First Record of Lyconus brachycolus (Gadiformes: Lyconidae) in Spanish Waters: An Update on Taxonomic Knowledge
Abstract
1. Introduction
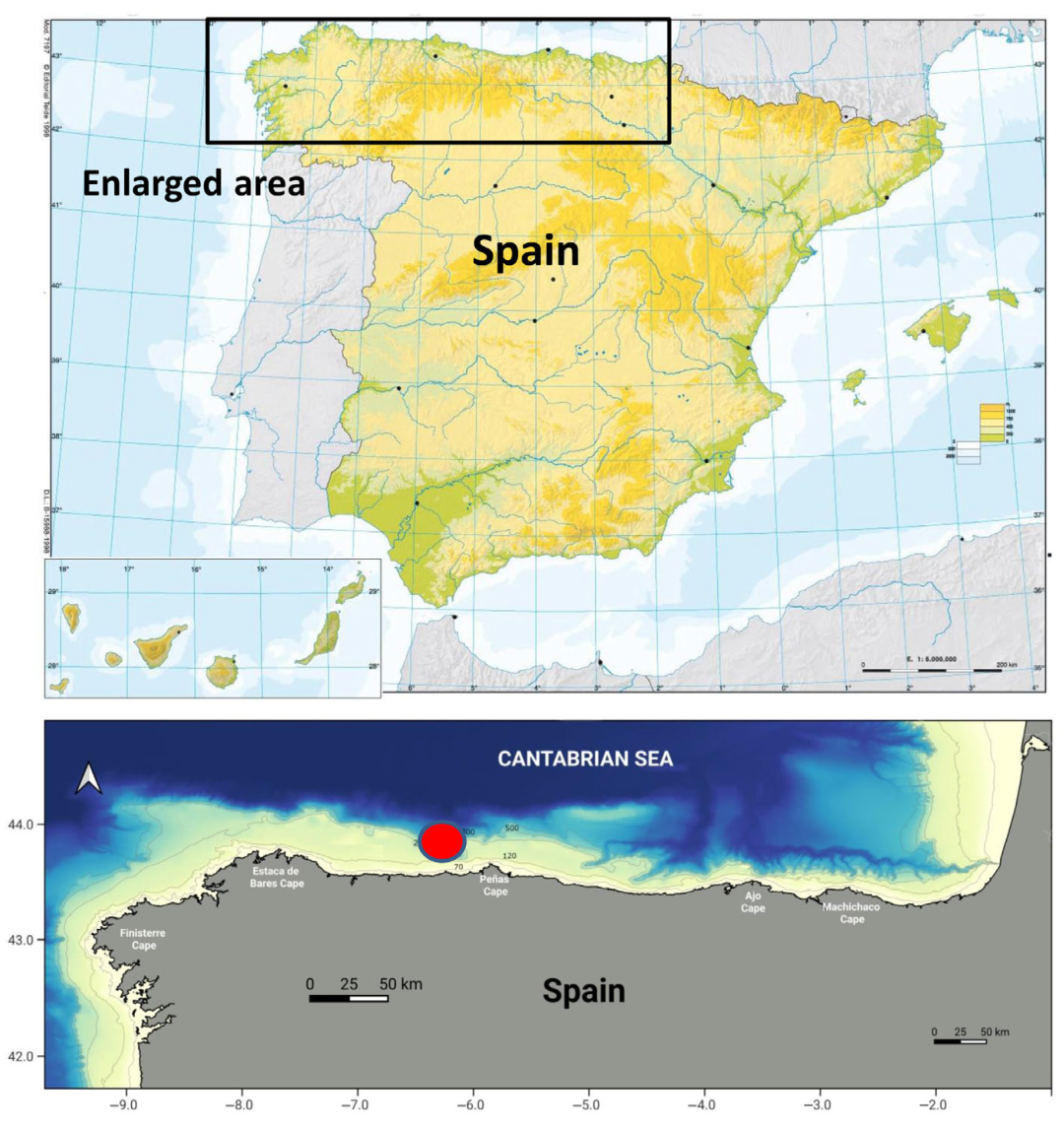
2. Materials and Methods
2.1. Sampling Data and Morphological Analysis
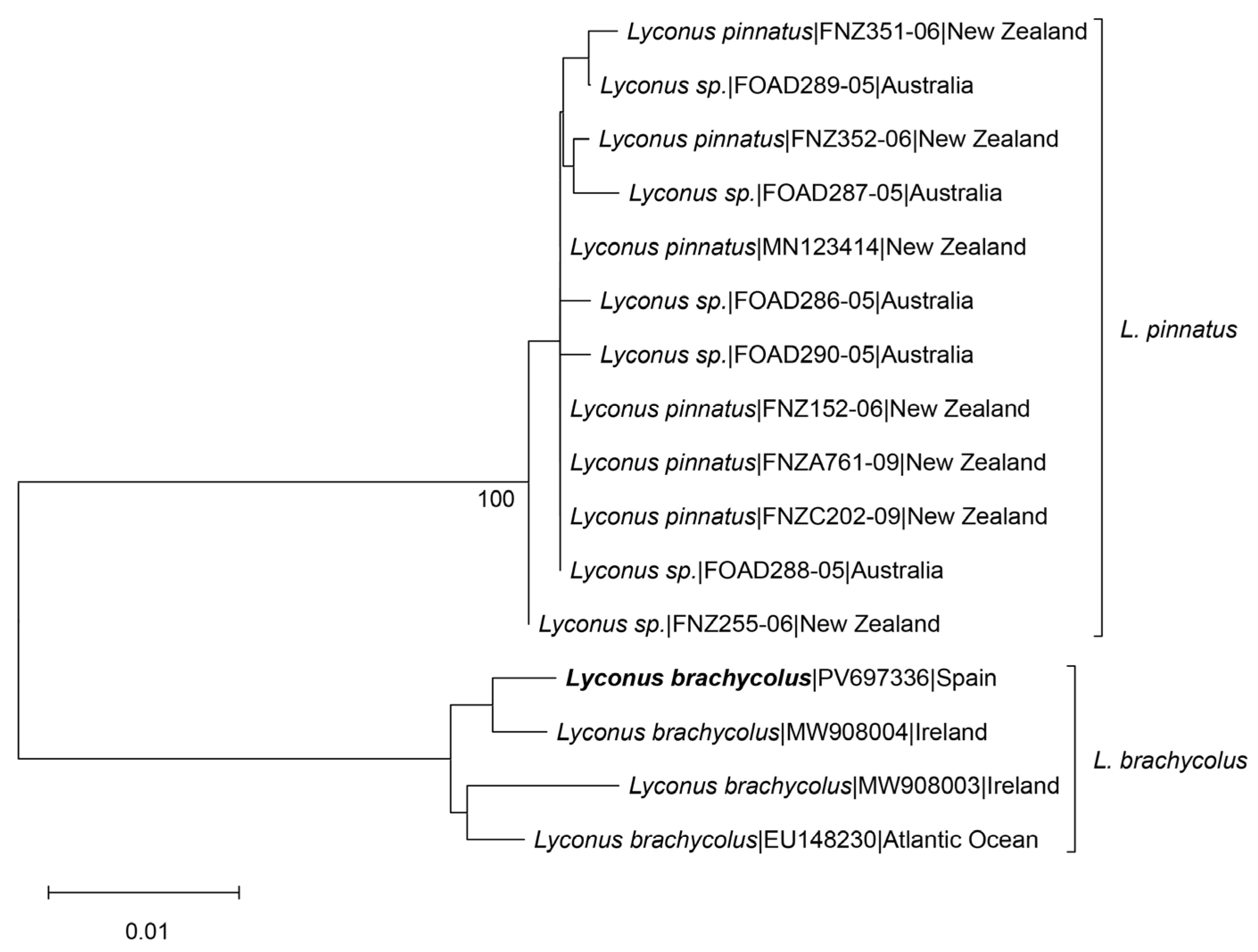
2.2. Molecular Analysis
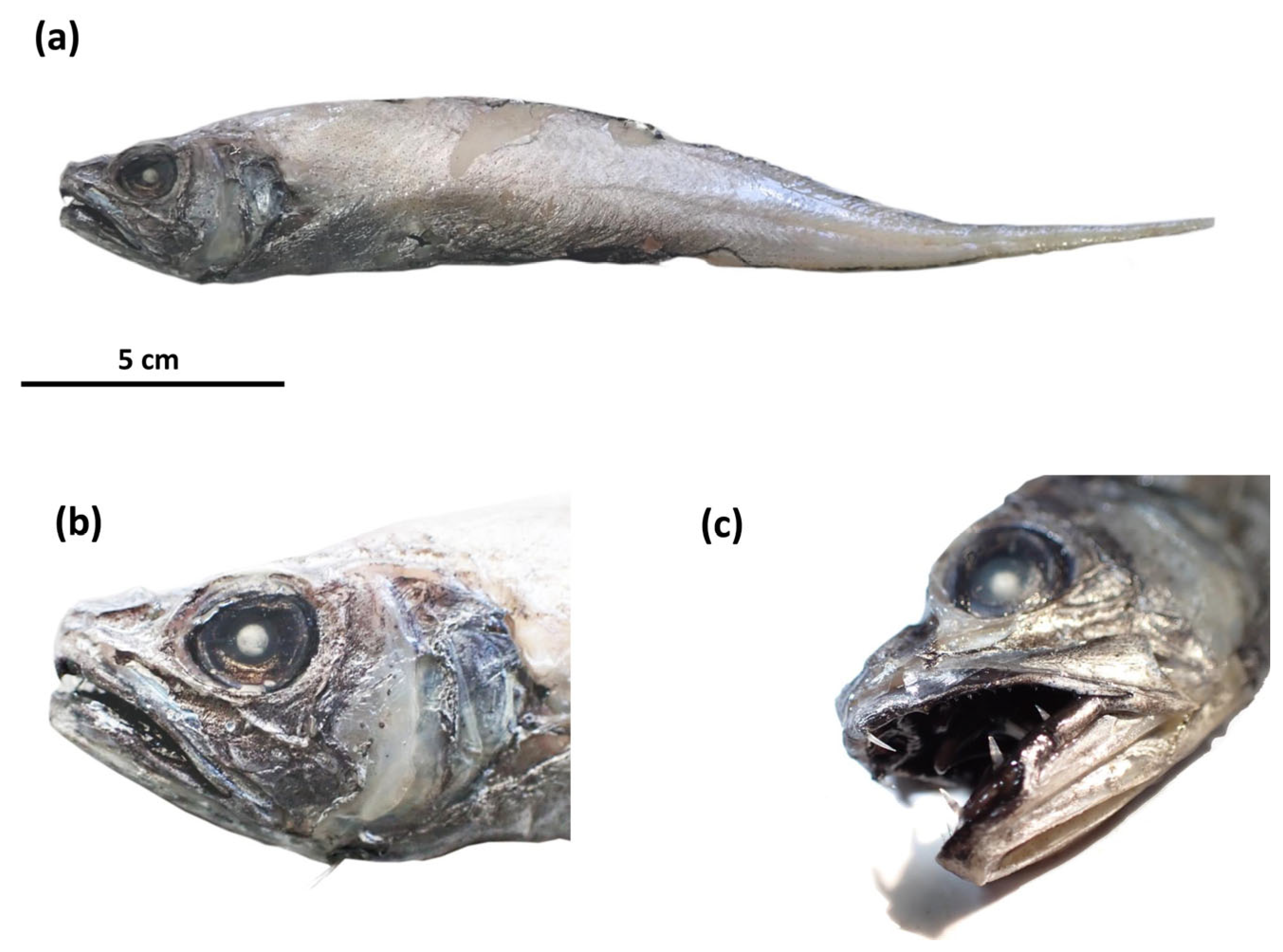
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roa-Varón, A.; Dikow, R.B.; Carnevale, G.; Tornabene, L.; Baldwin, C.C.; Li, C.; Hilton, E.J. Confronting sources of systematic error to resolve historically contentious relationships: A case study using gadiform fishes (Teleostei, Paracanthopterygii, Gadiformes). Syst. Biol. 2021, 70, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.R.; Braga, A.C.; Nunan, G.W.; Costa, P.A. On new collections of deep-sea Gadiformes (Actinopterygii: Teleostei) from the Brazilian continental slope, between 11 and 23 S. Zootaxa 2010, 2433, 25–46. [Google Scholar] [CrossRef]
- Roa-Varón, A.; Ortí, G. Phylogenetic relationships among families of Gadiformes (Teleostei, Paracanthopterygii) based on nuclear and mitochondrial data. Mol. Phylogenetics Evol. 2009, 52, 688–704. [Google Scholar] [CrossRef] [PubMed]
- von der Heyden, S.; Matthee, C.A. Towards resolving familial relationships within the Gadiformes, and the resurrection of the Lyconidae. Mol. Phylogenetics Evol. 2008, 48, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Inada, T. Merlucciidae. In FAO Species Catalogue: Gadiform Fishes of the World (Order Gadiformes). An