Evolutionary Dynamics and Functional Conservation of amh Signaling in Teleost Lineages
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of amh Signaling Genes
2.2. Phylogenetic and Conserved Domain Analysis of amh Signaling Genes
2.3. Synteny Analysis of amh Signaling Genes
2.4. Molecular Evolution Analysis of amh Signaling Genes
2.5. Expression Patterns of amh Signaling Genes
2.6. Weighted Gene Co-Expression Network Analysis of amh Signaling Genes
3. Results and Discussion
3.1. Copy Number of amh Signaling Genes in Teleosts
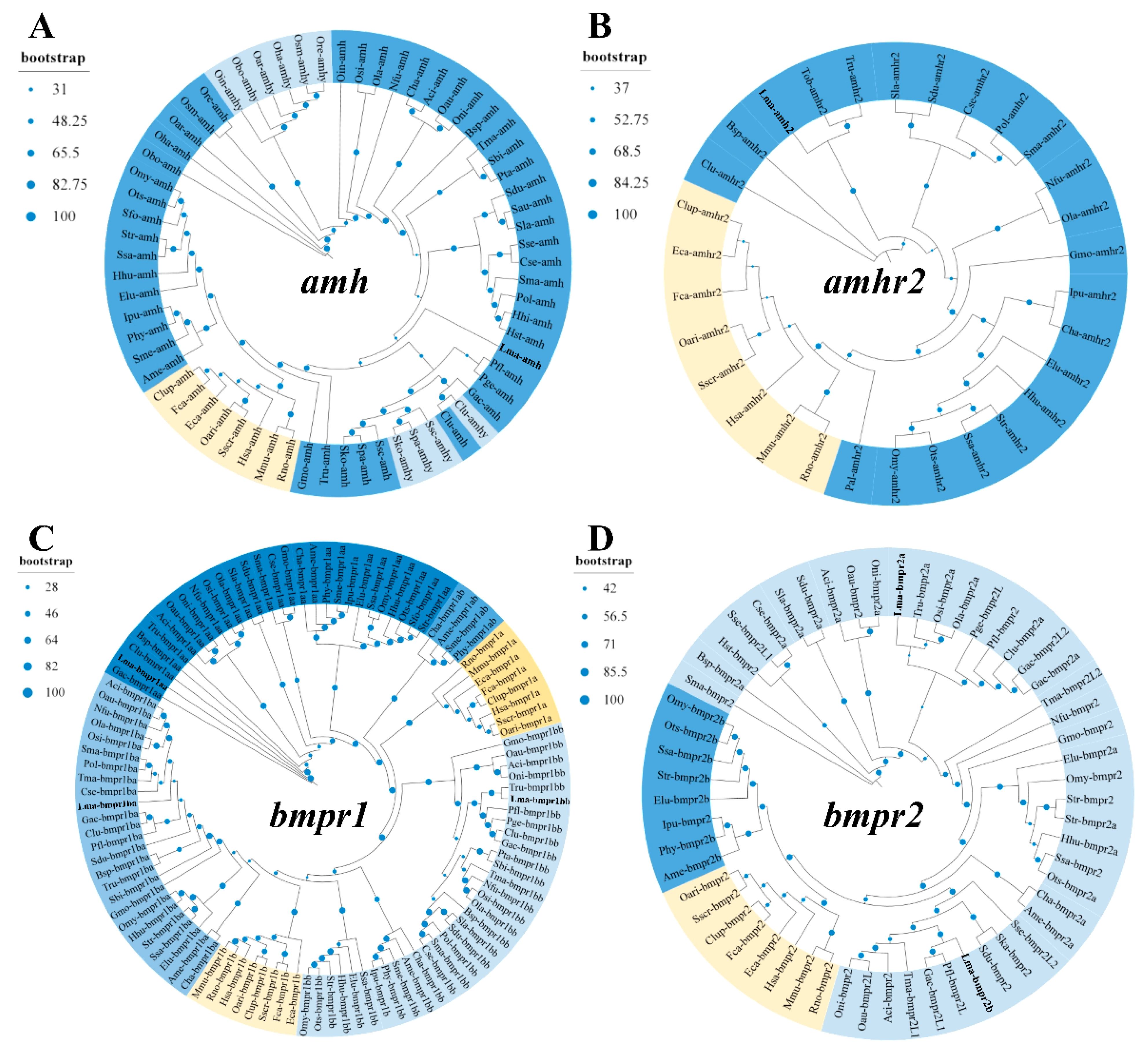
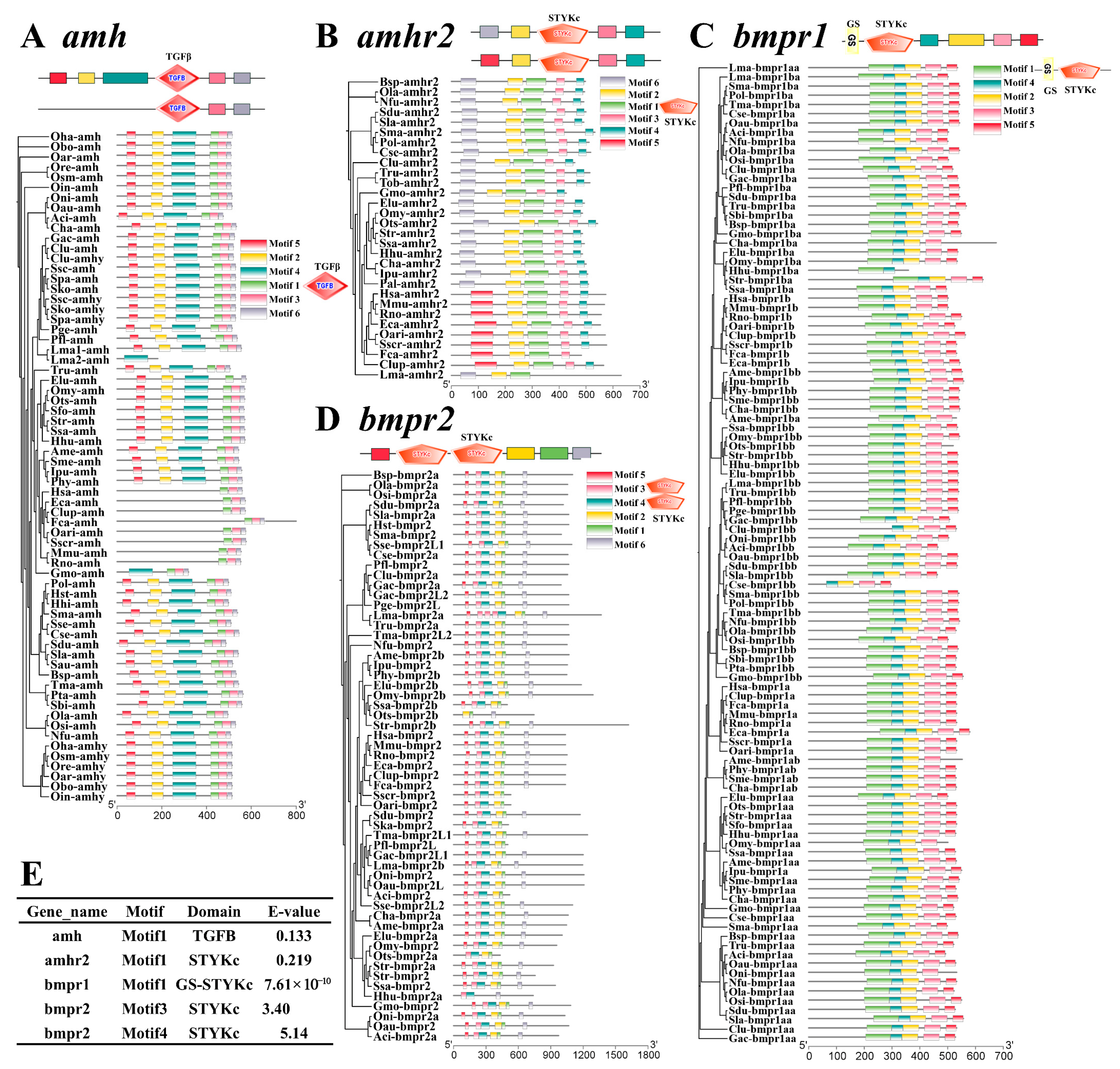
3.2. Phylogeny and Functional Domain of amh Signaling Genes
3.3. Conserved Synteny of amh Signaling Genes
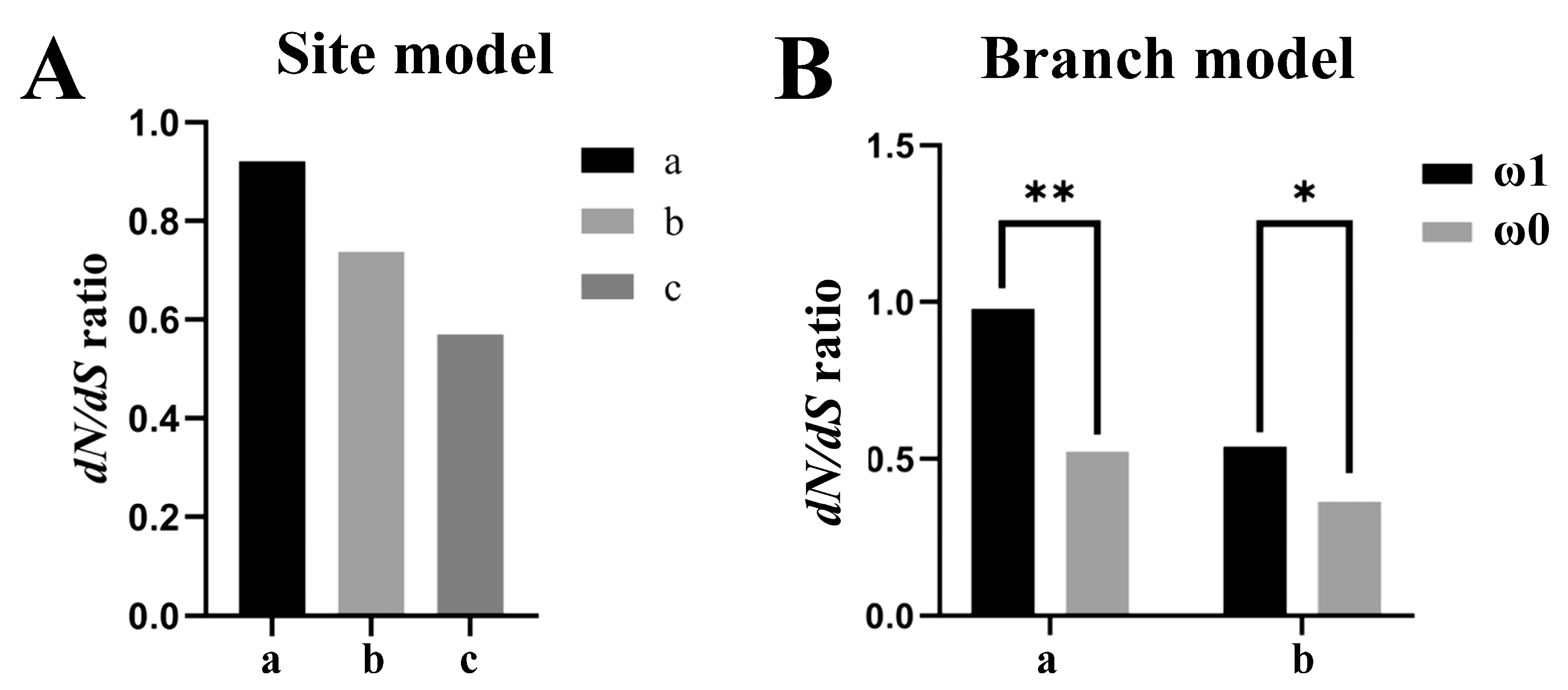
3.4. Molecular Evolution of amh Signaling Genes
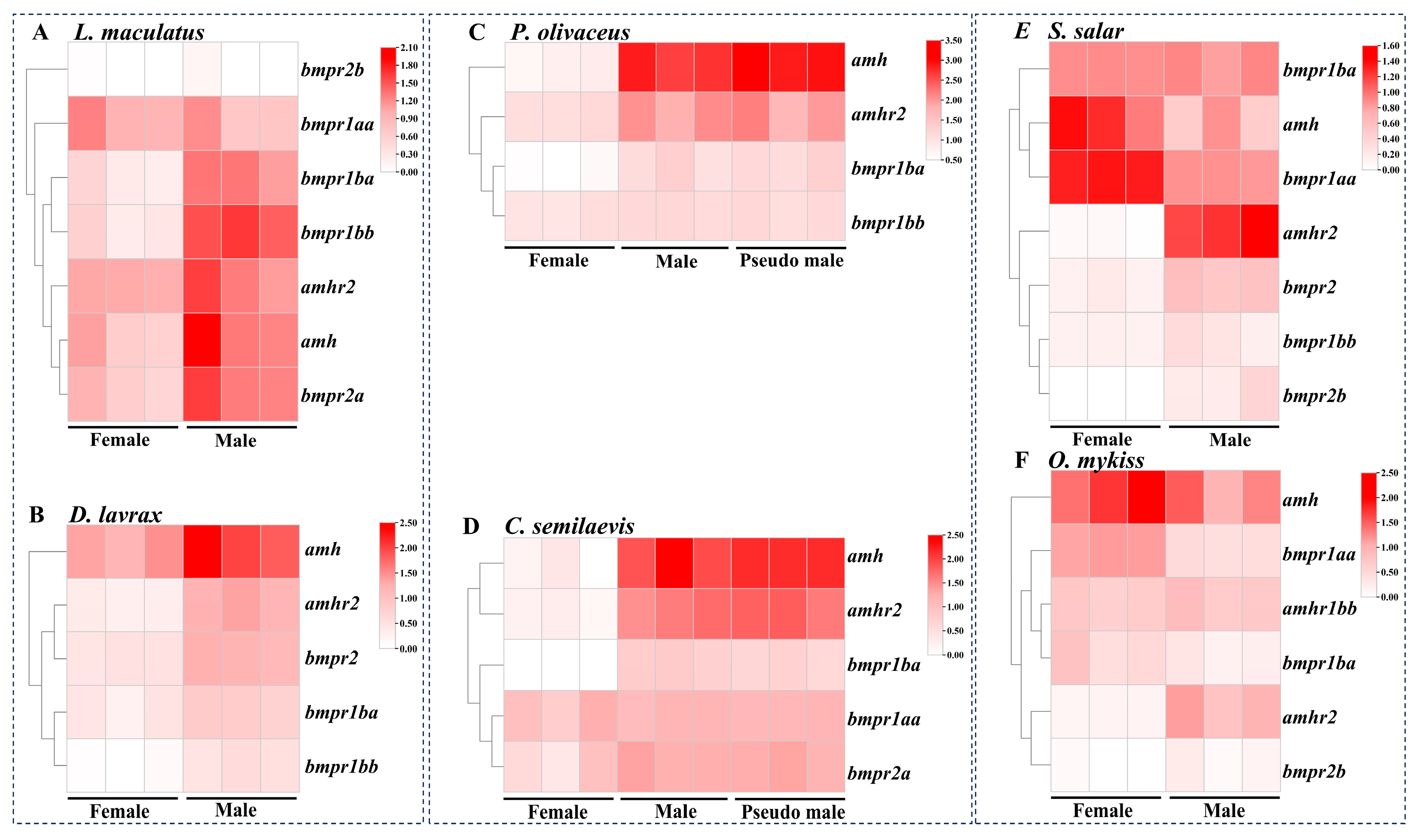
3.5. Expression Profiles of amh Signaling Genes in Teleost Species
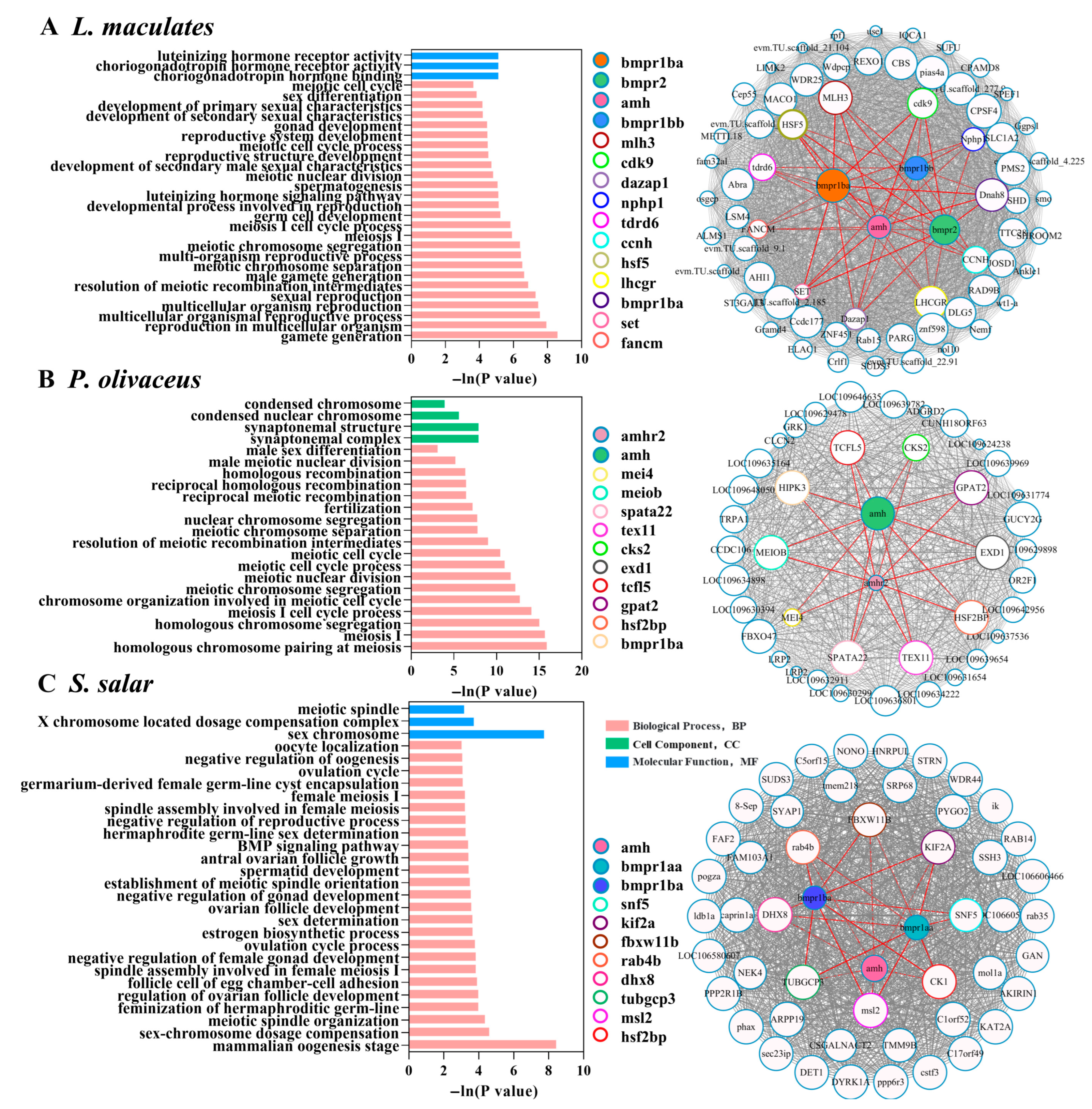
3.6. Gene Co-Expression Network of amh Signaling Genes in Teleost Species
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Z.; Gao, D. The Cause–Effect model of master sex determination gene acquisition and the evolution of sex chromosomes. Int. J. Mol. Sci. 2025, 26, 3282. [Google Scholar] [CrossRef] [PubMed]
- Kitano, J.; Ansai, S.; Takehana, Y.; Yamamoto, Y. Diversity and convergence of sex-determination mechanisms in teleost fish. Annu. Rev. Anim. Biosci. 2024, 12, 233–259. [Google Scholar] [CrossRef]
- Wang, J.; Tao, W.; Kocher, T.D.; Wang, D. Sex chromosome turnover and biodiversity in fishes. J. Genet. Genom. 2024, 51, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Kay, T.; Depincé, A.; Adolfi, M.; Schartl, M.; Guiguen, Y.; Herpin, A. Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200091. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef] [PubMed]
- Taslima, K.; Kikuchi, K.; Hosoya, S. Genomic architecture and sex chromosome systems of commercially important fish species in Asia—Current status, knowledge gaps and future prospects. Rev. Aquac. 2024, 16, 1918–1946. [Google Scholar] [CrossRef]
- Zhu, Z.; Younas, L.; Zhou, Q. Evolution and regulation of animal sex chromosomes. Nat. Rev. Genet. 2025, 26, 59–74. [Google Scholar] [CrossRef]
- Mullen, R.D.; Ontiveros, A.E.; Moses, M.M.; Behringer, R.R. AMH and AMHR2 mutations: A spectrum of reproductive phenotypes across vertebrate species. Dev. Biol. 2019, 455, 1–9. [Google Scholar] [CrossRef]
- Moses, M.M.; Behringer, R.R. A gene regulatory network for Müllerian duct regression. Environ. Epigenet. 2019, 5, dvz017. [Google Scholar] [CrossRef]
