Insights into the Process of Fish Diversity Pattern Changes and the Current Status of Spatiotemporal Dynamics in the Three Gorges Reservoir Area Using eDNA
Abstract
1. Introduction
2. Materials and Methods
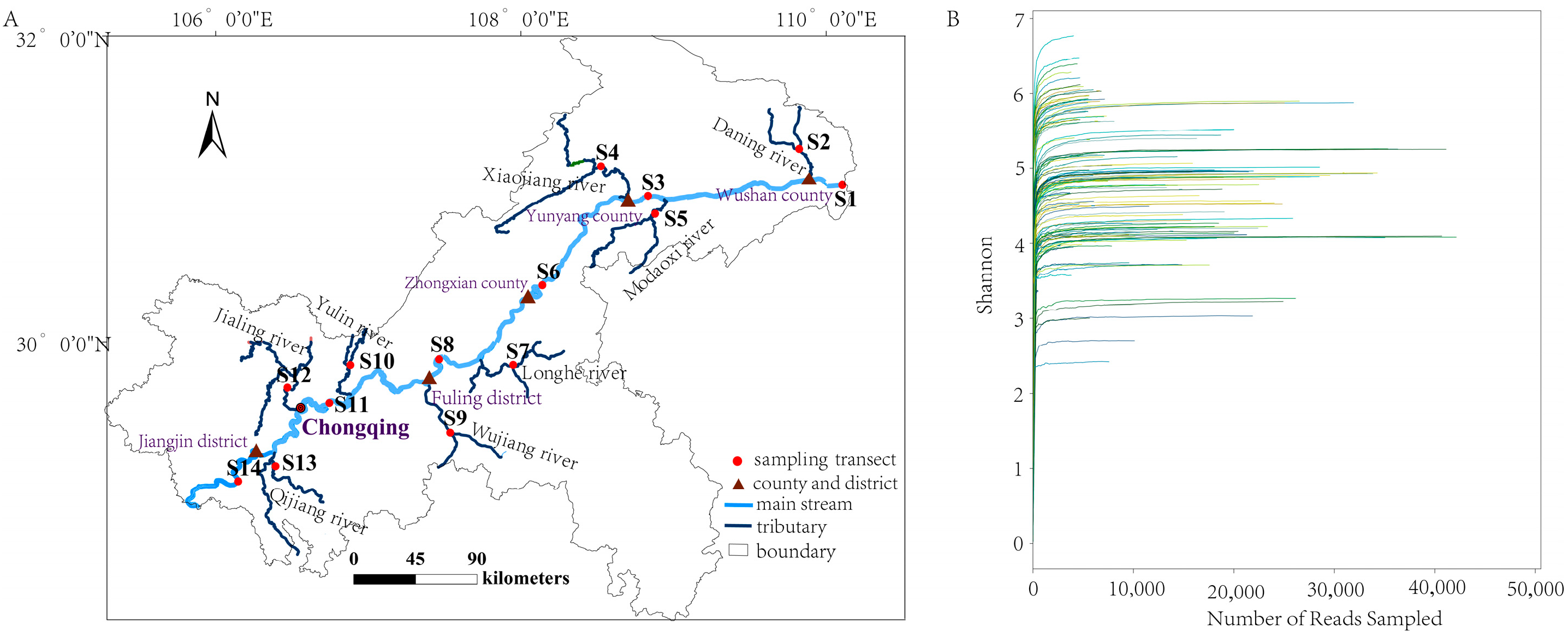
2.1. Study Area and Historical Data Collection
2.2. eDNA Water Sampling and Environmental Properties Measurements
2.3. eDNA Processing, PCR Amplification, and Sequencing
2.4. Bioinformatics Analysis
2.5. Analysis of Temporal and Spatial Changes
2.6. Correlation Analysis of Environmental Factors
3. Results
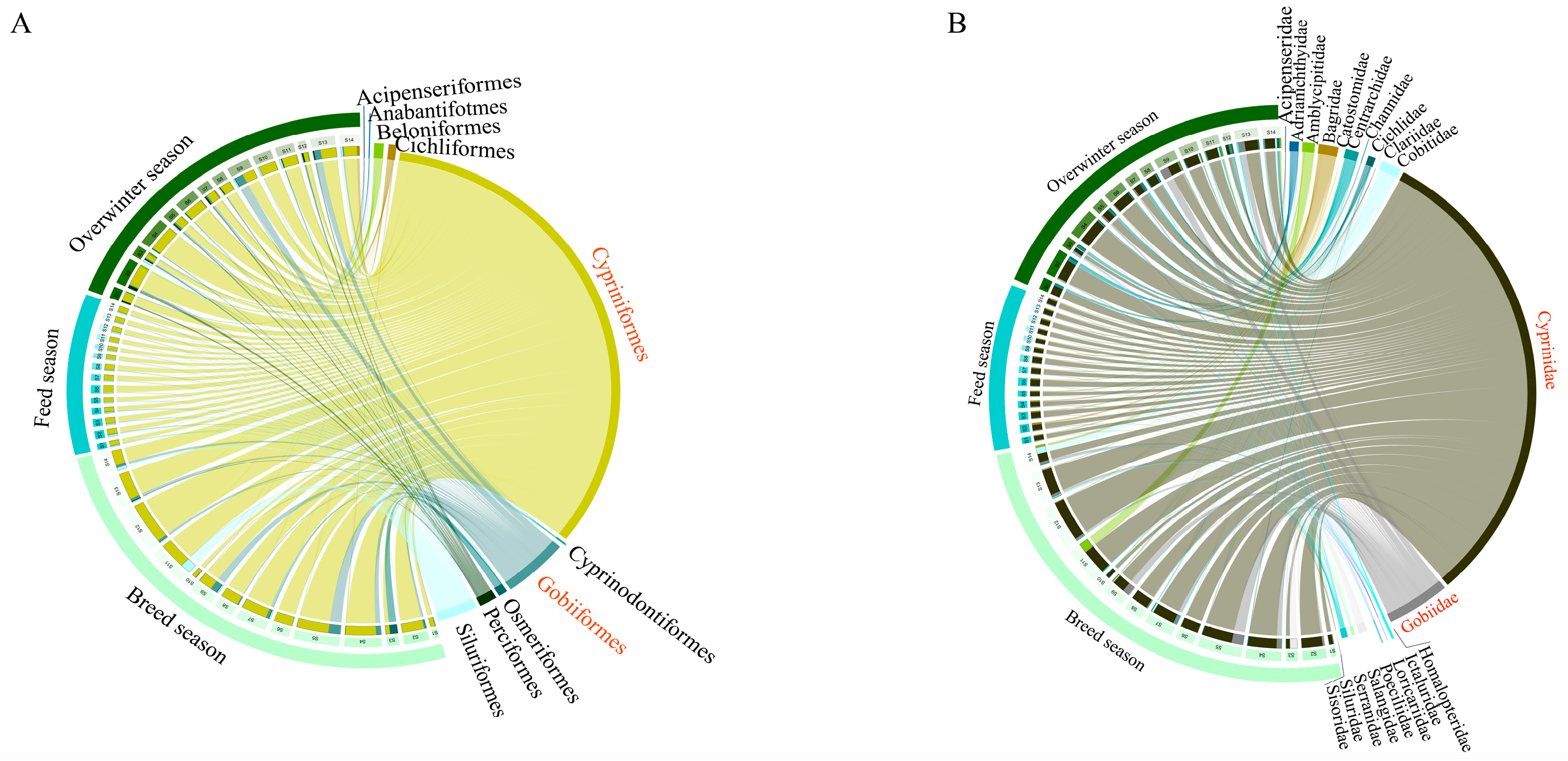
3.1. Current Status of Fish Species Composition
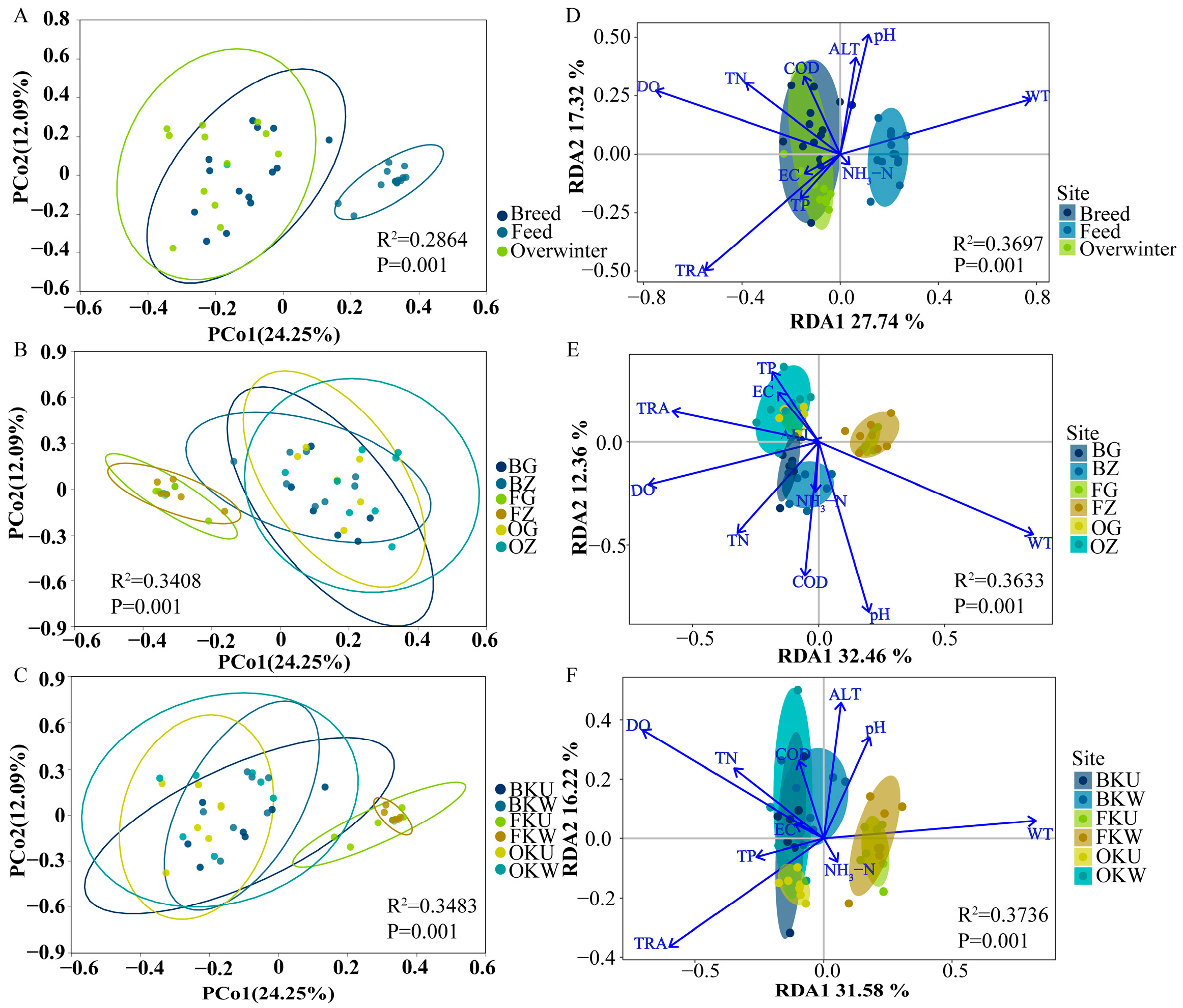
3.2. Changes in Patterns of Fish Community
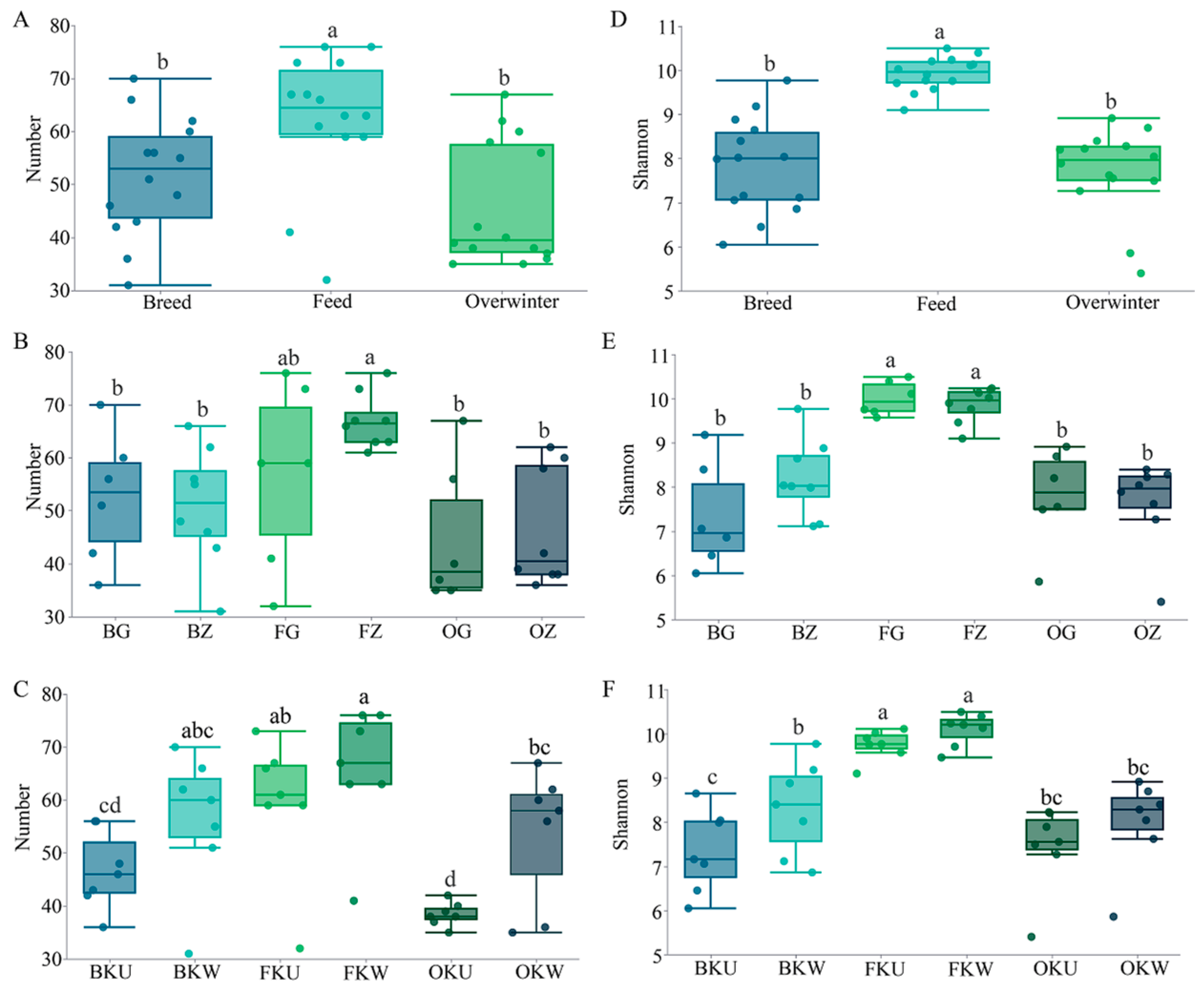
3.3. Current Status of Spatiotemporal Dynamics of Fish Diversity
3.4. Effects of Environmental Factors on Fish Communities
4. Discussion
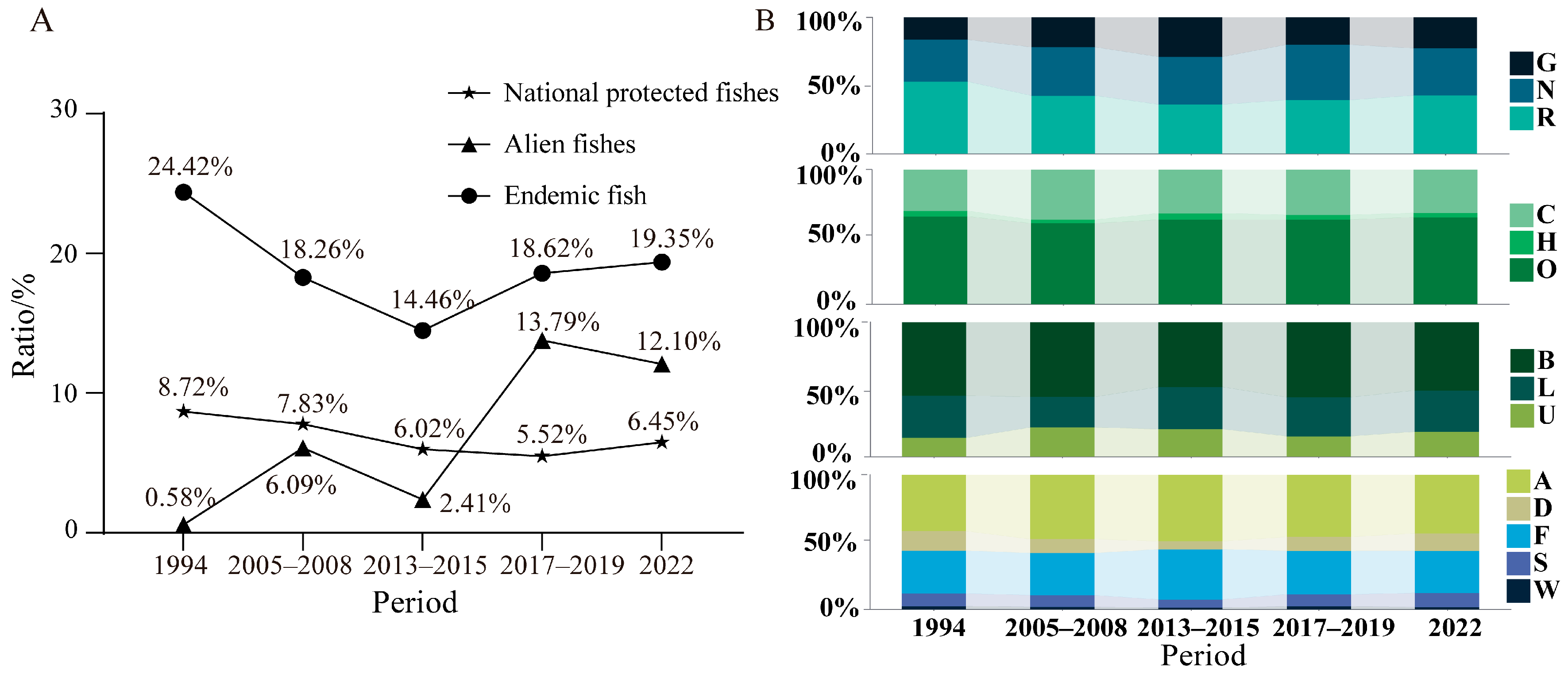
4.1. Reservoir Construction Has Significantly Altered Fish Community Structure
4.2. Significant Differences Between Seasons in Regard to the Fish Community
4.3. The Influence of Environmental Factors on the Spatiotemporal Distribution of Fish
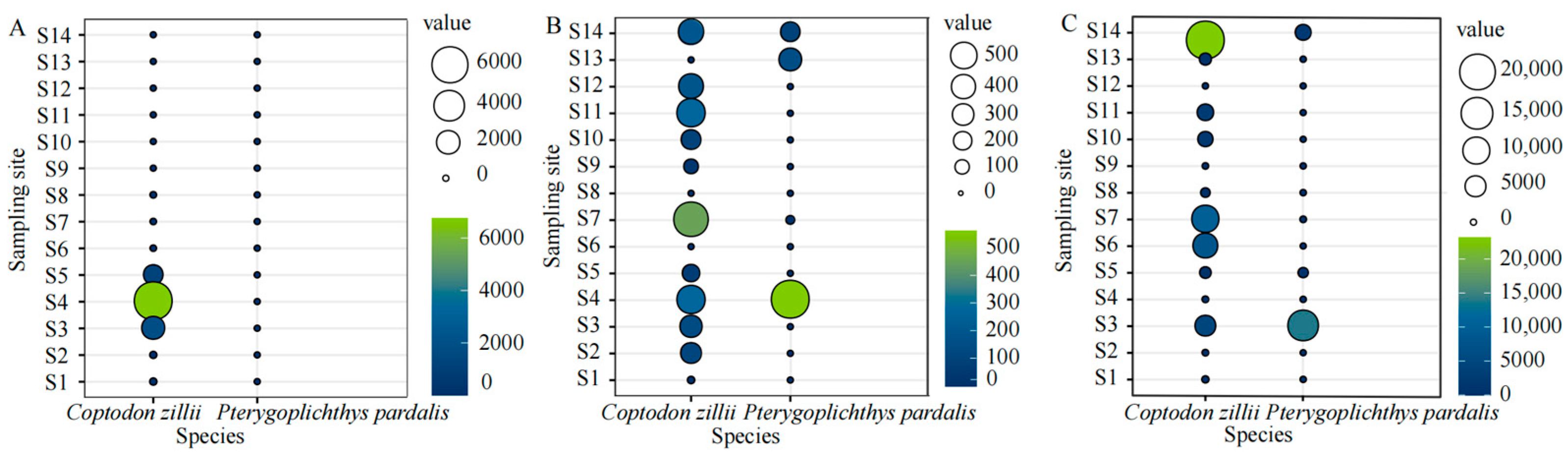
4.4. Alien Fish Need to Be Controlled
4.5. The Limitations of the eDNA Method
4.6. Recommendations for the Conservation of Fish Diversity in Large Reservoirs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Z.; Yu, Y. Investigation of Fish Biodiversity in Baima Lake Based on Environmental DNA Metabarcoding. Environ. Monit. Forewarning 2024, 16, 57–69. [Google Scholar] [CrossRef]
- Zhou, Z.; Tong, A.; Wu, Y.; Shui, B.; Hu, C.; Wang, J. The impact of rare species loss on species diversity and functional diversity of fish communities. Acta Ecol. Sin. 2025, 45, 252–264. [Google Scholar] [CrossRef]
- Gao, X.; Zeng, Y.; Wang, J.; Liu, H. Immediate impacts of the second impoundment on fish communities in the Three Gorges Reservoir. Environ. Biol. Fishes 2010, 87, 163–173. [Google Scholar] [CrossRef]
- Xiang, R.; Wang, L.; Li, H.; Tian, Z.; Zheng, B. Temporal and spatial variation in water quality in the Three Gorges Reservoir from 1998 to 2018. Sci. Total Environ. 2021, 768, 144866. [Google Scholar] [CrossRef]