Annotated and Illustrated Catalogue of Cods, Hakes, Grenadiers and Other Gadiform Fishes Known to Date; Cohen, D.M., Inada, T., Iwamoto, T., Scialabba, N., Eds.; FAO Fisheries Synopsis. No. 125; FAO: Rome, Italy, 1990; Volume 10, pp. 319–346. [Google Scholar]
- Lloris, D.; Matallanas, J.; Oliver, P. Hakes of the World (Family Merlucciidae). An Annotated and Illustrated Catalogue of Hake Species Known to Date; FAO Species Catalogue for Fishery Purposes: Rome, Italy, 2005; pp. 1–57. [Google Scholar]
- Bolshakova, Y.Y.; Evseenko, S.A. First finding of larvae of an undescribed species of the genus Lyconus (Lyconidae) in the North Pacific Ocean. J. Ichthyol. 2023, 63, 371–375. [Google Scholar] [CrossRef]
- Báez, C.; Rodríguez-Cabello, C.; Bañón, R.; Brito, A.; Falcón, J.M.; Maño, T.; Baro, J.; Macías, D.; Meléndez, M.J.; Camiñas, J.A.; et al. Updating the national checklist of marine fishes in Spanish waters: An approach to priority hotspots and lessons for conservation. Mediterr. Mar. Sci. 2019, 20, 260–270. [Google Scholar] [CrossRef]
- Bañón, R.; de Carlos, A.; Farias, C.; Vilas-Arrondo, N.; Baldó, F. Exploring deep-sea biodiversity in the Porcupine Bank (NE Atlantic) through fish integrative taxonomy. J. Mar. Sci. Eng. 2021, 9, 1075. [Google Scholar] [CrossRef]
- Meyer, A. Evolution of mitochondrial DNA in fishes. In Biochemistry and Molecular Biology of Fishes; Hochachka, P.W., Mommsen, T.P., Eds.; Elsevier: Hague, The Netherlands, 1993; pp. 1–38. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- ICES. SISP 15—Manual of the IBTS North Eastern Atlantic Surveys. Series of ICES Survey Protocols (2012–2020); Report; ICES: Nelson, New Zealand, 2017. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; pp. 1–333. [Google Scholar]
