Comparison of Bioinformatic Pipelines for eDNA Metabarcoding Data Analysis of Fish Populations †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Bioinformatic Processing
3. Results
3.1. eDNA Metabarcoding
3.2. Number of Taxa Detected and Assigned Reads
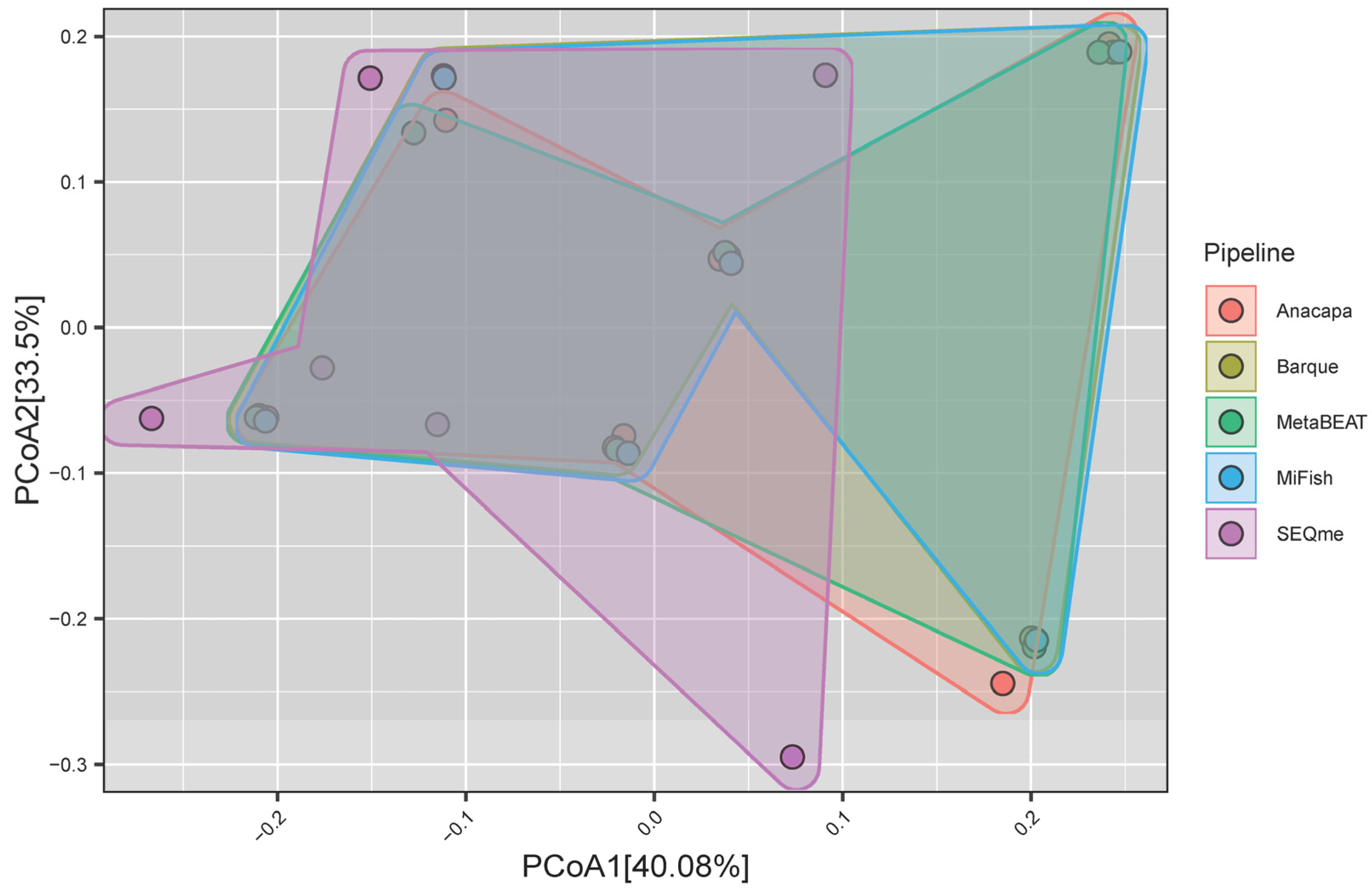
3.3. Alpha and Beta Diversities
4. Discussion
4.1. Comparison of the Detection Among Bioinformatic Pipelines and Conventional Methods
4.2. Comparison of Alpha and Beta Diversities Among the Bioinformatic Pipelines
4.3. Bioinformatic Pipelines Analogy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seymour, M. Rapid Progression and Future of Environmental DNA Research. Commun. Biol. 2019, 2, 80. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef]
- Hänfling, B.; Lawson Handley, L.; Read, D.S.; Hahn, C.; Li, J.; Nichols, P.; Blackman, R.C.; Oliver, A.; Winfield, I.J. Environmental DNA Metabarcoding of Lake Fish Communities Reflects Long-term Data from Established Survey Methods. Mol. Ecol. 2016, 25, 3101–3119. [Google Scholar] [CrossRef] [PubMed]
- Kottelat, M.; Freyhof, J. Handbook of European Freshwater Fishes; Publications Kottelat: Cornol, Switzerland, 2007. [Google Scholar]
- Dejean, T.; Valentini, A.; Duparc, A.; Pellier-Cuit, S.; Pompanon, F.; Taberlet, P.; Miaud, C. Persistence of Environmental DNA in Freshwater Ecosystems. PLoS ONE 2011, 6, e23398. [Google Scholar] [CrossRef] [PubMed]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying Effects of UV-B, Temperature, and pH on eDNA Degradation in Aquatic Microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport Distance of Invertebrate Environmental DNA in a Natural River. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation Monitoring of Aquatic Biodiversity Using Environmental DNA Metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a Set of Universal PCR Primers for Metabarcoding Environmental DNA from Fishes: Detection of More than 230 Subtropical Marine Species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Quail, M.; Smith, M.E.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A Tale of Three next Generation Sequencing Platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq Sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef]
- Stoler, N.; Nekrutenko, A. Sequencing Error Profiles of Illumina Sequencing Instruments. NAR Genom. Bioinform. 2021, 3, lqab019. [Google Scholar] [CrossRef]
- Laehnemann, D.; Borkhardt, A.; McHardy, A.C. Denoising DNA Deep Sequencing Data—High-Throughput Sequencing Errors and Their Correction. Brief. Bioinform. 2016, 17, 154–179. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring, 1st ed.; Oxford University Press: Oxford, UK, 2018. [Google Scholar] [CrossRef]
