Application of Flow Cytometry to Determine Cell DNA Content in the Genetic Breeding of Fish
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Methods
2.2.1. Selection of Internal Standards
2.2.2. Preparation of Nuclear Suspension
- (1)
- Embryos: Take one (or more) embryos from the gastrulation stage to the caudal stage and place them in a small culture dish. Cut them into pieces with small scissors, add a small amount of PBS, transfer them to a sterilized 1.5 mL Eppendorf tube (EP tube), and keep them stationary for 2 min. Use a pipette to aspirate the bottom sediment and reserve the cell suspension for later use.
- (2)
- Fry: Take a single fry within 15 days after hatching (or 3 to 5 mixed fry), peel the abdominal cavity in a small Petri dish with fine tweezers, remove intestinal feces and other debris, cut it with small scissors, add a small amount of PBS and transfer it to a 1.5 mL EP tube, keep it stationary for 2 min, and then take the upper cell suspension for later use.
- (3)
- Blood: Collect 0.2 mL ACD anticoagulant solution with a syringe, and, after the fish tail is disinfected with complex iodine, take 0.2–0.3 mL blood from below the lateral line. Store the sample in a 1.5 mL EP tube and dilute it with PBS for later use.
- (4)
- Sperm: Aspirate 10 μL of sperm from the cloaca of adult fish with a 10–200 μL pipette and store in a 1.5 mL EP tube, diluted with PBS for later use.
- (5)
- Caudal fins: Use ophthalmic scissors to cut a part of the caudal fin tissue and place it in a 1.5 mL EP tube, add 50 μL of ACD, cut the tissue into a cell suspension with ophthalmic scissors, keep it stationary for 2 min, and then take the upper cell suspension for later use.
- (6)
- Gonadal tissues: In this study, the early ovaries of female fish before stage II development and the sperms of males at each stage of development were all suitable for FCM for ploidy detection. Via ophthalmology, extract ovarian or testicular tissues of the size of a mung bean and cut the tissues into pieces with small scissors in a small Petri dish. Add a small amount of PBS and transfer the sample to a 1.5 mL EP tube, keep it stationary for 2 min, and take the upper cell suspension for later use.
- (7)