- Saitou, N.; Nei, M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. Molecular Evolutionary Genetics Analysis Version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Holt, E.W.L.; Byrne, L.W. On a new species of Lyconus from the North-east Atlantic. Ann. Mag. Nat. Hist. 1906, 7, 423–426. [Google Scholar] [CrossRef]
- Maul, G.E. Monografia dos peixos do Museu Municipal do Funchal. Familias Macrounidae e Merlucciidae. Bol. Mus. Munic. Funchal 1951, 5, 5–55. [Google Scholar]
- Golovan, G.A. Rare and newly recorded chondricthyian and teleostean fishes of the continental slope of West Africa. Tr.-Ta Okeanol. SSSR 1976, 104, 277–317. [Google Scholar]
- Matallanas, J.; Lloris, D. Primera cita de Lyconus brachycolus Holt &Byrne, 1906 (Pisces, Merlucciidae) en el Atlántico sur. Misc. Zool. 1987, 11, 257–261. [Google Scholar]
- Evseenko, S.A.; Suntsov, A.V. Larvae of Lyconus pinnatus (Merlucciidae) from the Southwestern Pacific, with comments on the diagnostic characters of species of the genus Lyconus. J. Ichthyol. 1991, 35, 98–110. [Google Scholar]
- Bañón, R.; Conde-Pardo, P.; Álvarez-Salgado, X.A.; de Carlos, A.; Arronte, J.C.; Piedracoba, S. Tropicalization of fish fauna of Galician coastal waters, in the NW Iberian upwelling system. Reg. Stud. Mar. Sci. 2024, 70, 103369. [Google Scholar] [CrossRef]