- Adolfi, M.C.; Nakajima, R.T.; Nóbrega, R.H.; Schartl, M. Intersex, hermaphroditism, and gonadal plasticity in vertebrates: Evolution of the Müllerian duct and amh/amhr2 signaling. Annu. Rev. Anim. Biosci. 2019, 7, 149–172. [Google Scholar] [CrossRef]
- Hattori, R.S.; Strüssmann, C.A.; Fernandino, J.I.; Somoza, G.M. Genotypic sex determination in teleosts: Insights from the testis-determining amhy gene. Gen. Comp. Endocrinol. 2013, 192, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dai, S.; Wu, J.; Wei, X.; Zhou, X.; Chen, M.; Tan, D.; Pu, D.; Li, M.; Wang, D. Roles of anti-Müllerian hormone and its duplicates in sex determination and germ cell proliferation of Nile tilapia. Genetics 2022, 220, iyab237. [Google Scholar] [CrossRef] [PubMed]
- Hattori, R.S.; Somoza, G.M.; Fernandino, J.I.; Colautti, D.C.; Miyoshi, K.; Gong, Z.; Yamamoto, Y.; Strüssmann, C.A. The duplicated Y-specific amhy gene is conserved and linked to maleness in silversides of the genus Odontesthes. Genes 2019, 10, 679. [Google Scholar] [CrossRef]
- Jeffries, D.L.; Mee, J.A.; Peichel, C.L. Identification of a candidate sex determination gene in Culaea inconstans suggests convergent recruitment of an Amh duplicate in two lineages of stickleback. J. Evol. Biol. 2022, 35, 1683–1695. [Google Scholar] [CrossRef]
- Song, W.; Xie, Y.; Sun, M.; Li, X.; Fitzpatrick, C.K.; Vaux, F.; O’Malley, K.G.; Zhang, Q.; Qi, J.; He, Y. A duplicated amh is the master sex-determining gene for Sebastes rockfish in the Northwest Pacific. Open Biol. 2021, 11, 210063. [Google Scholar] [CrossRef]
- Hattori, R.S.; Kumazawa, K.; Nakamoto, M.; Nakano, Y.; Yamaguchi, T.; Kitano, T.; Yamamoto, E.; Fuji, K.; Sakamoto, T. Y-specific amh allele, amhy, is the master sex-determining gene in Japanese flounder Paralichthys olivaceus. Front. Genet. 2022, 13, 1007548. [Google Scholar] [CrossRef]
- Rafati, N.; Chen, J.; Herpin, A.; Pettersson, M.E.; Han, F.; Feng, C.; Wallerman, O.; Rubin, C.J.; Péron, S.; Cocco, A.; et al. Reconstruction of the birth of a male sex chromosome present in Atlantic herring. Proc. Natl. Acad. Sci. USA 2020, 117, 24359–24368. [Google Scholar] [CrossRef] [PubMed]
- Jasonowicz, A.J.; Simeon, A.; Zahm, M.; Cabau, C.; Klopp, C.; Roques, C.; Iampietro, C.; Lluch, J.; Donnadieu, C.; Parrinello, H.; et al. Generation of a chromosome-level genome assembly for Pacific halibut (Hippoglossus stenolepis) and characterization of its sex-determining genomic region. Mol. Ecol. Resour. 2022, 22, 2685–2700. [Google Scholar] [CrossRef]