- Livers, gills, spleens, and other tissues: Use ophthalmic scissors to cut liver, gill, and spleen tissues of the size of a mung bean, place them in a small Petri dish, cut them into pieces with small scissors, add a small amount of PBS, transfer them to a 1.5 mL EP tube, keep them stationary for 2 min, and take the upper cell suspension for later use.
2.2.3. Specific Staining of DNA in Nuclear Suspension
2.2.4. Detection via FCM
3. Results and Analysis
3.1. Detection at the Embryo and Fry Stages
3.2. In Vivo Detection of Adult Fish
3.3. Detection of Specific Organizations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wade, D.; Diaw, P.A.; Daneau, G.; Camara, M.; Dieye, T.N.; Mboup, S.; Kestens, L. CD4 T-cell enumeration in a field setting: Evaluation of CyFlow counter using the CD4 easy count kit-dry and Pima CD4 systems. PLoS ONE 2013, 8, e75484. [Google Scholar] [CrossRef] [PubMed]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Drescher, H.; Weiskirchen, S.; Weiskirchen, R. Flow cytometry: A blessing and a curse. Biomedicines 2021, 9, 1613. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.P. The evolution of spectral flow cytometry. Cytom. Part A 2022, 101, 812–817. [Google Scholar] [CrossRef]
- Fleisher, T.A.; Oliveira, J.B. Flow cytometry. In Clinical Immunology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1239–1251.e1. [Google Scholar]
- Lu, Y.K.; Liu, W.L.; Hu, Z.X. Progress in Standardization of Fluorescence Flow Cytometer. Laser Optoelectron. Prog. 2023, 60, 38–50. [Google Scholar]
- Wilkinson, M.G. Flow Cytometry in Microbiology; Wilkinson Department of Life Sciences, University of Limerick: Limerick, Ireland, 2015. [Google Scholar] [CrossRef]
- Lei, T.J.; Chen, W.; Chen, Z.C.; Tang, X.H.; Zhang, Z.H.; Tan, C.; Zhu, F.Y.; Liao, A.W.; Xiao, C.L.; Yin, S.M.; et al. Thermal Analysis and Design of Multichannel Flow Cytometry Laser Platform. Laser Optoelectron. Prog. 2024, 61, 326–334. [Google Scholar]
- Hu, Z.B.; Li, B.; Qu, S.F. Analysis to the Key Points of Quality Control for Flow Cytometry Detection Reagents. Chin. Pharm. Aff. 2021, 35, 1142–1148. [Google Scholar]
- Gonneau, C.; Wang, L.; Mitra-Kaushik, S.; Trampont, P.; Litwin, V. Progress towards global standardization for quantitative flow cytometry. Bioanalysis 2021, 13, 1591–1595. [Google Scholar] [CrossRef]
