Expression Conditions of Melanogenic Enzymes and Immune Molecular Markers in Atlantic Salmon Muscle During Different Productive Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Sampling
2.2. Tissue Homogenization and RNA Isolation
2.3. Quantitative Real-Time PCR and Statistical Analysis
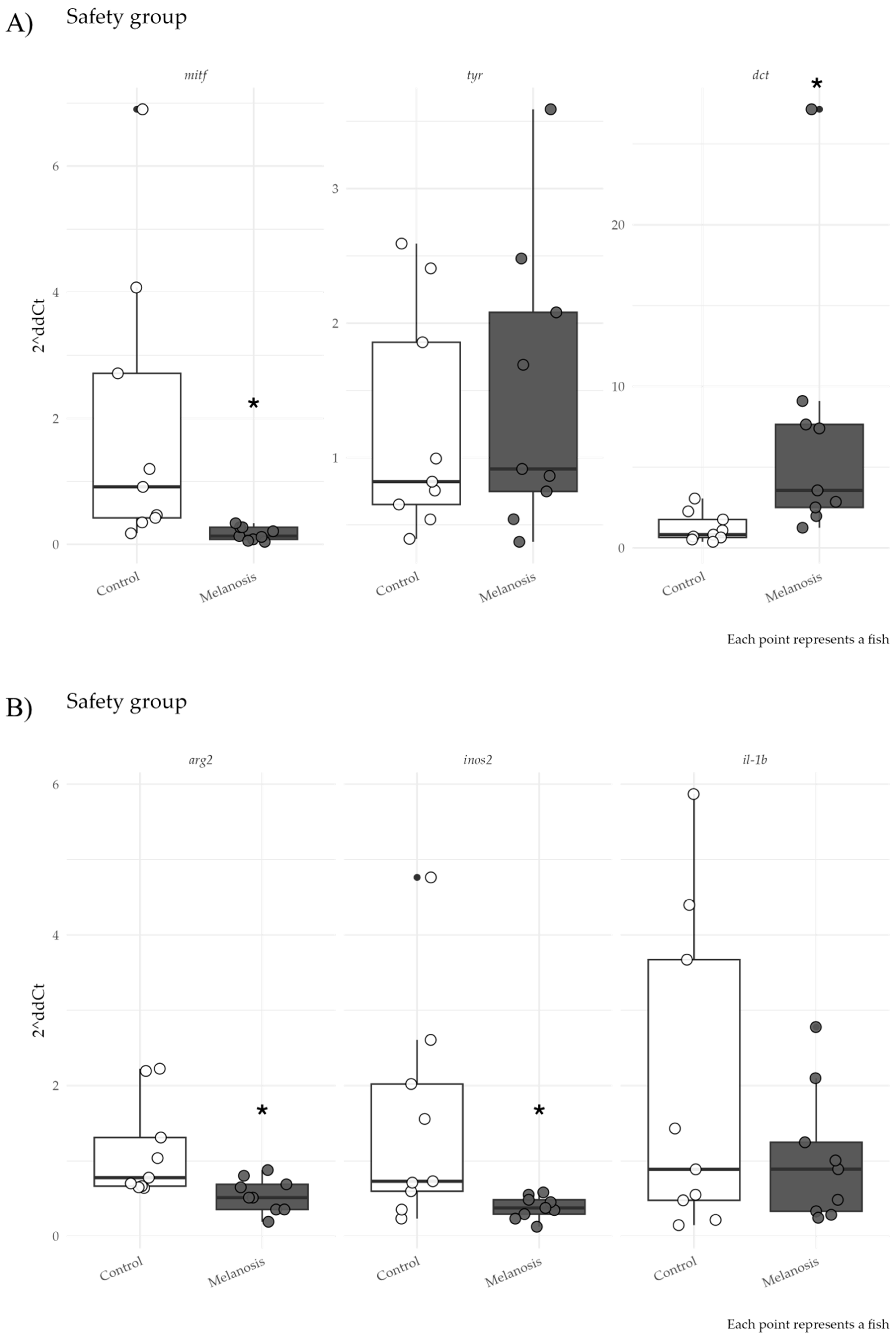
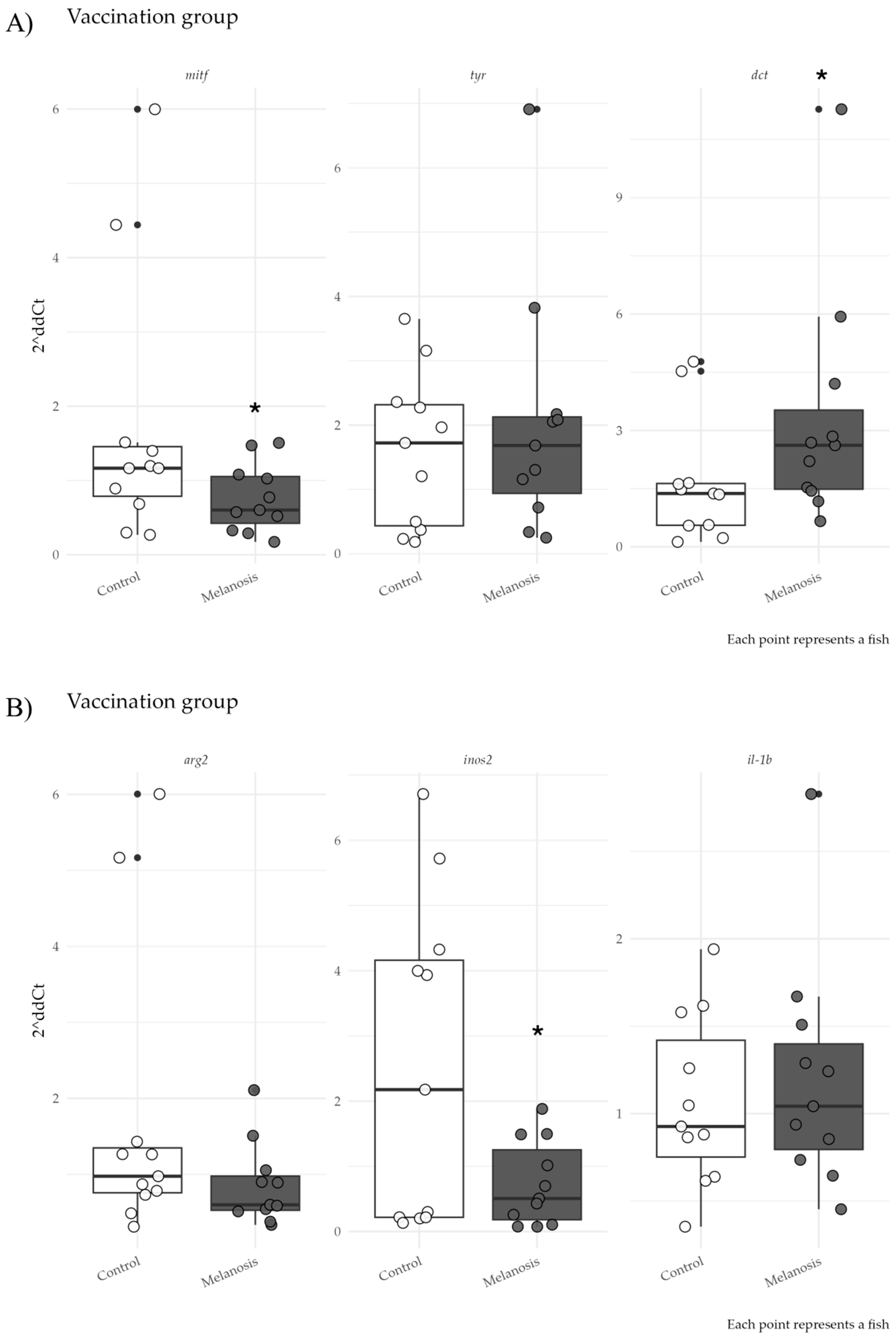
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bjorgen, H.; Kumar, S.; Gunnes, G.; Press, C.M.; Rimstad, E.; Koppang, E.O. Immunopathological characterization of red focal changes in Atlantic salmon (Salmo salar) white muscle. Vet. Immunol. Immunopathol. 2020, 222, 110035. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Moghadam, H.; Larsson, T.; Afanasyev, S.; Morkore, T. Gene expression profiling in melanised sites of Atlantic salmon fillets. Fish Shellfish Immunol. 2016, 55, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Koppang, E.O.; Haugarvoll, E.; Hordvik, I.; Aune, L.; Poppe, T.T. Vaccine-associated granulomatous inflammation and melanin accumulation in Atlantic salmon, Salmo salar L., white muscle. J. Fish Dis. 2005, 28, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Bjorgen, H.; Wessel, O.; Fjelldal, P.G.; Hansen, T.; Sveier, H.; Saebo, H.R.; Enger, K.B.; Monsen, E.; Kvellestad, A.; Rimstad, E.; et al. Piscine orthoreovirus (PRV) in red and melanised foci in white muscle of Atlantic salmon (Salmo salar). Vet. Res. 2015, 46, 89. [Google Scholar] [CrossRef]
- Moretro, T.; Moen, B.; Heir, E.; Hansen, A.A.; Langsrud, S. Contamination of salmon fillets and processing plants with spoilage bacteria. Int. J. Food Microbiol. 2016, 237, 98–108. [Google Scholar] [CrossRef]
- Malik, M.S.; Bjorgen, H.; Nyman, I.B.; Wessel, O.; Koppang, E.O.; Dahle, M.K.; Rimstad, E. PRV-1 Infected Macrophages in Melanized Focal Changes in White Muscle of Atlantic Salmon (Salmo salar) Correlates with a Pro-Inflammatory Environment. Front. Immunol. 2021, 12, 664624. [Google Scholar] [CrossRef]