- Klüver, N.; Pfennig, F.; Pala, I.; Storch, K.; Schlieder, M.; Froschauer, A.; Gutzeit, H.O.; Schartl, M. Differential expression of anti-Müllerian hormone (AMH) and anti-Müllerian hormone receptor type II (AMHRII) in the teleost medaka. Dev. Dyn. 2007, 236, 271–281. [Google Scholar] [CrossRef]
- Wang, W.; Liang, S.; Zou, Y.; Wu, Z.; Wang, L.; Liu, Y.; You, F. Amh dominant expression in Sertoli cells during the testicular differentiation and development stages in the olive flounder Paralichthys olivaceus. Gene 2020, 755, 144906. [Google Scholar] [CrossRef]
- Lin, C.J.; Jeng, S.R.; Lei, Z.Y.; Yueh, W.S.; Dufour, S.; Wu, G.C.; Chang, C.F. Involvement of transforming growth factor beta family genes in gonadal differentiation in Japanese eel, Anguilla japonica, according to sex-Related gene expressions. Cells 2021, 10, 3007. [Google Scholar] [CrossRef]
- Pfennig, F.; Standke, A.; Gutzeit, H.O. The role of Amh signaling in teleost fish--Multiple functions not restricted to the gonads. Gen. Comp. Endocrinol. 2015, 223, 87–107. [Google Scholar] [CrossRef]
- Shanshan, L.; Bing, S.; Zhuo, L.; Jing, Z.; Songlin, C. Cloning and expression of anti-Müllerian hormone gene in half- smooth tongue-sole, Cynoglossus semilaevis. J. Fish. Sci. China 2013, 20, 35–43, (In Chinese with English Abstract). [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, F.; Liu, S.; Zhao, H.; Lu, W.; Zhang, Q. Transcriptomic analysis reveals functional interaction of mRNA–lncRNA–miRNA in steroidogenesis and spermatogenesis of gynogenetic Japanese flounder (Paralichthys olivaceus). Biology 2022, 11, 213. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Li, M.; Lu, W.; Wang, Y.; Li, X.; Cheng, J. transcriptomic modulation reveals the specific cellular response in Chinese sea bass (Lateolabrax maculatus) gills under salinity change and alkalinity stress. Int. J. Mol. Sci. 2023, 24, 5877. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology in 2010: Extensions and refinements. Nucleic Acids Res. 2010, 38, D331–D335. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Sharman, A.C.; Holland, P.W. Conservation, duplication, and divergence of developmental genes during chordate evolution. Neth. J. Zool. 1995, 46, 47–67. [Google Scholar] [CrossRef]
- Razavi, S.M.; Sabbaghian, M.; Jalili, M.; Divsalar, A.; Wolkenhauer, O.; Salehzadeh-Yazdi, A. Comprehensive functional enrichment analysis of male infertility. Sci. Rep. 2017, 7, 15778. [Google Scholar] [CrossRef]