- Sato, Y.; Miya, M.; Fukunaga, T.; Sado, T.; Iwasaki, W. MitoFish and MiFish Pipeline: A Mitochondrial Genome Database of Fish with an Analysis Pipeline for Environmental DNA Metabarcoding. Mol. Biol. Evol. 2018, 35, 1553–1555. [Google Scholar] [CrossRef] [PubMed]
- Curd, E.E.; Gold, Z.; Kandlikar, G.S.; Gomer, J.; Ogden, M.; O’Connell, T.; Pipes, L.; Schweizer, T.M.; Rabichow, L.; Lin, M.; et al. Anacapa Toolkit: An Environmental DNA Toolkit for Processing Multilocus Metabarcode Datasets. Methods Ecol. Evol. 2019, 10, 1469–1475. [Google Scholar] [CrossRef]
- Zafeiropoulos, H.; Viet, H.Q.; Vasileiadou, K.; Potirakis, A.; Arvanitidis, C.; Topalis, P.; Pavloudi, C.; Pafilis, E. PEMA: A Flexible Pipeline for Environmental DNA Metabarcoding Analysis of the 16S/18S Ribosomal RNA, ITS, and COI Marker Genes. GigaScience 2020, 9, giaa022. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, W.; Jiang, Y.; Wen, Y.; Li, M.; Liu, L.; Zou, K. New Insights into Biologic Interpretation of Bioinformatic Pipelines for Fish eDNA Metabarcoding: A Case Study in Pearl River Estuary. J. Environ. Manag. 2024, 368, 122136. [Google Scholar] [CrossRef]
- Gardner, P.P.; Watson, R.J.; Morgan, X.C.; Draper, J.L.; Finn, R.D.; Morales, S.E.; Stott, M.B. Identifying Accurate Metagenome and Amplicon Software via a Meta-Analysis of Sequence to Taxonomy Benchmarking Studies. PeerJ 2019, 7, e6160. [Google Scholar] [CrossRef]
- Van Den Berg, C.P.; Troscianko, J.; Endler, J.A.; Marshall, N.J.; Cheney, K.L. Quantitative Colour Pattern Analysis (QCPA): A Comprehensive Framework for the Analysis of Colour Patterns in Nature. Methods Ecol. Evol. 2020, 11, 316–332. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Gao, X.; Lin, H.; Revanna, K.; Dong, Q. A Bayesian Taxonomic Classification Method for 16S rRNA Gene Sequences with Improved Species-Level Accuracy. BMC Bioinform. 2017, 18, 247. [Google Scholar] [CrossRef]
- Normandeau, E. Environmental DNA Metabarcoding Analysis. Available online: https://github.com/enormandeau/barque (accessed on 3 December 2020).
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.; Lund, D. metaBEAT—metaBarcoding and Environmental DNA Analysis Tool. Available online: https://github.com/HullUni-bioinformatics/metaBEAT (accessed on 3 January 2021).
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- SEQme. Microbiome and Metagenome Data Analysis Workshop, České Budějovice, Czech Republic. 2018. Available online: https://www.seqme.eu/en/ (accessed on 4 June 2018).