- Duan, X. Studies on Fish Resources and the Fishes of Early Life History Stage in the Upper Yangtze River. Master’s Thesis, Huazhong Agricultural University, Hangzhou, China, 2008. [Google Scholar]
- Pereira, H.R.; Gomes, L.F.; Soares, P.T.; Martins, P.R.; Pelicice, F.M.; Teresa, F.B.; Vieira, L.C.G. Long-term responses of fish diversity to river regulation: A multi-metric approach. Environ. Biol. Fishes 2021, 104, 71–84. [Google Scholar] [CrossRef]
- Zhang, Q.; Lou, Z. The environmental changes and mitigation actions in the Three Gorges Reservoir region, China. Environ. Sci. Policy 2011, 14, 1132–1138. [Google Scholar] [CrossRef]
- He, Y.; Chen, W.; Chen, J.; Zeng, W.; Zhang, X.; Yang, T.; Zhai, D.; Liu, H.; Xiong, F.; Wang, Y. Application of DNA Barcode for Fish Diversity in the Three Gorges Reservoir Area. Sichuan J. Zool. 2023, 42, 508–518. [Google Scholar] [CrossRef]
- Bao, Y.; Gao, P.; He, X. The water-level fluctuation zone of Three Gorges Reservoir—A unique geomorphological unit. Earth-Sci. Rev. 2015, 150, 14–24. [Google Scholar] [CrossRef]
- Zheng, B.; Zhao, Y.; Qin, Y.; Ma, Y.; Han, C. Input characteristics and sources identification of nitrogen in the three main tributaries of the Three Gorges Reservoir, China. Environ. Earth Sci. 2016, 75, 1219. [Google Scholar] [CrossRef]
- Li, M.; Ma, Q.; Chen, L.; Liu, H. Natural reproduction and hydrologic requirements of pelagophil fish in the three gorges reservoir. Acta Hydrobiol. Sin. 2019, 43, 84–96. [Google Scholar] [CrossRef]
- Wei, N.; Zhang, Y.; Wu, F.; Shen, Z.; Ru, H.; Ni, Z. Current Status and Changes in Fish Assemblages in the Three Gorges Reservoir. Resour. Environ. Yangtze Basin 2021, 30, 1858–1869. [Google Scholar]
- Wu, X. Fish Diversity in Tributaries of the Three Gorges Reservoir and Its Responses to the Reservoir Formation. Master’s Thesis, Southwest University, Chongqing, China, 2021. [Google Scholar] [CrossRef]
- Wei, X.; Hu, H.; Liu, L.; Wang, Y.; Pei, D. Influence of large reservoir operation on change patterns of fish community assembly and growth adaption at low and high-water levels. Ecol. Indic. 2023, 155, 111019. [Google Scholar] [CrossRef]