- Bjarne, H.; Thomas, L.; Tone-Kari, Ø.; Odd Helge, R.; Laura Martinez, R.; Bente, R. Improved fillet quality in harvest-size Atlantic salmon fed high n-3 canola oil as a DHA-source. Aquaculture 2022, 560, 738555. [Google Scholar] [CrossRef]
- Sissener, N.H.; Waagbø, R.; Rosenlund, G.; Tvenning, L.; Susort, S.; Lea, T.B.; Oaland, Ø.; Chen, L.; Breck, O. Reduced n-3 long chain fatty acid levels in feed for Atlantic salmon (Salmo salar L.) do not reduce growth, robustness or product quality through an entire full scale commercial production cycle in seawater. Aquaculture 2016, 464, 236–245. [Google Scholar] [CrossRef]
- Larsen, H.A.; Austbo, L.; Morkore, T.; Thorsen, J.; Hordvik, I.; Fischer, U.; Jirillo, E.; Rimstad, E.; Koppang, E.O. Pigment-producing granulomatous myopathy in Atlantic salmon: A novel inflammatory response. Fish Shellfish Immunol. 2012, 33, 277–285. [Google Scholar] [CrossRef]
- Bjorgen, H.; Haldorsen, R.; Oaland, O.; Kvellestad, A.; Kannimuthu, D.; Rimstad, E.; Koppang, E.O. Melanized focal changes in skeletal muscle in farmed Atlantic salmon after natural infection with Piscine orthoreovirus (PRV). J. Fish Dis. 2019, 42, 935–945. [Google Scholar] [CrossRef]
- Berg, A.; Yurtseva, A.; Hansen, T.; Lajus, D.; Fjelldal, P.G. Vaccinated farmed Atlantic salmon are susceptible to spinal and skull deformities. J. Appl. Ichthyol. 2012, 28, 446–452. [Google Scholar] [CrossRef]
- Brimsholm, M.; Ronning, L.; Rimstad, E.; Koppang, E.O.; Bjorgen, H. Diffuse melanization of the red skeletal musculature in farmed Atlantic salmon (Salmo salar L.). J. Fish Dis. 2023, 46, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Bjorgen, H.; Brimsholm, M.; Asserson, C.F.; Skaar, K.; Knutsen, G.M.; Oaland, O.; Haldorsen, R.; Fjelldal, P.G.; Hansen, T.; Rimstad, E.; et al. Deciphering the pathogenesis of melanized focal changes in the white skeletal muscle of farmed Atlantic salmon (Salmo salar). J. Fish Dis. 2024, 47, e13988. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, K.; Keselman, H.; Algina, J.; Lix, L.; Wilcox, R. Conventional and Robust Paired and Independent-Samples t Tests: Type I Error and Power Rates. J. Mod. Appl. Stat. Methods 2003, 2, 481–496. [Google Scholar] [CrossRef]
- Bjorgen, H.; Rimstad, E.; Koppang, E.O. Melanisation in Salmonid Skeletal Muscle: A Review. J. Fish Dis. 2025, 48, e14063. [Google Scholar] [CrossRef]
- Edvardsen, R.B.; Leininger, S.; Kleppe, L.; Skaftnesmo, K.O.; Wargelius, A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS ONE 2014, 9, e108622. [Google Scholar] [CrossRef]
- Guyonneau, L.; Murisier, F.; Rossier, A.; Moulin, A.; Beermann, F. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol. Cell Biol. 2004, 24, 3396–3403. [Google Scholar] [CrossRef]
- Wang, F.; Ma, W.; Fan, D.; Hu, J.; An, X.; Wang, Z. The biochemistry of melanogenesis: An insight into the function and mechanism of melanogenesis-related proteins. Front. Mol. Biosci. 2024, 11, 1440187. [Google Scholar] [CrossRef]
- Rozanowska, M.; Sarna, T.; Land, E.J.; Truscott, T.G. Free radical scavenging properties of melanin interaction of eu- and pheo-melanin models with reducing and oxidising radicals. Free Radic. Biol. Med. 1999, 26, 518–525. [Google Scholar] [CrossRef]
- Jennings, M.J.; Zumbo, B. The Robustness of Validity and Efficiency of the Related Samples t-Test in the Presence of Outliers. Psicológica 2002, 23, 415–450. [Google Scholar]
- Thorsen, J.; Hoyheim, B.; Koppang, E.O. Isolation of the Atlantic salmon tyrosinase gene family reveals heterogenous transcripts in a leukocyte cell line. Pigment Cell Res. 2006, 19, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Larsen, H.A.; Austbo, L.; Konig, M.; Sorum, H.; Rimstad, E.; Koppang, E.O. Transcription of the tyrosinase gene family in an Atlantic salmon leukocyte cell line (SHK-1) is influenced by temperature, but not by virus infection or bacterin stimulation. Dev. Comp. Immunol. 