- Zhao, S.T.; Wu, X.D.; Wang, C.; Chen, Y.Q.; Tang, Y.G. Principles, applications and latest developments of flow cytometer. Progress. Mod. Biomed. 2011, 11, 4378–4381. [Google Scholar]
- Wang, S.; Tang, C.; Tao, M.; Qin, Q.; Zhang, C.; Luo, K.; Zhao, R.; Wang, J.; Ren, L.; Xiao, J.; et al. Establishment and application of distant hybridization technology in fish. Sci. China Life Sci. 2019, 62, 22–45. [Google Scholar] [CrossRef]
- Gala de Pablo, J.; Lindley, M.; Hiramatsu, K.; Goda, K. High-throughput Raman flow cytometry and beyond. Acc. Chem. Res. 2021, 54, 2132–2143. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, Y.; Ning, H.X.; Wang, T.; Yin, S. Review on genetic breeding progress of Bagridae family in China. J. Fish. China 2024, 48, 93–109. [Google Scholar]
- Yang, S.; Zeng, K.; Luo, L.; Qian, W.; Wang, Z.; Doležel, J.; Zhang, M.; Gao, X.; Deng, Z. A flow cytometry-based analysis to establish a cell cycle synchronization protocol for Saccharum spp. Sci. Rep. 2020, 10, 5016. [Google Scholar] [CrossRef]
- Feng, X.; Chen, H.; Xiao, B.; Wu, Q.; Zhang, J.; Zhang, N.; Li, P.; Wang, L.; Yin, J.; Sui, Z. Ploidy Identification by Flow Cytometry and Application of the Method to Characterize Seasonal Ploidy Variation of Wild Populations of the Red Alga Gracilariopsis lemaneiformis. Mar. Biotechnol. 2022, 24, 1073–1083. [Google Scholar] [CrossRef]
- Chen, H.; Feng, X.; Jiang, M.; Xiao, B.; Zhang, J.; Zhang, W.; Hu, Y.; Sui, Z. Estimating the ploidy of Gracilariopsis lemaneiformis at both the cellular and genomic level1. J. Phycol. 2020, 56, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Iturri, L.; Coronilla, J.S.; Lallemand, Y.; Perdiguero, E.G. Identification of erythromyeloid progenitors and their progeny in the mouse embryo by flow cytometry. J. Vis. Exp. JoVE 2017, 125, 55305. [Google Scholar]
- Yu, Y.-R.A.; O’koren, E.G.; Hotten, D.F.; Kan, M.J.; Kopin, D.; Nelson, E.R.; Que, L.; Gunn, M.D. A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS ONE 2016, 11, e0150606. [Google Scholar] [CrossRef]
- Doležel, J.; Lucretti, S.; Molnár, I.; Cápal, P.; Giorgi, D. Chromosome analysis and sorting. Cytom. Part A 2021, 99, 328–342. [Google Scholar] [CrossRef]