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; DiBattista, J.D.; Fielder, S.; Wilkes Walburn, J.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2022, 4, 9–33. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.; Gough, K.C. The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight–unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Wang, M.; Yang, S.; Wang, W.; Duan, C.; Liu, Z.; Chen, Q.; Li, Y.; Shen, Y. Fish diversity in chongqing section of the national nature reserve for rare and endemic fish in the upper yangtze river based on eDNA technology. Acta Hydrobiol. Sin. 2022, 46, 2–16. [Google Scholar] [CrossRef]
- Xu, N.; Chang, J. Preliminary Study on Fish Species Detection in the Middle and Lower Yangtze River Using Environmental DNA. J. Hydroecol. 2016, 37, 49–55. [Google Scholar] [CrossRef]
- He, S.; Ni, D.; Xie, W.; Tang, C.; Deng, H.; Duan, X.; Tian, H.; Shao, J.; Wang, K.; Zhang, H. Characteristics and changes of fish distribution in the mainstem of the three gorges reservoir area at the beginning of the ten-year fishing ban. Acta Hydrobiol. Sin. 2024, 49, 022502-1–022502-12. [Google Scholar] [CrossRef]
- Ding, R. Sichuan Fish Chronicles; Sichuan Science and Technology Press: Chengdu, China, 1994. [Google Scholar]
- Shao, X.; Li, D.; Cai, Q. The composition of the fish community in xiangxi bay and resources evaluation. Acta Hydrobiol. Sin. 2006, 30, 70–74. [Google Scholar]
- Wu, Q.; Duan, X.; Xu, S.; Xiong, C.; Chen, D. Studies on Fishery Resources in the Three Gorges Reservoir of the Yangtze River. Freshw. Fish. 2007, 37, 70–75. [Google Scholar] [CrossRef]
- Yang, S.; Gao, X.; Ma, B.; Kong, Y.; Liu, H. Seasonal Dynamics of Fish Community in Mudong Section of the Three Gorges Reservoir of the Yangtze River, China. Chin. J. Appl. Env. Biol. 2010, 16, 555–560. [Google Scholar]
- Dong, C.; Yang, Z.; Gong, Y.; Tang, H. Fish resource status and biodiversity conservation in the main channel of Three Gorges Reservoir. J. Hydroecol. 2019, 40, 15–21. [Google Scholar]