- Arronte, J.C.; Antolínez, A.; Bañón, R.; Heredia, J.; de Carlos, A. First record of Diapterus brevirostris (Teleostei: Gerridae) in Atlantic European waters: A case of introduced species. J. Mar. Biol. Assoc. U. K. 2024, 104, e61. [Google Scholar] [CrossRef]
- Rodríguez-Cabello, C.; Pérez, M.; Bañón, R. Occurrence of Apristurus species in the Galicia Bank Seamount (NE Atlantic). J. Appl. Ichthyol. 2014, 30, 906–915. [Google Scholar] [CrossRef]
- Danovaro, R.; Company, J.B.; Corinaldesi, C.; D’Onghia, G.; Galil, B.; Gambi, C.; Gooday, A.J.; Lampadariou, N.; Luna, G.M.; Morigi, C.; et al. Deep-sea biodiversity in the Mediterranean Sea: The known, the unknown, and the unknowable. PLoS ONE 2010, 5, e11832. [Google Scholar] [CrossRef] [PubMed]
- McClenaghan, B.; Fahner, N.; Cote, D.; Chawarski, J.; McCarthy, A.; Rajabi, H.; Singer, G.; Hajibabaei, M. Harnessing the power of eDNA metabarcoding for the detection of deep-sea fishes. PLoS ONE 2020, 15, e0236540. [Google Scholar] [CrossRef] [PubMed]
- Porteiro, F.M.; Sutton, T.; Byrkjedal, I.; Orlov, A.M.; Heino, M.; Menezes, G.; Bergstad, O.A. Fishes of the Northern Mid-Atlantic Ridge Collected During the MAR-ECO Cruise in June–July 2004: An Annotated Checklist; Arquipélago. Life and Marine Sciences; Universidade dos Açores: Ponta Delgada, Portugal, 2017; Volume 10, 126p. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D. bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Pavan-Kumar, A.; Gireesh-Babu, P.; Jaiswar, A.K.; Chaudhari, A.; Krishna, G.; Lakra, W.S. DNA Barcoding of Marine Fishes: Prospects and Challenges. In DNA Barcoding in Marine Perspectives; Trivedi, S., Ansari, A., Ghosh, S., Rehman, H., Eds.; Springer: Cham, Switzerland, 2016; pp. 285–299. [Google Scholar] [CrossRef]
- Hubert, N.; Hanner, R. DNA barcoding, species delineation and taxonomy: A historical perspective. DNA Barcodes 2015, 3, 44–58. [Google Scholar] [CrossRef]
- Phillips, J.D.; Gillis, D.J.; Hanner, R.H. Incomplete estimates of genetic diversity within species: Implications for DNA barcoding. Ecol. Evol. 2019, 9, 2996–3010. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
| Lyconusbrachycolus | ||||
|---|---|---|---|---|
| MHN USC 25186-3 | Holotype | Previous Studies Range | Mean ± SE (n) | |
| Total length (mm) | 216 | 237 | ||
| Standard length (mm) | 210 | 232 | 180–349 | |
| As % SL | ||||
| Head length | 17.9 | 17.9 | 17.1–20.6 | 18.1 ± 1.6 (9) |
| Preorbital length | 5 | 5.2 | 4.8–6.1 | 5.1 ± 0.5 (7) |
| Postorbital length | 8.1 | 7.8–10 | ||
| Eye diameter | 4.5 | 4.5 | 3.2–5.1 | 4.4 ± 0.6 (7) |
| Interorbital width | 6.2 | 5.6 | 5–6.6 | 5.8 ± 0.5 (7) |
| Upper jaw length | 8.9 | |||
| Lower jaw length | 8.4 | |||
| Predorsal1 fin length | 21.1 | 19.4 | 18.3–23.9 | 21 ± 2 (9) |
| Preanal fin length | 42.1 | 40.5 | 35.5–50.6 | 43 ± 4.3 (9) |
| Prepectoral fin length | 20.9 | 17.8–21.1 | 19.9 ± 1.8 (3) | |
| Prepelvic fin length | 20 | 18.5–22.5 | 20.3 ± 2 (3) | |
| Dorsal1 fin base | 5.7 | 4.3–5 | 5 ± 0.7 (3) | |
| Dorsal2 fin base | 72 | |||
| Anal fin base | 58.1 | |||