2013, 41, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Pena, A.; Maldonado, L. Gene expression associated with immune response in Atlantic salmon head-kidney vaccinated with inactivated whole-cell bacterin of Piscirickettsia salmonis and pathogenic isolates. Fish Shellfish Immunol. 2019, 93, 789–795. [Google Scholar] [CrossRef]
- Oberfeld, D.; Franke, T. Evaluating the robustness of repeated measures analyses: The case of small sample sizes and nonnormal data. Behav. Res. Methods 2013, 45, 792–812. [Google Scholar] [CrossRef] [PubMed]
- Vachtenheim, J.; Borovansky, J. “Transcription physiology” of pigment formation in melanocytes: Central role of MITF. Exp. Dermatol. 2010, 19, 617–627. [Google Scholar] [CrossRef]
- de la Serna, I.L.; Ohkawa, Y.; Higashi, C.; Dutta, C.; Osias, J.; Kommajosyula, N.; Tachibana, T.; Imbalzano, A.N. The microphthalmia-associated transcription factor requires SWI/SNF enzymes to activate melanocyte-specific genes. J. Biol. Chem. 2006, 281, 20233–20241. [Google Scholar] [CrossRef]
- Lee, A.; Lim, J.; Lim, J.S. Emerging roles of MITF as a crucial regulator of immunity. Exp. Mol. Med. 2024, 56, 311–318. [Google Scholar] [CrossRef]
- Wan, P.; Hu, Y.; He, L. Regulation of melanocyte pivotal transcription factor MITF by some other transcription factors. Mol. Cell Biochem. 2011, 354, 241–246. [Google Scholar] [CrossRef]
- Lee, W.H.; Ha, Y.; Park, J.I.; Joh, W.B.; Park, M.; Kim, J.K.; Jeon, H.K.; Kim, Y.J. Triglochin maritima Extracts Exert Anti-Melanogenic Properties via the CREB/MAPK Pathway in B16F10 Cells. Mar. Drugs 2024, 22, 532. [Google Scholar] [CrossRef]
- Fang, D.; Tsuji, Y.; Setaluri, V. Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 2002, 30, 3096–3106. [Google Scholar] [CrossRef]