- Luo, J. Using Environmental DNA to Explore Fish Diversity in Lakes of the Central Yunnan Plateau. Master’s Thesis, Yunnan University, Kunming, China, 2019. [Google Scholar]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Chen, Y. Research on the Application of Environmental DNA Technology in the Investigation of Fish Resources in the Upper Liuchong River. Master’s Thesis, Southwest University, Chongqing, China, 2020. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, R. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Schnell, I.B.; Bohmann, K.; Gilbert, M.T.P. Tag jumps illuminated–reducing sequence–to–sample misidentifications in metabarcoding studies. Mol. Ecol. Resour. 2015, 15, 1289–1303. [Google Scholar] [CrossRef]
- Shu, L. Assessment of Fish Diversity Based on Environmental DNA Technology: Kinetics of Embryonic Development, Primer Evaluation and Monitoring of Fish Diversity in Erhai Lake. Ph.D. Thesis, Southwest University, Chongqing, China, 2022. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Li, T.; Tang, L.; Wang, L.; An, L.; Wang, J.; Mo, K.; Chen, Q. Impacts of hydropower development on the distribution of fish populations and changes in ecotypes—A case study of the Xiluodu to Xiangjiaba river section. Acta Ecol. Sin. 2020, 40, 13. [Google Scholar]
- Yang, H.; Shen, L.; He, Y.; Tian, H.; Gao, L.; Wu, J.; Mei, Z.; Wei, N.; Wang, L.; Zhu, T. Status of aquatic organisms resources and their environments in Yangtze River system (2017–2021). Aquac. Fish. 2024, 9, 833–850. [Google Scholar] [CrossRef]
- Yang, M.; Sheng, P.; Zhang, Y.; Guo, L.; Wang, H.; Gao, X.; Chen, Y. Characteristics of fish assemblages of poyang lake at the initial stage of the fishing ban. Acta Hydrobiol. Sin. 2022, 46, 1569–1579. [Google Scholar] [CrossRef]
- Surhone, L.M.; Tennoe, M.T.; Henssonow, S.F. Scheffé’s Method; Wiley: New York, NY, USA, 2010. [Google Scholar]
- Williams, L.J.; Abdi, H. Fisher’s least significant difference (LSD) test. Encycl. Res. Des. 2010, 218, 840–853. [Google Scholar]
- Lai, J.; Zou, Y.; Zhang, J.; Peres-Neto, P.R. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca. hp R package. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Zhang, W. Temporal and Spatial Changes of Fish Community Structure and Diversity in the Three Gorges Reservoir Area. Master’s Thesis, Jianghan University, Wuhan, China, 2023. [Google Scholar]