| Pectoral fin length | 13.8 | 11.2 | 11–24.4 | 14.7 ± 4.9 (7) |
| Pelvic fin length | 5 | 7.3 | 4.6–8.6 | 6.7 ± 1.6 (6) |
| Body depth at pectoral fin | 13.8 | 14.2 | 12.9–15 | 14.1 ± 0.9 (9) |
| Body width | 5.8 | 7.8 | 5.9–7.2 | 6.7± 0.9 |
| Meristic | ||||
| Dorsal1 fin rays | 8 | 9–10 | ||
| Dorsal2 fin rays | 107 | 105–111 | ||
| Anal fin rays | 87 | 86–96 | ||
| Pectoral fin rays | 13 | 13 | 13–14 | |
| Pelvic fin rays | 8 | 9 | 8–9 | |
| Branchiostegal rays | 7 | 7 | ||
| Gill rakers | 3 + 10 | 3–5 + 9–12 | ||
| Vomer teeth | 3 + 4 | 4 + 4 | 2–4 + 2–4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bañón, R.; de Carlos, A.; Arronte, J.C. First Record of Lyconus brachycolus (Gadiformes: Lyconidae) in Spanish Waters: An Update on Taxonomic Knowledge. Fishes 2025, 10, 351. https://doi.org/10.3390/fishes10070351
Bañón R, de Carlos A, Arronte JC. First Record of Lyconus brachycolus (Gadiformes: Lyconidae) in Spanish Waters: An Update on Taxonomic Knowledge. Fishes. 2025; 10(7):351. https://doi.org/10.3390/fishes10070351
Chicago/Turabian StyleBañón, Rafael, Alejandro de Carlos, and Juan Carlos Arronte. 2025. "First Record of Lyconus brachycolus (Gadiformes: Lyconidae) in Spanish Waters: An Update on Taxonomic Knowledge" Fishes 10, no. 7: 351. https://doi.org/10.3390/fishes10070351
APA StyleBañón, R., de Carlos, A., & Arronte, J. C. (2025). First Record of Lyconus brachycolus (Gadiformes: Lyconidae) in Spanish Waters: An Update on Taxonomic Knowledge. Fishes, 10(7), 351. https://doi.org/10.3390/fishes10070351