- Kanegane, H.; Hoshino, A.; Okano, T.; Yasumi, T.; Wada, T.; Takada, H.; Okada, S.; Yamashita, M.; Yeh, T.-W.; Nishikomori, R.; et al. Flow cytometry-based diagnosis of primary immunodeficiency diseases. Allergol. Int. 2018, 67, 43–54. [Google Scholar] [CrossRef]
- de Azambuja, A.P.; Mion, A.L.V.; Schluga, Y.C.; Beltrame, M.P.; Senegaglia, A.C.; Funke, V.A.M.; Bonfim, C.; Pasquini, R. Comprehensive Analysis of High-Sensitive Flow Cytometry and Molecular Mensurable Residual Disease in Philadelphia Chromosome-Positive Acute Leukemia. Int. J. Mol. Sci. 2025, 26, 2116. [Google Scholar] [CrossRef]
- Bonar, M.M.; Tilton, J.C. High sensitivity detection and sorting of infectious human immunodeficiency virus (HIV-1) particles by flow virometry. Virology 2017, 505, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Ikoma-Colturato, M.R.V.; Bertolucci, C.M.; Conti-Spilari, J.E.; Oliveira, E.; Simioni, A.J.; Figueredo-Pontes, L.L.; Furtado, F.M.; Alegretti, A.P.; Azambuja, A.P.; Gevert, F.; et al. Multicentric standardization of minimal/measurable residual disease in B-cell precursor acute lymphoblastic leukaemia using next-generation flow cytometry in a low/middle-level income country. Br. J. Haematol. 2022, 200, 381. [Google Scholar] [CrossRef] [PubMed]
- Musich, T.; Jones, J.C.; Keele, B.F.; Jenkins, L.M.M.; Robert-Guroff, M. Flow virometric sorting and analysis of HIV quasispecies from plasma. JCI Insight 2017, 2, e90626. [Google Scholar] [CrossRef]
- Longin, C.; Petitgonnet, C.; Guilloux-Benatier, M.; Rousseaux, S.; Alexandre, H. Application of flow cytometry to wine microorganisms. Food Microbiol. 2017, 62, 221–231. [Google Scholar] [CrossRef]
- Zand, E.; Froehling, A.; Schoenher, C.; Zunabovic-Pichler, M.; Schlueter, O.; Jaeger, H. Potential of flow cytometric approaches for rapid microbial detection and characterization in the food industry—A review. Foods 2021, 10, 3112. [Google Scholar] [CrossRef]
- Serpaggi, V.; Remize, F.; Grand, A.S.; Alexandre, H. Specific identification and quantification of the spoilage microorganism Brettanomyces in wine by flow cytometry: A useful tool for winemakers. Cytom. Part A 2010, 77, 497–499. [Google Scholar] [CrossRef]