- Luo, Z.; Zhang, Y.; Zheng, B.; Fu, G.; Lu, Z. Analysis of aquatic environmental characters in Three Gorges Reservoir of China at early reserving stage. Resour. Environ. Yangtze Basin 2005, 14, 780–785. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Y.; Yang, W.; Li, W.; Yang, S. Effect of water temperature on the life activities of temperate freshwater fish. J. Hydroecol. 2021, 42, 117–122. [Google Scholar] [CrossRef]
- Dai, L.; Li, H.; Chen, X. Study on Water Temperature Structure and Its Impact on Water Quality at Reservoir Area. HongShui River 2010, 29, 30–35. [Google Scholar]
- Luo, Z.-X.; Bo, Z.; Zheng, B.-H.; Zhang, Y. Nitrogen and phosphorus loadings in branch backwater reaches and the reverse effects in the main stream in Three Gorges Reservoir. China Environ. Sci. 2007, 27, 208–212. [Google Scholar]
- Wang, X.; Yang, X.; Peng, H.; Dai, J.; Yang, T. Research Progress on Effects of Ammonia Nitrogen Stress on Life Activities of Fish. J. Aquac. 2024, 8, 18–24. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Younes, A.; Saito, H.; Tani, S.; Ikeo, R.; Kawai, K. Metagenomic analysis of gut microbiome from tilapia species across several regions in Japan. Aquaculture 2023, 576, 739809. [Google Scholar] [CrossRef]
- Xing, Y.; Tang, S.; Temesgen, T.; Zhao, J. Invasion and management implications of Coptodon zillii in China. Chin. J. Ecol. 2024, 44, 645–653. [Google Scholar]
- Liu, F.; Wei, H.; Hu, Y.; Gu, D.; Mou, X.; Luo, D.; Xu, M. Age and growth characteristic of invasive fish Pterygoplichthys pardalis in the Liuxi River. Freshw. Fish. 2019, 49, 22–30. [Google Scholar] [CrossRef]
- Zhang, S.; Zheng, Y.; Zhan, A.; Dong, C.; Zhao, J.; Yao, M. Environmental DNA captures native and non-native fish community variations across the lentic and lotic systems of a megacity. Sci. Adv. 2022, 8, eabk0097. [Google Scholar] [CrossRef]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef]
- Shogren, A.J.; Tank, J.L.; Egan, S.P.; Bolster, D.; Riis, T. Riverine distribution of mussel environmental DNA reflects a balance among density, transport, and removal processes. Freshw. Biol. 2019, 64, 1467–1479. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]

| Order | Family | Species (Proportion) | Sequences (Proportion) |
|---|---|---|---|
| Cypriniformes | Cyprinidae | 70 (56.45%) | 4,872,927 (74.12%) |