- Jiang, S.T.; Chiou, Y.Y.; Wang, E.; Lin, H.K.; Lee, S.P.; Lu, H.Y.; Wang, C.K.; Tang, M.J.; Li, H. Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Hum. Mol. Genet. 2008, 17, 3368–3379. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, Y.; Li, N.; Hu, X.; Li, G.; Ding, Y.; Li, J.; Shen, Y.; Wang, X.; Wang, J. Novel compound heterozygous variants in the LHCGR gene identified in a subject with Leydig cell hypoplasia type 1. J. Pediatr. Endocrinol. Metab. 2018, 31, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Akpınar, M.; Lesche, M.; Fanourgakis, G.; Fu, J.; Anastassiadis, K.; Dahl, A.; Jessberger, R. TDRD6 mediates early steps of spliceosome maturation in primary spermatocytes. PLoS Genet. 2017, 13, e1006660. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Li, Y.; Zhi, E.; Shen, G.; Jiang, X.; Li, D.; Zhao, X.; Ruan, T.; Jiang, C.; et al. HSF5 deficiency causes male infertility involving spermatogenic arrest at meiotic prophase i in humans and mice. Adv. Sci. 2024, 11, e2402412. [Google Scholar] [CrossRef]
- Whitfield, M.; Thomas, L.; Bequignon, E.; Schmitt, A.; Stouvenel, L.; Montantin, G.; Tissier, S.; Duquesnoy, P.; Copin, B.; Chantot, S.; et al. Mutations in DNAH17, encoding a sperm-specific axonemal outer dynein arm heavy chain, cause isolated male infertility due to asthenozoospermia. Am. J. Hum. Genet. 2019, 105, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Tsui, V.; Lyu, R.; Novakovic, S.; Stringer, J.M.; Dunleavy, J.E.M.; Granger, E.; Semple, T.; Leichter, A.; Martelotto, L.G.; Merriner, D.J.; et al. Fancm has dual roles in the limiting of meiotic crossovers and germ cell maintenance in mammals. Cell Genom. 2023, 3, 100349. [Google Scholar] [CrossRef]
- Petrillo, C.; Barroca, V.; Ribeiro, J.; Lailler, N.; Livera, G.; Keeney, S.; Martini, E.; Jain, D. shani mutation in mouse affects splicing of Spata22 and leads to impaired meiotic recombination. Chromosoma 2020, 129, 161–179. [Google Scholar] [CrossRef]
- Ghieh, F.; Passet, B.; Poumerol, E.; Castille, J.; Calvel, P.; Vilotte, J.L.; Sellem, E.; Jouneau, L.; Mambu-Mambueni, H.; Garchon, H.J.; et al. A partial deletion within the meiosis-specific sporulation domain SPO22 of Tex11 is not associated with infertility in mice. PLoS ONE 2024, 19, e0309974. [Google Scholar] [CrossRef]
- Zhu, X.; Hu, K.; Cheng, H.; Wu, H.; Li, K.; Gao, Y.; Lv, M.; Xu, C.; Geng, H.; Shen, Q.; et al. Novel MEIOB pathogenic variants including a homozygous non-canonical splicing variant, cause meiotic arrest and human non-obstructive azoospermia. Clin. Genet. 2024, 105, 99–105. [Google Scholar] [CrossRef]
- Kumar, R.; Ghyselinck, N.; Ishiguro, K.; Watanabe, Y.; Kouznetsova, A.; Höög, C.; Strong, E.; Schimenti, J.; Daniel, K.; Toth, A.; et al. MEI4—A central player in the regulation of meiotic DNA double-strand break formation in the mouse. J. Cell Sci. 2015, 128, 1800–1811. [Google Scholar] [CrossRef]