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Lobl, R.T.; Maenza, R.M. Androgenization: Alterations in Uterine Growth and Morphology. Biol. Reprod. 1975, 13, 255–268. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Whittier, T.R. Development of IBI Metrics for Lakes in Southern New England. In Assessing the Sustainability and Biological Integrity of Water Resources Using Fish Communities; Simon, T.P., Ed.; CRC Press: Boca Raton, FL, USA, 2020; pp. 563–582. [Google Scholar] [CrossRef]
- Koch, L.F. Index of Biotal Dispersity. Ecology 1957, 38, 145–148. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Říha, M.; Ricard, D.; Vašek, M.; Prchalová, M.; Mrkvička, T.; Jůza, T.; Čech, M.; Draštík, V.; Muška, M.; Kratochvíl, M.; et al. Patterns in Diel Habitat Use of Fish Covering the Littoral and Pelagic Zones in a Reservoir. Hydrobiologia 2015, 747, 111–131. [Google Scholar] [CrossRef]
- Blabolil, P.; Ricard, D.; Peterka, J.; Říha, M.; Jůza, T.; Vašek, M.; Prchalová, M.; Čech, M.; Muška, M.; Seďa, J.; et al. Predicting Asp and Pikeperch Recruitment in a Riverine Reservoir. Fish. Res. 2016, 173, 45–52. [Google Scholar] [CrossRef]
- Říha, M.; Jůza, T.; Prchalová, M.; Mrkvička, T.; Čech, M.; Draštík, V.; Muška, M.; Kratochvíl, M.; Peterka, J.; Tušer, M.; et al. The Size Selectivity of the Main Body of a Sampling Pelagic Pair Trawl in Freshwater Reservoirs during the Night. Fish. Res. 2012, 127–128, 56–60. [Google Scholar] [CrossRef]
- Jůza, T.; Ricard, D.; Blabolil, P.; Čech, M.; Draštík, V.; Frouzová, J.; Muška, M.; Peterka, J.; Prchalová, M.; Říha, M.; et al. Species-Specific Gradients of Juvenile Fish Density and Size in Pelagic Areas of Temperate Reservoirs. Hydrobiologia 2015, 762, 169–181. [Google Scholar] [CrossRef]
- Vejřík, L.; Vejříková, I.; Kočvara, L.; Blabolil, P.; Peterka, J.; Sajdlová, Z.; Jůza, T.; Šmejkal, M.; Kolařík, T.; Bartoň, D.; et al. The Pros and Cons of the Invasive Freshwater Apex Predator, European Catfish Silurus glanis, and Powerful Angling Technique for Its Population Control. J. Environ. Manag. 2019, 241, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Blabolil, P.; Čech, M.; Draštík, V.; Holubová, M.; Kočvara, L.; Kubečka, J.; Muška, M.; Prchalová, M.; Říha, M.; Sajdlová, Z.; et al. Less Is More—Basic Quantitative Indices for Fish Can Be Achieved with Reduced Gillnet Sampling. Fish. Res. 2021, 240, 105983. [Google Scholar] [CrossRef]
- Sellers, G.S.; Di Muri, C.; Gómez, A.; Hänfling, B. Mu-DNA: A Modular Universal DNA Extraction Method Adaptable for a Wide Range of Sample Types. MBMG 2018, 2, e24556. [Google Scholar] [CrossRef]