| Cobitidae | 13 (10.48%) | 200,496 (3.05%) | |
| Homalopteridae | 3 (2.42%) | 22,907 (0.35%) | |
| Catostomidae | 1 (0.81%) | 362 (0.01%) | |
| Siluriformes | Bagridae | 9 (7.26%) | 216,718 (3.3%) |
| Amblycipitidae | 3 (2.42%) | 118,661 (1.8%) | |
| Sisoridae | 3 (2.42%) | 3072 (0.05%) | |
| Siluridae | 2 (1.61%) | 61,210 (0.93%) | |
| Clariidae | 1 (0.81%) | 1018 (0.02%) | |
| Ictaluridae | 1 (0.81%) | 1589 (0.02%) | |
| Loricariidae | 1 (0.81%) | 17,188 (0.26%) | |
| Perciformes | Serranidae | 4 (3.22%) | 33,800 (0.51%) |
| Centrarchidae | 2 (1.61%) | 133,513 (2.03%) | |
| Gobiiformes | Gobiidae | 3 (2.42%) | 613,293 (9.33%) |
| Cichliformes | Cichlidae | 2 (1.61%) | 73,640 (1.12%) |
| Osmeriformes | Salangidae | 2 (1.61%) | 81,411 (1.24%) |
| Acipenseriformes | Acipenseridae | 1 (0.81%) | 6257 (0.09%) |
| Anabantiformes | Channidae | 1 (0.81%) | 8824 (0.13%) |
| Beloniformes | Adrianichthyidae | 1 (0.81%) | 98,840 (1.5%) |
| Cyprinodontiformes | Poeciliidae | 1 (0.81%) | 8973 (0.14%) |
| Sample | Chao1 | Richness | Shannon | Simpson | ACE |
|---|---|---|---|---|---|
| BS1 | 8525.22 | 3822 | 6.0484 | 0.1154 | 8574.17 |
| BS2 | 56,014.62 | 19,850 | 7.1589 | 0.1246 | 65,521.95 |
| BS3 | 29,719.98 | 10,190 | 7.0566 | 0.1104 | 30,245.73 |
| BS4 | 93,441.31 | 27,964 | 7.9901 | 0.0500 | 97,345.04 |
| BS5 | 140,481.70 | 37,240 | 8.6479 | 0.0323 | 156,216.50 |
| BS6 | 49,424.59 | 15,135 | 6.4519 | 0.1638 | 59,649.74 |
| BS7 | 86,236.62 | 23,858 | 8.0394 | 0.0515 | 91,036.82 |
| BS8 | 36,861.74 | 13,016 | 6.8607 | 0.0917 | 38,379.46 |
| BS9 | 34,133.69 | 16,639 | 8.0203 | 0.0308 | 34,761.52 |
| BS10 | 18,585.19 | 9069 | 8.8822 | 0.0117 | 19,828.23 |
| BS11 | 101,692.90 | 34,211 | 8.3998 | 0.0455 | 108,518.10 |
| BS12 | 75,562.51 | 32,580 | 7.1142 | 0.1240 | 81,265.38 |
| BS13 | 93,942.10 | 35,307 | 9.7713 | 0.0150 | 91,855.06 |
| BS14 | 63,992.52 | 24,364 | 9.1828 | 0.0261 | 62,782.31 |
| FS1 | 43,252.42 | 11,782 | 10.1120 | 0.0053 | 46,333.38 |
| FS2 | 36,191.81 | 11,743 | 9.0999 | 0.0254 | 40,027.40 |
| FS3 | 51,309.32 | 12,817 | 9.7572 | 0.0184 | 53,520.79 |
| FS4 | 43,607.63 | 13,004 | 9.9042 | 0.0075 | 47,677.50 |
| FS5 | 29,127.04 | 11,552 | 9.7682 | 0.0060 | 32,985.07 |