- Wang, P.; Li, Y.; Hong, W.; Zhen, J.; Ren, J.; Li, Z.; Xu, A. The changes of microRNA expression profiles and tyrosinase related proteins in MITF knocked down melanocytes. Mol. Biosyst. 2012, 8, 2924–2931. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, C.A.; Zuloaga, R.; Poblete-Morales, M.; Vera-Tobar, T.; Mercado, L.; Avendano-Herrera, R.; Valdes, J.A.; Molina, A. Fish skeletal muscle tissue is an important focus of immune reactions during pathogen infection. Dev. Comp. Immunol. 2017, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wentzel, A.S.; Petit, J.; van Veen, W.G.; Fink, I.R.; Scheer, M.H.; Piazzon, M.C.; Forlenza, M.; Spaink, H.P.; Wiegertjes, G.F. Transcriptome sequencing supports a conservation of macrophage polarization in fish. Sci. Rep. 2020, 10, 13470. [Google Scholar] [CrossRef] [PubMed]
- Wiegertjes, G.F.; Wentzel, A.S.; Spaink, H.P.; Elks, P.M.; Fink, I.R. Polarization of immune responses in fish: The ‘macrophages first’ point of view. Mol. Immunol. 2016, 69, 146–156. [Google Scholar] [CrossRef]
- Ho, P.-Y.; Byadgi, O.; Wang, P.-C.; Tsai, M.-A.; Liaw, L.-L.; Chen, S.-C. Identification, Molecular Cloning of IL-1? and Its Expression Profile during Nocardia seriolae Infection in Largemouth Bass, Micropterus salmoides. Int. J. Mol. Sci. 2016, 17, 1670. [Google Scholar] [CrossRef]
- Hu, B.; Chen, B.; Mao, M.; Chen, M.; Liu, X.; Cui, Q.; Liu, Y.; Jiang, C. Molecular characterization and expression analysis of the interleukin 1b gene in Pacific cod (Gadus macrocephalus). Dev. Comp. Immunol. 2018, 88, 213–218. [Google Scholar] [CrossRef]
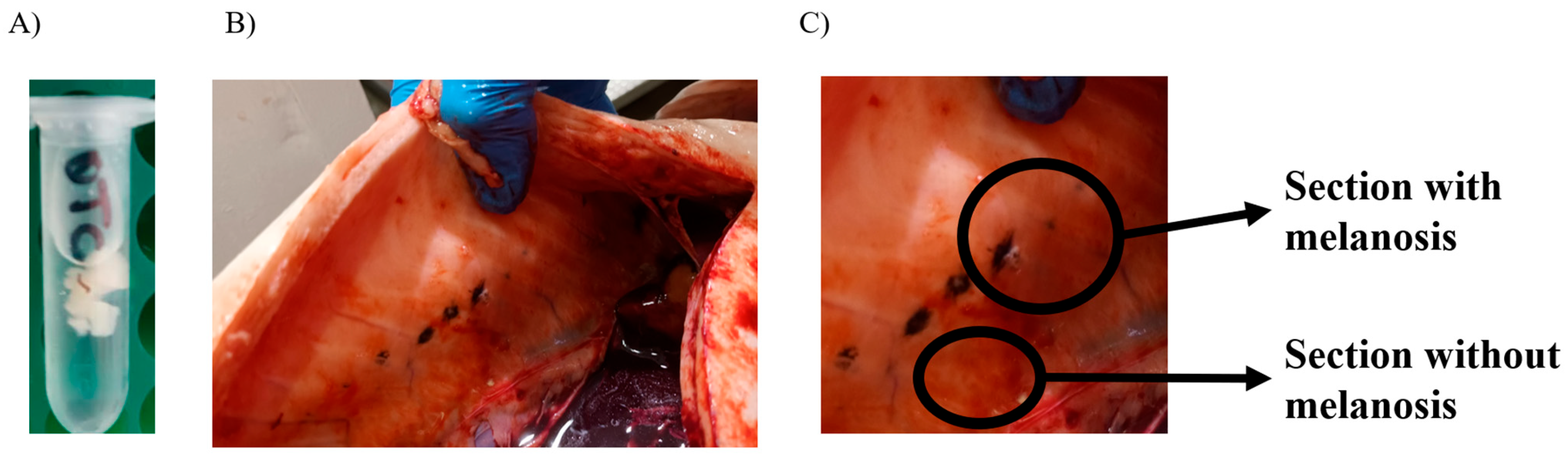

| Group | n | Stage | Weight | Coefficient of Variation | K |
|---|---|---|---|---|---|
| Safety | 9 | Freshwater | 144.61 | 10% | 0.81 |
| Vaccination | 11 | Freshwater | 209.09 | 22% | 1.20 |
| Harvest | 10 | Seawater | 6058.20 | 18% | 1.29 |
| Gene | Primers Sequence (5′→3′) | Product bp | Accession Number or Reference |