- Atanasova, M.; Yordanova, G.; Nenkova, R.; Ivanov, Y.; Godjevargova, T.; Dinev, D. Brewing yeast viability measured using a novel fluorescent dye and image cytometer. Biotechnol. Biotechnol. Equip. 2019, 33, 548–558. [Google Scholar] [CrossRef]
- Wang, Y.; Hammes, F.; De Roy, K.; Verstraete, W.; Boon, N. Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol. 2010, 28, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.C.; Gomes, T.; Habuda-Stanić, M.; Lomba, J.A.B.; Romić, Ž.; Turkalj, J.V.; Lillicrap, A. Characterization of multiple biomarker responses using flow cytometry to improve environmental hazard assessment with the green microalgae Raphidocelis subcapitata. Sci. Total environment 2019, 687, 827–838. [Google Scholar] [CrossRef]
- Wang, D.; Reyes, F.L.d.L.; Ducoste, J.J. Enhancing Microalgae Cell Inactivation Through Hydrodynamic Cavitation: Insights from Flow Cytometry Analysis. Water Res. 2025, 277. [Google Scholar] [CrossRef]
- Pulikkot, S.; Paul, S.; Hall, A.; Gardner, B.; Liu, W.; Hu, L.; Vella, A.T.; Chen, Y.; Fan, Z. Monitoring Circulating Myeloid Cells in Peritonitis with an In Vivo Imaging Flow Cytometer. Biomolecules 2024, 14, 886. [Google Scholar] [CrossRef] [PubMed]
- Linzke, M.; Yan, S.L.R.; Tárnok, A.; Ulrich, H.; Groves, M.R.; Wrenger, C. Live and let dye: Visualizing the cellular compartments of the malaria parasite Plasmodium falciparum. Cytom. Part A 2020, 97, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Lal, J.; Vaishnav, A.; Singh, S.K.; Meena, D.K.; Biswas, P.; Mehta, N.K.; Priyadarshini, M.B. Biotechnological innovation in fish breeding: From marker assisted selection to genetic modification. Discov. Biotechnol. 2024, 1, 7–21. [Google Scholar] [CrossRef]
- Manohar, S.M.; Shah, P.; Nair, A. Flow cytometry: Principles, applications and recent advances. Bioanalysis 2022, 13, 185–202. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, F.; Li, B.; Duan, L.; Yu, J.; Huang, Z.; Huang, S.; Liu, H.; Liu, Q.; Liu, S. Multi-omics reveals the molecular mechanisms of rapid growth in distant hybrid fish. Aquaculture 2025, 596, 741783. [Google Scholar] [CrossRef]