| FS6 | 43,148.62 | 15,491 | 9.5746 | 0.0091 | 47,538.16 |
| FS7 | 41,431.73 | 13,807 | 10.0231 | 0.0066 | 47,616.64 |
| FS8 | 40,782.17 | 13,886 | 9.7092 | 0.0068 | 47,026.40 |
| FS9 | 27,423.30 | 10,647 | 9.4643 | 0.0097 | 31,990.83 |
| FS10 | 34,713.30 | 9343 | 10.2063 | 0.0064 | 38,112.42 |
| FS11 | 39,974.01 | 11,501 | 10.3964 | 0.0052 | 40,562.15 |
| FS12 | 51,773.67 | 12,874 | 10.2381 | 0.0056 | 55,564.40 |
| FS13 | 46,715.31 | 12,553 | 10.1364 | 0.0070 | 50,341.60 |
| FS14 | 55,802.94 | 15,064 | 10.4963 | 0.0060 | 60,927.68 |
| OS1 | 19,451.27 | 9771 | 7.4945 | 0.0471 | 18,882.00 |
| OS2 | 51,074.54 | 19,503 | 7.8920 | 0.0449 | 48,498.48 |
| OS3 | 21,029.19 | 10,491 | 8.2038 | 0.0236 | 20,231.17 |
| OS4 | 24,421.12 | 11,635 | 5.4016 | 0.2207 | 23,544.02 |
| OS5 | 24,640.14 | 11,612 | 8.2268 | 0.0255 | 23,687.44 |
| OS6 | 43,024.33 | 16,245 | 7.5536 | 0.0762 | 42,510.28 |
| OS7 | 20,677.22 | 9145 | 7.2673 | 0.0533 | 19,769.00 |
| OS8 | 18,092.01 | 8625 | 5.8583 | 0.1736 | 19,465.20 |
| OS9 | 37,733.27 | 16,087 | 7.6211 | 0.0515 | 37,498.15 |
| OS10 | 39,206.78 | 16,664 | 8.2844 | 0.0381 | 38,735.13 |
| OS11 | 35,626.10 | 17,224 | 8.6965 | 0.0222 | 35,421.48 |
| OS12 | 17,542.52 | 8614 | 8.0434 | 0.0326 | 17,289.62 |
| OS13 | 48,119.58 | 19,116 | 8.3985 | 0.0323 | 47,007.66 |
| OS14 | 45,194.01 | 19,690 | 8.9154 | 0.0206 | 46,244.19 |
| Pairs | R2 | p |
|---|---|---|
| Breed vs. Feed | 0.2895 | 0.0010 ** |
| Breed vs. Overwinter | 0.0981 | 0.0010 ** |
| Feed vs. Overwinter | 0.3501 | 0.0010 ** |
| BG vs. BZ | 0.0862 | 0.3508 |
| BG vs. FG | 0.3399 | 0.0050 ** |
| BG vs. FZ | 0.3463 | 0.0025 ** |
| BG vs. OG | 0.1449 | 0.0538 |
| BG vs. OZ | 0.1230 | 0.0538 |
| BZ vs. FG | 0.3114 | 0.0025 ** |
| BZ vs. FZ | 0.3147 | 0.0025 ** |
| BZ vs. OG | 0.1506 | 0.0038 ** |
| BZ vs. OZ | 0.1340 | 0.0120 * |
| FG vs. FZ | 0.0494 | 0.8690 |
| FG vs. OG | 0.3721 | 0.0038 ** |
| FG vs. OZ | 0.3728 | 0.0025 ** |
| FZ vs. OG | 0.3768 | 0.0025 ** |
| FZ vs. OZ | 0.3831 | 0.0025 ** |
| OG vs. OZ | 0.0693 | 0.6332 |
| BKU vs. BKW | 0.0831 | 0.3490 |
| BKU vs. FKU | 0.2829 | 0.0025 ** |
| BKU vs. FKW | 0.3231 | 0.0038 ** |
| BKU vs. OKU | 0.1351 | 0.0135 * |
| BKU vs. OKW | 0.1231 | 0.0259 * |
| BKW vs. FKU | 0.3412 | 0.0025 ** |