|---|---|---|---|
| mitf | gattgagagaagacggaggttt | 100 bp | XM_014136388.2 |
| gcctttgttccaacgcatac | |||
| tyr | tgggaaacaaggtcctgggctac | 111 bp | [21] |
| actgccagatcagctgagcctc | |||
| dct | tctcactctgcagccaatgac | 85 bp | [22] |
| cagacttcctcatccactcatcaaa | |||
| inos2 | catcggcaggattcagtggtccaat | 136 bp | [6] |
| ggtaatcgcagaccttaggtttcctc | |||
| arg2 | cctgaaggacttgggtgtccagta | 109 bp | [6] |
| ccgctgcttccttgacaagaggt | |||
| il-1β | atcaccatgcgtcacattgc | 90 bp | [23] |
| gtccttgaactcggttccca | |||
| β-actin | cccatctacgagggttacgc | 112 bp | AF012125.1 |
| tgaaactgtaaccgcgctct |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés, R.; Valenzuela, C.A.; Johnson, A.; Valenzuela, A.; Valdés, J.A.; Escobar-Aguirre, S. Expression Conditions of Melanogenic Enzymes and Immune Molecular Markers in Atlantic Salmon Muscle During Different Productive Stages. Fishes 2025, 10, 302. https://doi.org/10.3390/fishes10070302
Cortés R, Valenzuela CA, Johnson A, Valenzuela A, Valdés JA, Escobar-Aguirre S. Expression Conditions of Melanogenic Enzymes and Immune Molecular Markers in Atlantic Salmon Muscle During Different Productive Stages. Fishes. 2025; 10(7):302. https://doi.org/10.3390/fishes10070302
Chicago/Turabian StyleCortés, Raúl, Cristián A. Valenzuela, Andrés Johnson, Ariel Valenzuela, Juan Antonio Valdés, and Sebastián Escobar-Aguirre. 2025. "Expression Conditions of Melanogenic Enzymes and Immune Molecular Markers in Atlantic Salmon Muscle During Different Productive Stages" Fishes 10, no. 7: 302. https://doi.org/10.3390/fishes10070302
APA StyleCortés, R., Valenzuela, C. A., Johnson, A., Valenzuela, A., Valdés, J. A., & Escobar-Aguirre, S. (2025). Expression Conditions of Melanogenic Enzymes and Immune Molecular Markers in Atlantic Salmon Muscle During Different Productive Stages. Fishes, 10(7), 302. https://doi.org/10.3390/fishes10070302