- Manan, H.; Hidayati, A.N.; Lyana, N.A.; Amin-Safwan, A.; Ma, H.; Kasan, N.A.; Ikhwanuddin, M. A review of gynogenesis manipulation in aquatic animals. Aquac. Fish. 2022, 7, 1–6. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, S.; Tang, C.; Tao, M.; Zhang, C.; Zhou, Y.; Qin, Q.; Luo, K.; Wu, C.; Hu, F.; et al. The Research Advances in Distant Hybridization and Gynogenesis in Fish. Rev. Aquac. 2025, 17, e12972. [Google Scholar] [CrossRef]
- Liu, S.J.; Li, S.F.; Liu, Y. The Early Embryonic Development of Allotetraploid Hybrids of Red Crucian Carp (♀) × Common Carp (♂). J. Naturnal Sci. Hunan Norm. Univ. 2001, 24, 55–57. [Google Scholar]
- Xiao, J.; Kang, X.; Xie, L.; Qin, Q.; He, Z.; Hu, F.; Zhang, C.; Zhao, R.; Wang, J.; Luo, K.; et al. The fertility of the hybrid lineage derived from female Megalobrama amblycephala × male Culter alburnus. Anim. Reprod. Sci. 2014, 151, 61–70. [Google Scholar] [CrossRef]
- Liu, S.J.; Zhao, R.R.; Li, J.H.; Sun, Y.D.; Zhang, C.; Luo, K.K. Comparison of DNA content of blood cells and sperm among the different ploidy level fish. Acta Zool. Sin. 2005, 51, 360–364. [Google Scholar]
- Zhang, C.; Liu, S.J.; Wu, Y.H.; Liu, J. DNA contents and cytological analyses of oogenesis of the dipoid gynogenetic progeny of allotetraploid hybrids. J. Fish. China 2015, 39, 1–7. [Google Scholar]
- He, W.; Sun, Y.; Qiang, J.; Luo, X.; Zhang, H.; Yang, C.; Luo, K.; Zhao, R.; Qin, Q.; Zhang, C.; et al. Structural Abnormalities of Spermatozoa in Triploid Gynogenetic Crucian Carp (Carassius auratus). Front. Genet. 2021, 12, 783014. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Liu, S.J.; Zhang, X.J.; Feng, H.; Liu, Y. Studies on the ploymorphic of sperm of F2 Hybrids of Red Crucian Carp × Common Carp. Acta Biol. Exp. Sin. 2004, 37, 276–282. [Google Scholar]
- Gao, R.C.; Wu, L.Y. The Application of Flow Cytometry in Aquatic Animal Research. Anhui Agric. Sci. Bull. 2010, 16, 15–17+23. [Google Scholar]
- Mgwatyu, Y.; Stander, A.A.; Ferreira, S.; Williams, W.; Hesse, U. Rooibos (Aspalathus linearis) genome size estimation using flow cytometry and k-mer analyses. Plants 2020, 9, 270. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Chang, R.F.; Cheng, H.H. The Principle and Method of Ploidy Analysis by Flow Cytometr. J. Yunnan Agric. Univ. 2006, 407–409+414. [Google Scholar]
- Andrew, W. Duncan, Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol. 2013, 24, 347–356. [Google Scholar]