| BKW vs. FKW | 0.3776 | 0.0025 ** |
| BKW vs. OKU | 0.1950 | 0.0025 ** |
| BKW vs. OKW | 0.1169 | 0.1313 |
| FKU vs. FKW | 0.0978 | 0.2331 |
| FKU vs. OKU | 0.3986 | 0.0025 ** |
| FKU vs. OKW | 0.3414 | 0.0083 ** |
| FKW vs. OKU | 0.4492 | 0.0038 ** |
| FKW vs. OKW | 0.3723 | 0.0025 ** |
| OKU vs. OKW | 0.0892 | 0.3386 |
| Name | Unique | Average. Share | Individual | I. Perc (%) | p |
|---|---|---|---|---|---|
| WT | 0.0345 | 0.0287 | 0.0632 | 19.87 | 0.001 ** |
| DO | 0.0162 | 0.0250 | 0.0412 | 12.96 | 0.001 ** |
| TRA | 0.0265 | 0.0107 | 0.0372 | 11.70 | 0.003 ** |
| pH | 0.0107 | 0.0211 | 0.0318 | 10.00 | 0.001 ** |
| NH3-N | 0.0269 | 0.0014 | 0.0283 | 8.90 | 0.044 * |
| ALT | 0.0215 | 0.0043 | 0.0258 | 8.11 | 0.958 |
| TN | 0.0097 | 0.0143 | 0.0240 | 7.55 | 0.001 ** |
| COD | 0.0197 | 0.0030 | 0.0227 | 7.14 | 0.384 |
| TP | 0.0169 | 0.0056 | 0.0225 | 7.08 | 0.007 ** |
| EC | 0.0226 | 0.0008 | 0.0218 | 6.86 | 0.503 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhang, Y.; Dong, X.; Zhou, X.; Liu, Z.; Chen, Q.; Chen, F.; Shen, Y. Insights into the Process of Fish Diversity Pattern Changes and the Current Status of Spatiotemporal Dynamics in the Three Gorges Reservoir Area Using eDNA. Fishes 2025, 10, 295. https://doi.org/10.3390/fishes10060295
Huang J, Zhang Y, Dong X, Zhou X, Liu Z, Chen Q, Chen F, Shen Y. Insights into the Process of Fish Diversity Pattern Changes and the Current Status of Spatiotemporal Dynamics in the Three Gorges Reservoir Area Using eDNA. Fishes. 2025; 10(6):295. https://doi.org/10.3390/fishes10060295
Chicago/Turabian StyleHuang, Jiaxin, Yufeng Zhang, Xiaohan Dong, Xinxin Zhou, Zhihao Liu, Qiliang Chen, Fan Chen, and Yanjun Shen. 2025. "Insights into the Process of Fish Diversity Pattern Changes and the Current Status of Spatiotemporal Dynamics in the Three Gorges Reservoir Area Using eDNA" Fishes 10, no. 6: 295. https://doi.org/10.3390/fishes10060295
APA StyleHuang, J., Zhang, Y., Dong, X., Zhou, X., Liu, Z., Chen, Q., Chen, F., & Shen, Y. (2025). Insights into the Process of Fish Diversity Pattern Changes and the Current Status of Spatiotemporal Dynamics in the Three Gorges Reservoir Area Using eDNA. Fishes, 10(6), 295. https://doi.org/10.3390/fishes10060295







