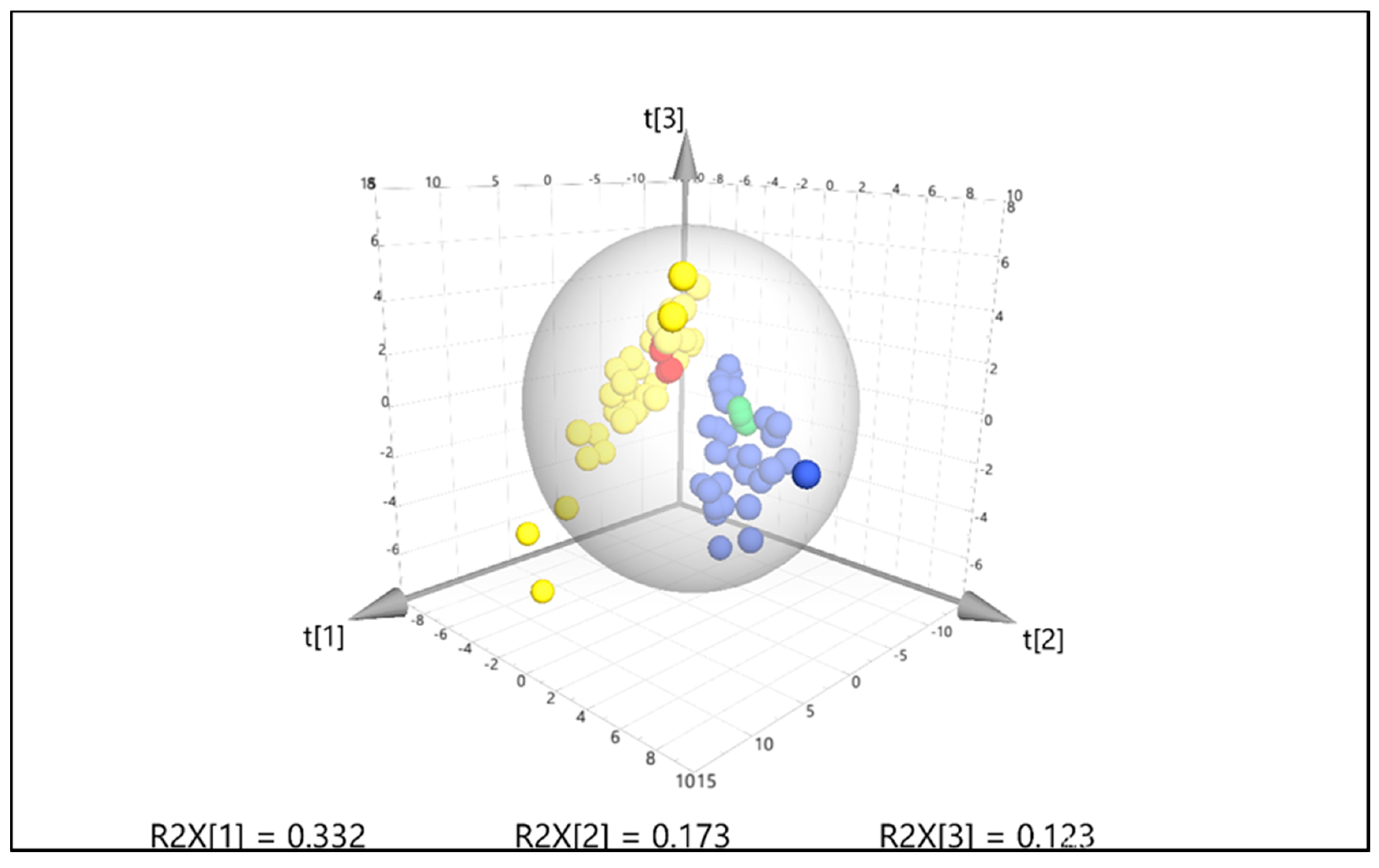
2.4. Targeted Feature Detection Using Thermo Xcalibur versus MZmine
2.5. Significance of Verified Mucosal Metabolites
3. Materials and Methods
3.1. Materials
3.2. Fish Breeding
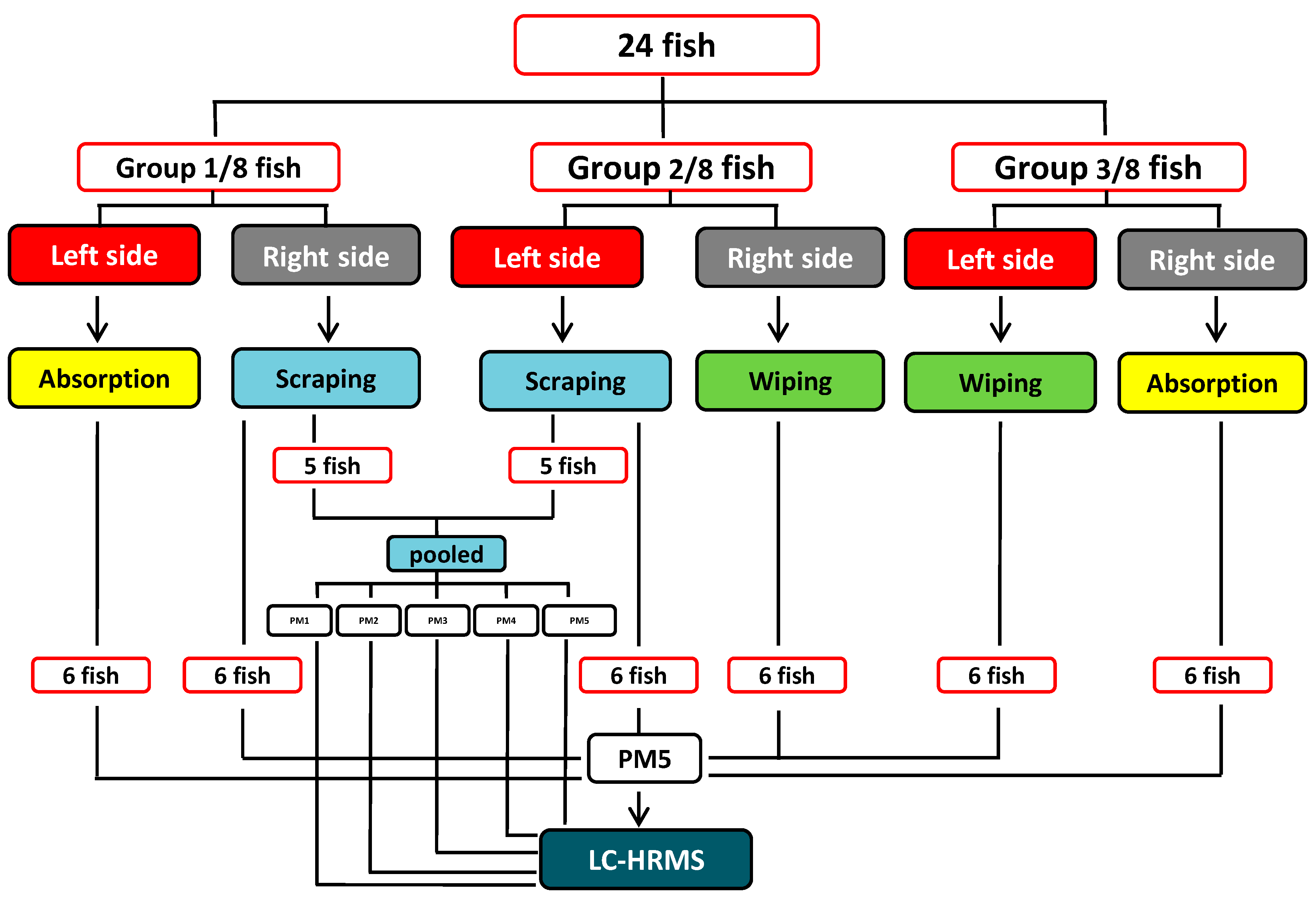
3.3. General Mucus Sampling Procedure
3.4. Epidermal Mucus Sampling
3.4.1. Absorption Method
3.4.2. Wiping Method
3.4.3. Scraping Method
3.5. Collection of Mucus Fluid
3.6. Sample Preparation
3.7. Liquid Chromatography–High-Resolution Mass SpectrometryData Acquisition
3.8. Raw Data Processing Using MZmine
3.9. Multivariate Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. The State of World Fisheries and Aquaculture 2016: Contributing to Food Security and Nutrition for All. Available online: http://wwwfaoorg/3/a-i5555epdf (accessed on 7 July 2016).