- Zhang, H.; Luo, M.; Tai, Y.; Li, M.; Cui, J.; Gao, X.; Ren, L.; Liu, S. mtDNA copy number contributes to growth diversity in allopolyploid fish. Reprod. Breed. 2024, 4, 55–60. [Google Scholar] [CrossRef]
- Wu, L.; Li, L.; Gao, A.; Ye, J.; Li, J. Antimicrobial roles of phagocytosis in teleost fish: Phagocytic B cells vs professional phagocytes. Aquac. Fish. 2024, 9, 105–114. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Zhang, C.; Luo, K.; Liu, Y. Production of gynogenetic progeny from allotetraploid hybrids red crucian carp × common carp. Aquaculture 2004, 236, 193–200. [Google Scholar] [CrossRef]
- Yang, H.X.; Zhang, J.; Pan, H.J. Feasibility of flow cytometry experiments based on fin. South China Fish. Sci. 2017, 13, 121–127. [Google Scholar]
- Geng, B.; Sun, X.W. The application of Flow cytometry on the DNA content and ploidy analysis of hydrophily organisms. Chin. J. Fish. 2008, 21, 21–24. [Google Scholar]
- Krasznai, Z.; Goda, K. Can flow cytometric DNA content analysis become a routine procedure in aquaculture? Cytom. Part A 2021, 99, 668–670. [Google Scholar] [CrossRef]
- Gong, R.; Zhang, J.; Dan, C.; Gui, G.J.; Mei, M.L. Identification of pelteobagrus fulvidraco, pelteobaggrus vachelli and their interspecific hybrid by dna markers. Acta Hydrobiol. Sin. 2017, 41, 321–325. [Google Scholar]
- Gui, J.F.; Bao, Z.M.; Zhang, X.J. Development Strategy for Aquaculture Genetic Breeding and Seed Industry. Eng. Sci. China 2016, 18, 8–14. [Google Scholar]
- Lin, M.J. Detection technology research of ploidy on live abalone. J. Fish. Res. 2012, 34, 225–230. [Google Scholar]
- Jia, C.; Meng, Q.; Chen, S.; Sun, R.; Xu, D.; Zhu, F.; Gao, B.; Zhao, J.; Shen, H. Construction of the first high-density genetic linkage map and QTL mapping for growth traits in black seabream (Acanthopagrus schlegelii). Aquaculture 2025, 595, 741588. [Google Scholar] [CrossRef]
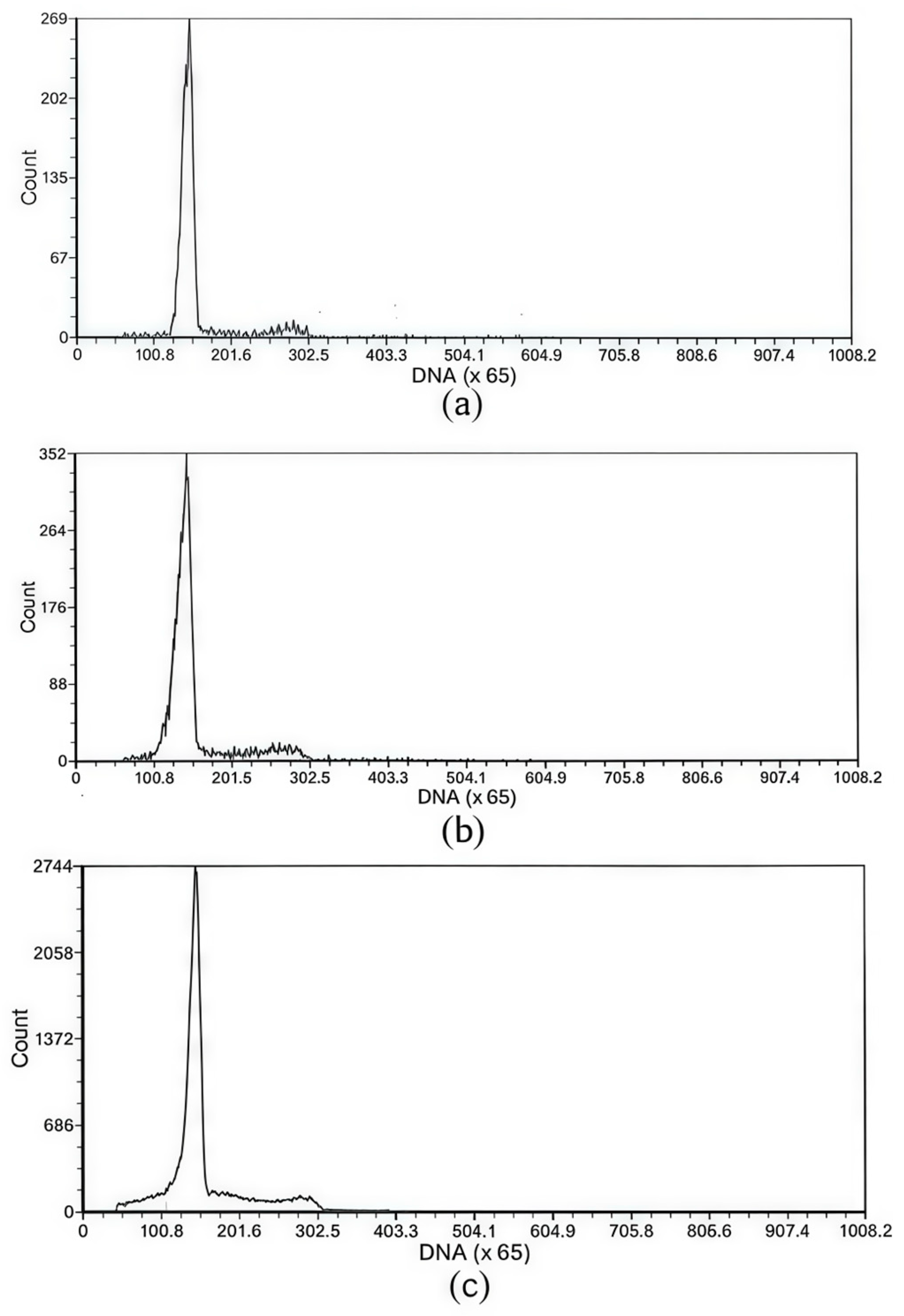
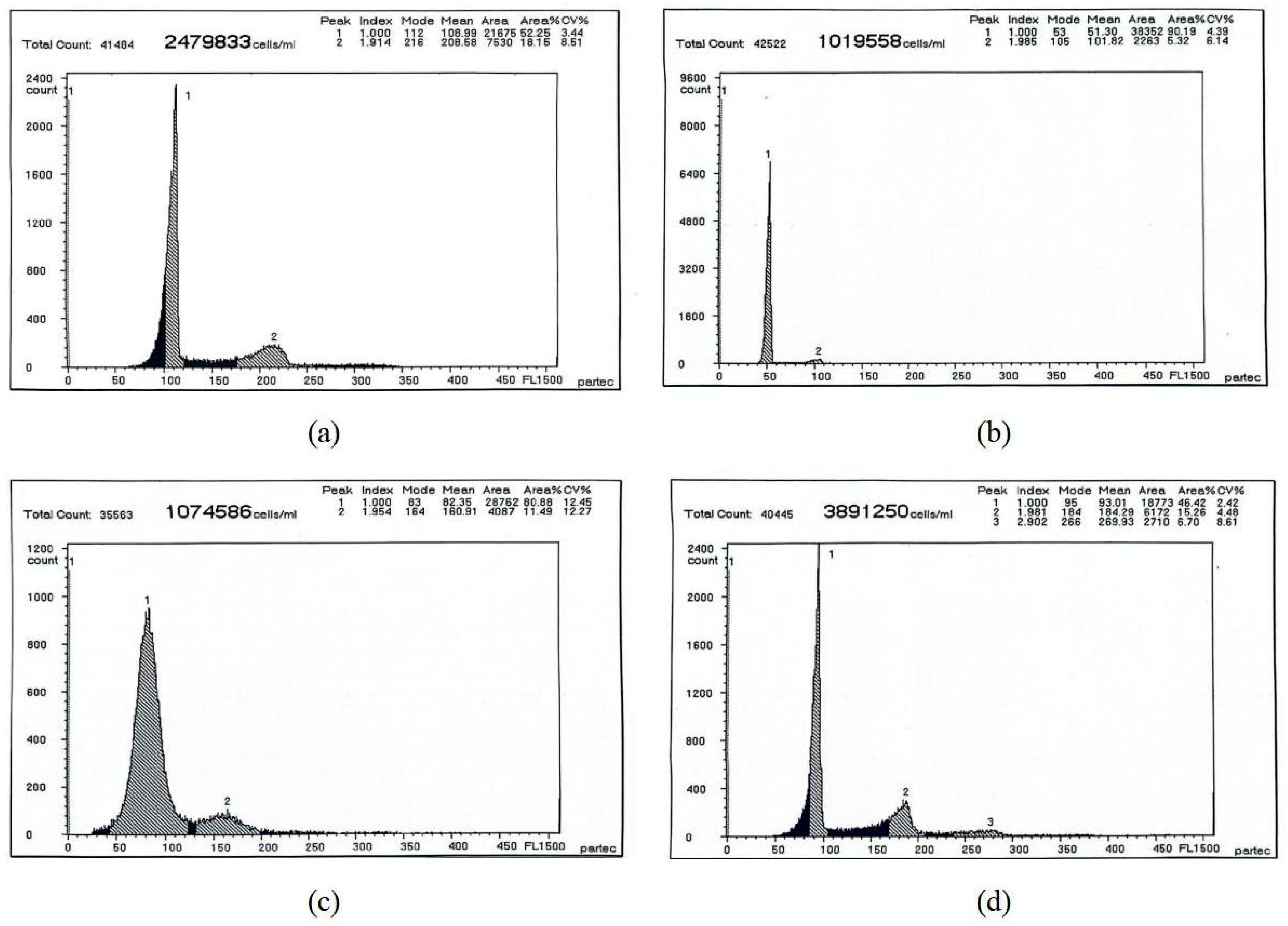
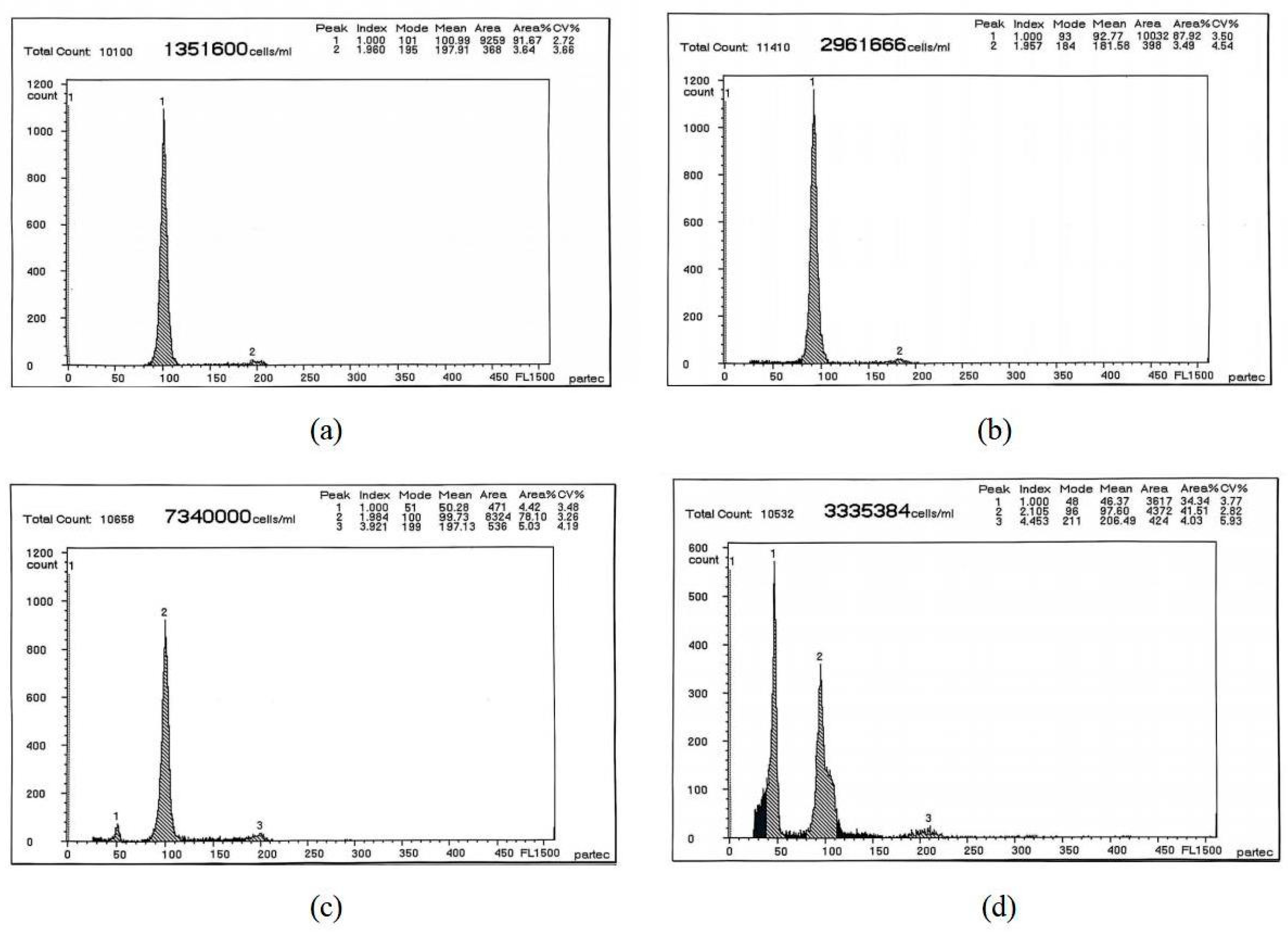
| Different Sample | Mean DNA Content | CV% |
|---|---|---|
| Blood cells of RCC | 108.99/208.58 | 3.44/8.51 |
| The sperm of diploid RCC | 51.30/101.82 | 4.19/6.34 |
| The sperm of 3nDTCC | 82.35/160.91 | 12.45/12.27 |
| The sperm of 4nAT | 90.01/184.29/269.93 | 2.42/4.48/8.61 |