- Brandsma, I.; Sato, K.; van Rossum-Fikkert, S.E.; van Vliet, N.; Sleddens, E.; Reuter, M.; Odijk, H.; van den Tempel, N.; Dekkers, D.H.W.; Bezstarosti, K.; et al. HSF2BP interacts with a conserved domain of BRCA2 and is required for mouse spermatogenesis. Cell Rep. 2019, 27, 3790–3798.e3797. [Google Scholar] [CrossRef]
- Säflund, M.; Özata, D.M. The MYBL1/TCFL5 transcription network: Two collaborative factors with central role in male meiosis. Biochem. Soc. Trans. 2023, 51, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- Shiromoto, Y.; Kuramochi-Miyagawa, S.; Nagamori, I.; Chuma, S.; Arakawa, T.; Nishimura, T.; Hasuwa, H.; Tachibana, T.; Ikawa, M.; Nakano, T. GPAT2 is required for piRNA biogenesis, transposon silencing, and maintenance of spermatogonia in mice. Biol. Reprod. 2019, 101, 248–256. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, K.M.; Pandey, R.R.; Homolka, D.; Reuter, M.; Janeiro, B.K.; Sachidanandam, R.; Fauvarque, M.O.; McCarthy, A.A.; Pillai, R.S. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol. Cell 2016, 61, 138–152. [Google Scholar] [CrossRef]
- Zong, W.; Wang, Y.; Zhang, L.; Lu, W.; Li, W.; Wang, F.; Cheng, J. DNA methylation mediates sperm quality via piwil1 and piwil2 regulation in Japanese flounder (Paralichthys olivaceus). Int. J. Mol. Sci. 2024, 25, 5935. [Google Scholar] [CrossRef]
- Perez, C.A.; Burocziova, M.; Jenikova, G.; Macurek, L. CK1-mediated phosphorylation of FAM110A promotes its interaction with mitotic spindle and controls chromosomal alignment. EMBO Rep. 2021, 22, e51847. [Google Scholar] [CrossRef]
- Ge, C.; Lin, C.; Zhang, M.; Yuan, J.; Feng, X.; Hao, Z.; Zhang, S.; Tian, Q. Tubgcp3 is a mitotic regulator of planarian epidermal differentiation. Gene 2021, 775, 145440. [Google Scholar] [CrossRef] [PubMed]
- Pagan, J.K.; Marzio, A.; Jones, M.J.; Saraf, A.; Jallepalli, P.V.; Florens, L.; Washburn, M.P.; Pagano, M. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat. Cell Biol. 2015, 17, 31–43. [Google Scholar] [CrossRef]
- Yi, Z.Y.; Ma, X.S.; Liang, Q.X.; Zhang, T.; Xu, Z.Y.; Meng, T.G.; Ouyang, Y.C.; Hou, Y.; Schatten, H.; Sun, Q.Y.; et al. Kif2a regulates spindle organization and cell cycle progression in meiotic oocytes. Sci. Rep. 2016, 6, 38574. [Google Scholar] [CrossRef] [PubMed]
- English, M.A.; Lei, L.; Blake, T.; Wincovitch, S.M.; Sood, R.; Azuma, M.; Hickstein, D.; Liu, P.P. Incomplete splicing, cell division defects, and hematopoietic blockage in dhx8 mutant zebrafish. Dev. Dyn. 2012, 241, 879–889. [Google Scholar] [CrossRef]