- Esteban, M.A. An overview of the immunological defenses in fish skin. ISRN Immunol. 2012, 2012, 853470. [Google Scholar]
- Easy, R.H.; Ross, N.W. Changes in Atlantic salmon (Salmo salar) epidermal mucus protein composition profiles following infection with sea lice (Lepeophtheirus salmonis). Comp. Biochem. Physiol. Part D Genom. Proteom. 2009, 4, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Guardiola, F.A.; Cuesta, A.; Abellán, E.; Meseguer, J.; Esteban, M.A. Comparative analysis of the humoral immunity of skin mucus from several marine teleost fish. Fish Shellfish Immunol. 2014, 40, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; MacKinnon, S.L.; Ross, N.W. A comparative study on innate immune parameters in the epidermal mucus of various fish species. Comp. Biochem. Physiol. Part. B. Biochem. Mol. Biol. 2007, 148, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Brinchmann, M.F. Immune relevant molecules identified in the skin mucus of fish using -omics technologies. Mol. Biosyst. 2016, 12, 2056–2063. [Google Scholar] [CrossRef] [PubMed]
- Bruce, T.J.; Brown, M.L. A review of immune system components, cytokines, and immunostimulants in cultured finfish species. Open J. Anim. Sci. 2017, 7, 267–288. [Google Scholar] [CrossRef]
- Guardiola, F.A.; Cuartero, M.; Del Mar Collado-Gonzalez, M.; Diaz Banos, F.G.; Cuesta, A.; Morinigo, M.A.; Esteban, M.A. Terminal carbohydrates abundance, immune related enzymes, bactericidal activity and physico-chemical parameters of the Senegalese sole (Solea senegalensis, Kaup) skin mucus. Fish Shellfish Immunol. 2017, 60, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Haniffa, M.A.; Viswanathan, S.; Jancy, D.; Poomari, K.; Manikandan, S. Antibacterial studies of fish mucus from two marketed air-breathing fishes—Channa striatus and Heteropneustes fossilis. Int. Res. J. Microbiol. 2014, 5, 22–27. [Google Scholar]
- Ekman, D.R.; Skelton, D.M.; Davis, J.M.; Villeneuve, D.L.; Cavallin, J.E.; Schroeder, A.; Jensen, K.M.; Ankley, G.T.; Collette, T.W. Metabolite profiling of fish skin mucus: A novel approach for minimally-invasive environmental exposure monitoring and surveillance. Environ. Sci. Technol. 2015, 49, 3091–3100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Watson, D.G.; Wang, L.J.; Westrop, G.D.; Coombs, G.H.; Zhang, T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A 2014, 1362, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Di Guida, R.; Engel, J.; Allwood, J.W.; Weber, R.J.; Jones, M.R.; Sommer, U.; Viant, M.R.; Dunn, W.B. Non-targeted UHPLC–MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics 2016, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; Van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Krauss, M.; Brack, W.; Schulze, T. Optimization of LC–Orbitrap-HRMS acquisition and MZmine 2 data processing for nontarget screening of environmental samples using design of experiments. Anal. Bioanal. Chem. 2016, 408, 7905–7915. [Google Scholar] [CrossRef] [PubMed]
- Shephard, K.L. Functions for fish mucus. Rev. Fish Biol. Fish. 1994, 4, 401–429. [Google Scholar] [CrossRef]
- Reverter, M.; Sasal, P.; Banaigs, B.; Lecchini, D.; Lecellier, G.; Tapissier-Bontemps, N. Fish mucus metabolome reveals fish life-history traits. Coral Reefs 2017, 36, 463–475. [Google Scholar] [CrossRef]
- Benhamed, S.; Guardiola, F.A.; Mars, M.; Esteban, M.Á. Pathogen bacteria adhesion to skin mucus of fishes. Vet. Microbiol. 2014, 171, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Maitra, S.K.; Nachum, R.; Pearson, F.C. Establishment of beta-hydroxy fatty acids as chemical marker molecules for bacterial endotoxin by gas chromatography-mass spectrometry. Appl. Environ. Microbiol. 1986, 52, 510–514. [Google Scholar] [PubMed]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.W. Fish cutaneous mucus: A new source of skin surface lipid. Lipids 1970, 5, 947–949. [Google Scholar] [CrossRef]
- Smeden, J.; Janssens, M.; Kaye, E.C.J.; Caspers, P.J.; Lavrijsen, A.P.; Vreeken, R.J.; Bouwstra, J.A. The importance of free fatty acid chain length for the skin barrier function in atopic eczema patients. Exp. Dermatol. 2014, 23, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Jais, A.M.M.; Matori, M.F.; Kittakoop, P.; Sowanborirux, K. Fatty acid compositions in mucus and roe of Haruan, Channa striatus, for wound healing. Gen. Pharmacol. 1998, 30, 561–563. [Google Scholar] [CrossRef]