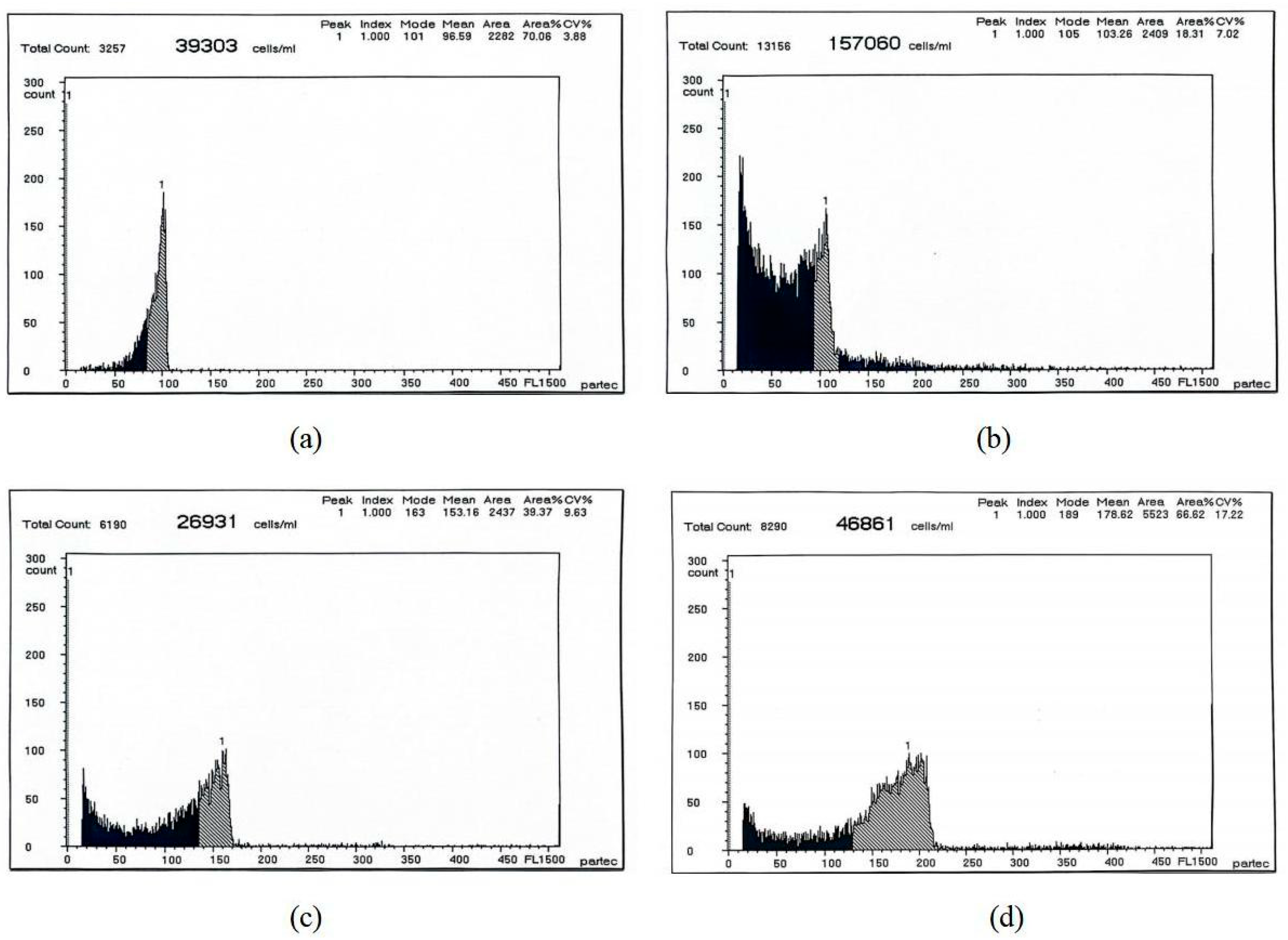
| Different Sample | Mean DNA Content | CV% |
|---|---|---|
| Blood cells of RCC | 96.59 | 3.88 |
| The caudal fin cells of RCC | 103.28 | 7.02 |
| The caudal fin cells of 3nDTCC | 153.16 | 9.63 |
| The caudal fin cells of 4nAT | 178.62 | 17.22 |
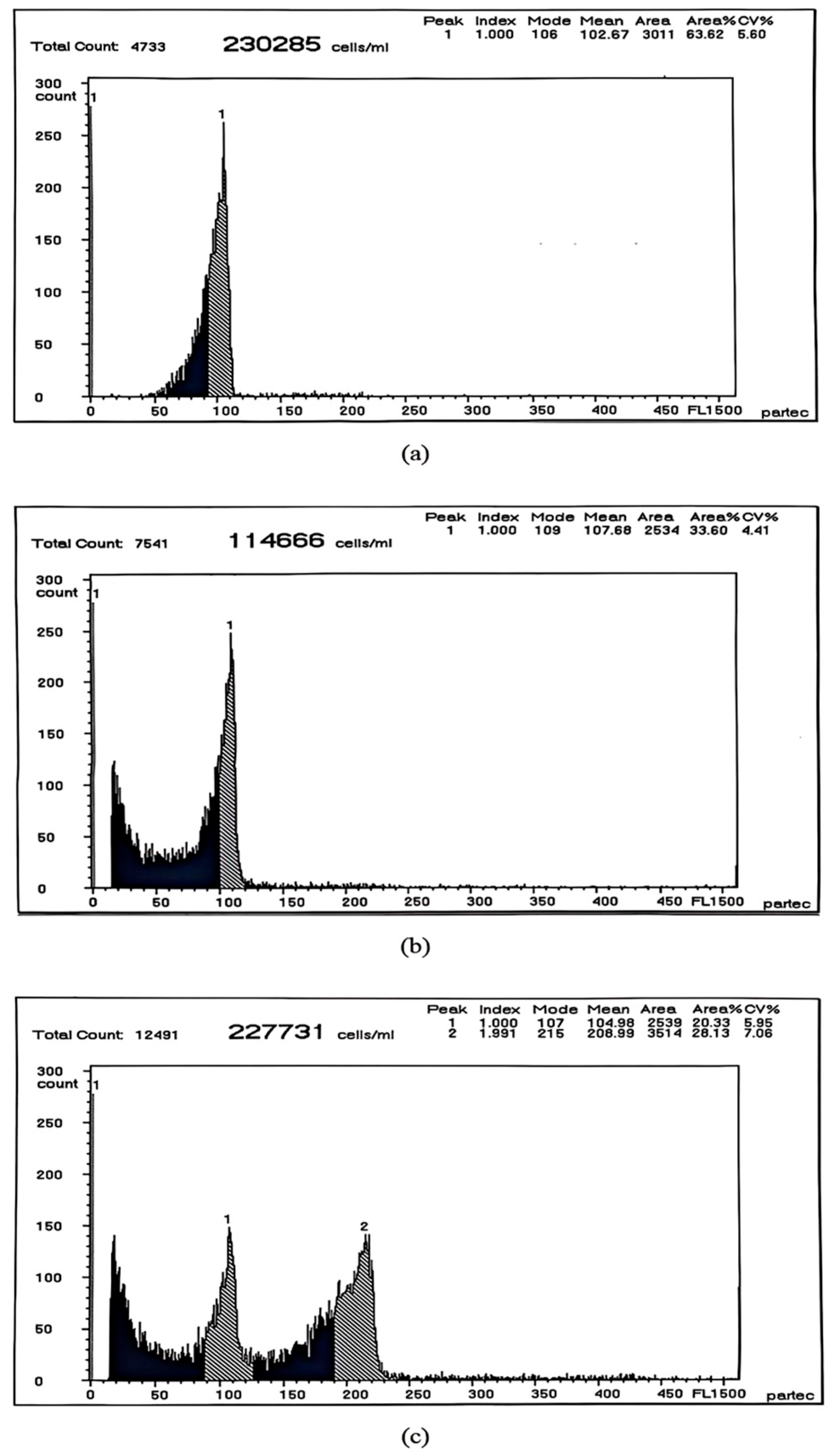
| Different Sample | Mean DNA Content | CV% |
|---|---|---|
| The blood cells of RCC | 102.67 | 5.60 |
| The ovary cells of JLF1 | 107.68 | 4.41 |
| The testis cells of JLF1 | 104.98/208.99 | 5.95/7.06 |
| Different Sample | Mean DNA Content | CV% |
|---|---|---|
| The blood cells of RCC | 100.99/197.91 | 2.72/3.66 |
| The gill tissues of RCC | 92.77/181.58 | 3.50/4.54 |
| The spleen tissues of RCC | 50.28/99.73/197.13 | 3.48/3.26/4.19 |
| The liver tissues of RCC | 46.37/97.60/206.49 | 3.77/2.82/5.93 |
| Different Tissues | Mean DNA Content | Experimental Ratio (Tissues/Blood) | Expected Ratio |
|---|---|---|---|
| Blood | 100.99 | ||
| Gill | 92.77 | 0.91 * | 1 * |
| Spleen | 99.73 | 0.98 * | 1 * |
| Liver | 46.37 | 0.45 * | 0.5 * |
| 97.60 | 0.96 * | 1 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Chen, Y.; Zhang, X.; Qiang, J.; Nie, L.; Luo, X.; Liang, B.; Chen, K.; Yang, F.; Zhao, R.; et al. Application of Flow Cytometry to Determine Cell DNA Content in the Genetic Breeding of Fish. Fishes 2025, 10, 227. https://doi.org/10.3390/fishes10050227
Zhu X, Chen Y, Zhang X, Qiang J, Nie L, Luo X, Liang B, Chen K, Yang F, Zhao R, et al. Application of Flow Cytometry to Determine Cell DNA Content in the Genetic Breeding of Fish. Fishes. 2025; 10(5):227. https://doi.org/10.3390/fishes10050227
Chicago/Turabian StyleZhu, Xinyan, Yang Chen, Xiaodie Zhang, Jiaxu Qiang, Lingtao Nie, Xinyue Luo, Binchao Liang, Kuo Chen, Fuzhong Yang, Rurong Zhao, and et al. 2025. "Application of Flow Cytometry to Determine Cell DNA Content in the Genetic Breeding of Fish" Fishes 10, no. 5: 227. https://doi.org/10.3390/fishes10050227
APA StyleZhu, X., Chen, Y., Zhang, X., Qiang, J., Nie, L., Luo, X., Liang, B., Chen, K., Yang, F., Zhao, R., & Zhang, C. (2025). Application of Flow Cytometry to Determine Cell DNA Content in the Genetic Breeding of Fish. Fishes, 10(5), 227. https://doi.org/10.3390/fishes10050227