- Hallacli, E.; Lipp, M.; Georgiev, P.; Spielman, C.; Cusack, S.; Akhtar, A.; Kadlec, J. Msl1-mediated dimerization of the dosage compensation complex is essential for male X-chromosome regulation in Drosophila. Mol. Cell 2012, 48, 587–600. [Google Scholar] [CrossRef]

| Order | Family | Genus | Species | amh | amhr2 | bmpr1 | bmpr2 |
|---|---|---|---|---|---|---|---|
| Anabantiformes | Osphronemidae | Betta | splendens | 1 | 1 | 3 | 1 |
| Cichliformes | Cichlidae | Amphilophus | citrinellus | 1 | 1 | 3 | 2 |
| Cichliformes | Cichlidae | Oreochromis | aureus | 1 | 0 | 1 | 2 |
| Cichliformes | Cichlidae | Oreochromis | niloticus | 1 | 0 | 2 | 2 |
| Cichliformes | Cichlidae | Haplochromis | burtoni | 1 | 1 | 3 | 2 |
| Cichliformes | Cichlidae | Pundamilia | nyererei | 1 | 1 | 3 | 2 |
| Cichliformes | Cichlidae | Maylandia | zebra | 1 | 1 | 2 | 1 |
| Cichliformes | Cichlidae | Astatotilapia | calliptera | 1 | 1 | 3 | 2 |
| Cichliformes | Cichlidae | Neolamprologus | brichardi | 0 | 0 | 3 | 1 |
| Perciformes Acropomatiformes | Lateolabracidae | Lateolabrax | maculatus | 1 | 1 | 3 | 2 |
| Perciformes Acanthuriformes | Moronidae | Dicentrarchus | labrax | 1 | 1 | 2 | 1 |
| Perciformes | Percidae | Perca | flavescens | 1 | 1 | 1 | 2 |
| Perciformes | Percidae | Pseudocaranx | georgianus | 1 | 0 | 1 | 1 |
| Perciformes | Percidae | Sander | lucioperca | 1 | 3 | 3 | 2 |
| Perciformes | Labridae | Labrus | bergylta | 1 | 1 | 3 | 2 |
| Perciformes | Centropomidae | Lates | calcarifer | 1 | 1 | 2 | 2 |
| Perciformes | Bovichthyidae | Cottoperca | gobio | 1 | 1 | 3 | 2 |
| Perciformes | Anabantidae | Anabas | testudineus | 2 | 1 | 3 | 2 |
| Perciformes | Pomacentridae | Amphiprion | ocellaris | 1 | 1 | 3 | 2 |
| Perciformes | Sciaenidae | Larimichthys | crocea | 1 | 1 | 2 | 1 |
| Perciformes | Pomacentridae | Amphiprion | percula | 1 | 1 | 2 | 1 |
| Perciformes | Pomacentridae | Acanthochromis | polyacanthus | 1 | 3 | 2 | 1 |
| Carangiformes | Carangidae | Seriola | dumerili | 1 | 1 | 3 | 2 |
| Carangiformes | Carangidae | Seriola | aureovittata | 1 | 0 | 0 | 0 |
| Carangiformes | Carangidae | Seriola | lalandi | 1 | 1 | 2 | 2 |
| Scorpaeniformes | Cyclopteridae | Cyclopterus | lumpus | 2 | 1 | 3 | 1 |
| Scorpaeniformes | Sebastidae | Sebastes | schlegelii | 2 | 1 | 1 | 2 |
| Scorpaeniformes | Sebastidae | Sebastes | pachycephalus | 2 | 1 | 1 | 1 |
| Scorpaeniformes | Sebastidae | Sebastes | koreanus | 2 | 1 | 1 | 1 |
| Scombriformes | Scombridae | Thunnus | maccoyii | 1 | 0 | 1 | 2 |