- Hara, T.J.; Macdonald, S.; Evans, R.E.; Marui, T.; Arai, S. Morpholine, bile acids and skin mucus as possible chemical cues in salmonid homing: Electrophysiological re-evaluation. In Mechanisms of Migration in Fishes; Springer: Berlin, Germany, 1984; pp. 363–378. [Google Scholar]
- Kallert, D.M.; Ponader, S.; Adelt, S.; Kaese, P.; Geyer, R.; Haas, W.; El-Matbouli, M. Analysis of rainbow trout Oncorhynchus mykiss epidermal mucus and evaluation of semiochemical activity for polar filament discharge in Myxobolus cerebralis actinospores. J. Fish Biol. 2010, 77, 1579–1598. [Google Scholar] [CrossRef] [PubMed]
- Saglio, P.; Fauconneau, B. Free amino acid content in the skin mucus of goldfish, Carassius auratus L.: Influence of feeding. Comp. Biochem. Physiol. Part A Physiol. 1985, 82, 67–70. [Google Scholar] [CrossRef]
- Stabell, O.B.; Selset, R. Comparison of mucus collecting methods in fish olfaction. Acta Physiol. 1980, 108, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Fauconneau, B.; Saglio, P. Protein-bound and free amino acid content in the skin mucus of the European eel, Anguilla anguilla (L.). Comp. Biochem. Physiol. B 1984, 77, 513–516. [Google Scholar] [CrossRef]
- Chong, K.; Ying, T.; Foo, J.; Jin, L.T.; Chong, A. Characterisation of proteins in epidermal mucus of discus fish (Symphysodon spp.) during parental phase. Aquaculture 2005, 249, 469–476. [Google Scholar] [CrossRef]
- Shiau, C.-Y.; Pong, Y.-J.; Chiou, T.-K.; Chai, T.-J. Effect of growth on the levels of free histidine and amino acids in white muscle of milkfish (Chanos chanos). J. Agric. Food Chem. 1997, 45, 2103–2106. [Google Scholar] [CrossRef]
- Shi, H.P.; Fishel, R.S.; Efron, D.T.; Williams, J.Z.; Fishel, M.H.; Barbul, A. Effect of supplemental ornithine on wound healing. J. Surg. Res. 2002, 106, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Wesley, J.A.; Supp, D.M. Role of arginine and omega-3 fatty acids in wound healing and infection. Adv. Wound. Care 2014, 3, 682–690. [Google Scholar]
- Button, B.; Boucher, R.C. Role of mechanical stress in regulating airway surface hydration and mucus clearance rates. Respir. Physiol. Neurobiol. 2008, 163, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Ueda, F.; Watanabe, M.; Hirata, Y.; Kyoi, T.; Kimura, K. Changes in cyclic AMP content of rat gastric mucosa induced by ulcerogenic stimuli—In relation to the antiulcer activity of irsogladine maleate. Jpn. J. Pharmacol. 1991, 55, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Van Waarde, A. Biochemistry of non-protein nitrogenous compounds in fish including the use of amino acids for anaerobic energy production. Comp. Biochem. Physiol. B 1988, 91, 207–228. [Google Scholar] [CrossRef]
- Ræder, I.L.U.; Paulsen, S.M.; Smalås, A.O.; Willassen, N.P. Effect of fish skin mucus on the soluble proteome of Vibrio salmonicida analysed by 2-D gel electrophoresis and tandem mass spectrometry. Microb. Pathog. 2007, 42, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to convert raw mass spectrometry data. Curr. Protoc. Bioinform. 2014, 46, 1–9. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- The R Project for Statistical Computing Website. Available online: https://www.r-project.org/ (accessed on 6 May 2018).
| PM | Number of Detected Metabolites | Maximum RSD [%] | Average RSD [%] |
|---|---|---|---|
| PM 1_Dilution with water | 52 | 30 | 9 |
| PM 2_Dilution with methanol | 52 | 62 | 8 |
| PM 3_Dilution with acetonitrile | 53 | 50 | 7 |
| PM 4_Freeze-drying, solution in 50% methanol | 54 | 160 | 57 |
| PM 5_Filtration only | 56 | 25 | 8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, L.; Tartor, H.; Grove, S.; Kristoffersen, A.B.; Uhlig, S. Workflow for the Targeted and Untargeted Detection of Small Metabolites in Fish Skin Mucus. Fishes 2018, 3, 21. https://doi.org/10.3390/fishes3020021
Ivanova L, Tartor H, Grove S, Kristoffersen AB, Uhlig S. Workflow for the Targeted and Untargeted Detection of Small Metabolites in Fish Skin Mucus. Fishes. 2018; 3(2):21. https://doi.org/10.3390/fishes3020021
Chicago/Turabian StyleIvanova, Lada, Haitham Tartor, Søren Grove, Anja B. Kristoffersen, and Silvio Uhlig. 2018. "Workflow for the Targeted and Untargeted Detection of Small Metabolites in Fish Skin Mucus" Fishes 3, no. 2: 21. https://doi.org/10.3390/fishes3020021
APA StyleIvanova, L., Tartor, H., Grove, S., Kristoffersen, A. B., & Uhlig, S. (2018). Workflow for the Targeted and Untargeted Detection of Small Metabolites in Fish Skin Mucus. Fishes, 3(2), 21. https://doi.org/10.3390/fishes3020021