| Pleuronectiformes | Soleidae | Solea | senegalensis | 1 | 0 | 0 | 2 |
| Pleuronectiformes | Cynoglossidae | Cynoglossus | semilaevis | 1 | 1 | 3 | 1 |
| Pleuronectiformes | Scophthalmidae | Scophthalmus | maximus | 1 | 1 | 3 | 1 |
| Pleuronectiformes | Paralichthyidae | Paralichthys | olivaceus | 2 | 1 | 2 | 0 |
| Pleuronectiformes | Pleuronectidae | Hippoglossus | hippoglossus | 1 | 0 | 0 | 0 |
| Pleuronectiformes | Pleuronectidae | Hippoglossus | stenolepis | 1 | 0 | 0 | 1 |
| Cyprinodontiformes | Nothobranchiidae | Nothobranchius | furzeri | 1 | 1 | 2 | 1 |
| Cyprinodontiformes | Nothobranchiidae | Nothobranchius | kadleci | 1 | 1 | 2 | 1 |
| Cyprinodontiformes | Poeciliidae | Poecilia | formosa | 1 | 1 | 2 | 2 |
| Cyprinodontiformes | Poeciliidae | Poecilia | reticulata | 1 | 1 | 2 | 1 |
| Cyprinodontiformes | Poeciliidae | Poecilia | latipinna | 1 | 1 | 2 | 2 |
| Cyprinodontiformes | Poeciliidae | Xiphophorus | couchianus | 1 | 0 | 1 | 1 |
| Cyprinodontiformes | Poeciliidae | Xiphophorus | maculatus | 1 | 1 | 2 | 2 |
| Cyprinodontiformes | Rivulidae | Kryptolebias | marmoratus | 1 | 1 | 2 | 1 |
| Cyprinodontiformes | Fundulidae | Fundulus | heteroclitus | 1 | 1 | 2 | 1 |
| Cyprinodontiformes | Cyprinodontidae | Cyprinodon | variegatus | 2 | 1 | 2 | 1 |
| Beloniformes | Adrianichthyidae | Oryzias | latipes | 1 | 1 | 3 | 1 |
| Beloniformes | Adrianichthyidae | Oryzias | sinensis | 1 | 0 | 3 | 1 |
| Beloniformes | Adrianichthyidae | Oryzias | melastigma | 1 | 1 | 2 | 1 |
| Beloniformes | Adrianichthyidae | Oryzias | javanicus | 1 | 1 | 2 | 1 |
| Atheriniformes | Atherinopsidae | Odontesthes | bonariensis | 2 | 1 | 2 | 1 |
| Atheriniformes | Atherinopsidae | Odontesthes | hatcheri | 2 | 1 | 0 | 0 |
| Atheriniformes | Atherinopsidae | Odontesthes | incisa | 2 | 0 | 0 | 0 |
| Gasterosteiformes | Gasterosteidae | Gasterosteus | aculeatus | 1 | 1 | 3 | 1 |
| Syngnathiformes | Syngnathidae | Syngnathoides | biaculeatus | 1 | 1 | 1 | 0 |
| Syngnathiformes | Syngnathidae | Phyllopteryx | taeniolatus | 1 | 1 | 1 | 0 |
| Syngnathiformes | Syngnathidae | Hippocampus | comes | 2 | 1 | 2 | 1 |
| Tetraodontiformes | Tetraodontidae | Takifugu | rubripes | 1 | 1 | 3 | 1 |
| Tetraodontiformes | Tetraodontidae | Takifugu | obscurus | 0 | 1 | 0 | 0 |
| Tetraodontiformes | Tetraodontidae | Tetraodon | nigroviridis | 1 | 0 | 2 | 1 |
| Clupeiformes | Clupeidae | Clupea | harengus | 1 | 1 | 4 | 1 |
| Clupeiformes | Clupeiformes | Denticeps | clupeoides | 2 | 1 | 2 | 2 |
| Gadiformes | Gadidae | Gadus | morhua | 1 | 1 | 3 | 1 |
| Gadiformes | Gadidae | Gadus | macrocephalus | 1 | 1 | 3 | 1 |
| Characiformes | Characidae | Pygocentrus | nattereri | 1 | 1 | 4 | 2 |
| Characiformes | Characidae | Astyanax | mexicanus | 1 | 0 | 4 | 2 |
| Siluriformes | Siluridae | Silurus | meridionalis | 1 | 1 | 1 | 0 |
| Siluriformes | Pangasiidae | Pangasianodon | hypophthalmus | 1 | 1 | 1 | 1 |
| Siluriformes | Ictaluridae | Ictalurus | punctatus | 1 | 1 | 2 | 1 |
| Salmoniformes | Salmonidae | Salvelinus | fontinalis | 1 | 0 | 1 | 0 |
| Salmoniformes | Salmonidae | Oncorhynchus | tshawytscha | 1 | 1 | 2 | 2 |
| Salmoniformes | Salmonidae | Oncorhynchus | mykiss | 1 | 1 | 3 | 2 |
| Salmoniformes | Salmonidae | Salmo | salar | 1 | 1 | 3 | 2 |
| Salmoniformes | Salmonidae | Salmo | trutta | 1 | 1 | 3 | 3 |
| Salmoniformes | Salmonidae | Hucho | hucho | 1 | 1 | 3 | 1 |
| Salmoniformes | Salmonidae | Oncorhynchus | kisutch | 1 | 1 | 4 | 3 |
| Esociformes | Esocidae | Esox | lucius | 1 | 1 | 3 | 2 |
| Osmeriformes | Osmeridae | Plecoglossus | altivelis | 1 | 1 | 0 | 0 |
| Beryciformes | Holocentridae | Myripristis | murdjan | 1 | 1 | 3 | 2 |
| Osteoglossiformes | Osteoglossidae | Arapaima | gigas | 1 | 0 | 2 | 1 |
| Osteoglossiformes | Mormyridae | Paramormyrops | kingsleyae | 1 | 2 | 3 | 2 |
| Cypriniformes | Cyprinidae | Cyprinus | carpio | 1 | 0 | 8 | 4 |
| Cypriniformes | Cyprinidae | Carassius | auratus | 1 | 0 | 8 | 4 |
| Cypriniformes | Cyprinidae | Danio | rerio | 1 | 0 | 4 | 2 |
| Lepisosteiformes | Lepisosteidae | Lepisosteus | oculatus | 1 | 1 | 2 | 1 |
| Synbranchiformes | Mastacembelidae | Mastacembelus | armatus | 1 | 1 | 2 | 1 |
| Gymnotiformes | Electrophoridae | Electrophorus | electricus | 1 | 1 | 4 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhang, Q.; Hu, K.; Lu, W.; Li, W.; Wang, F.; Cheng, J. Evolutionary Dynamics and Functional Conservation of amh Signaling in Teleost Lineages. Fishes 2025, 10, 327. https://doi.org/10.3390/fishes10070327
Zhang L, Zhang Q, Hu K, Lu W, Li W, Wang F, Cheng J. Evolutionary Dynamics and Functional Conservation of amh Signaling in Teleost Lineages. Fishes. 2025; 10(7):327. https://doi.org/10.3390/fishes10070327
Chicago/Turabian StyleZhang, Lingqun, Qingke Zhang, Kai Hu, Wei Lu, Weigang Li, Fengchi Wang, and Jie Cheng. 2025. "Evolutionary Dynamics and Functional Conservation of amh Signaling in Teleost Lineages" Fishes 10, no. 7: 327. https://doi.org/10.3390/fishes10070327
APA StyleZhang, L., Zhang, Q., Hu, K., Lu, W., Li, W., Wang, F., & Cheng, J. (2025). Evolutionary Dynamics and Functional Conservation of amh Signaling in Teleost Lineages. Fishes, 10(7), 327. https://doi.org/10.3390/fishes10070327