- Riaz, T.; Shehzad, W.; Viari, A.; Pompanon, F.; Taberlet, P.; Coissac, E. ecoPrimers: Inference of New DNA Barcode Markers from Whole Genome Sequence Analysis. Nucleic Acids Res. 2011, 39, e145. [Google Scholar] [CrossRef]
- Blabolil, P.; Harper, L.R.; Říčanová, Š.; Sellers, G.; Di Muri, C.; Jůza, T.; Vašek, M.; Sajdlová, Z.; Rychtecký, P.; Znachor, P.; et al. Environmental DNA Metabarcoding Uncovers Environmental Correlates of Fish Communities in Spatially Heterogeneous Freshwater Habitats. Ecol. Indic. 2021, 126, 107698. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Zhang, J.; Yilmaz, P.; Glöckner, F.O.; Stamatakis, A. Phylogeny-Aware Identification and Correction of Taxonomically Mislabeled Sequences. Nucleic Acids Res. 2016, 44, 5022–5033. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Hannon, G.J. Fastx-Toolkit. Available online: https://www.hannonlab.org/resources/ (accessed on 20 December 2020).
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 December 2020).
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-Glance Quality Assessment of Illumina Second-Generation Sequencing Data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Lim, Y.W.; Rohwer, F.; Edwards, R. TagCleaner: Identification and Removal of Tag Sequences from Genomic and Metagenomic Datasets. BMC Bioinform. 2010, 11, 341. [Google Scholar] [CrossRef]
- Aronesty, E. Comparison of Sequencing Utility Programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Hansen, M.A. Biopieces: A Bioinformatics Toolset and Framework. Available online: http://www.biopieces.org/ (accessed on 20 December 2020).
- R Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. Available online: https://cran.r-project.org/ (accessed on 1 December 2020).
- Lawson Handley, L.; Read, D.S.; Winfield, I.J.; Kimbell, H.; Johnson, H.; Li, J.; Hahn, C.; Blackman, R.; Wilcox, R.; Donnelly, R.; et al. Temporal and Spatial Variation in Distribution of Fish Environmental DNA in England’s Largest Lake. Environ. DNA 2019, 1, 26–39. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, 2001, 2.6-10. Available online: https://doi.org/10.32614/CRAN.package.vegan (accessed on 1 December 2020).
- IUCN. Gasterosteus Aculeatus; The IUCN Red List of Threatened Species. Available online: https://www.iucnredlist.org/ (accessed on 3 January 2021).
- O’Rourke, D.R.; Bokulich, N.A.; Jusino, M.A.; MacManes, M.D.; Foster, J.T. A Total Crapshoot? Evaluating Bioinformatic Decisions in Animal Diet Metabarcoding Analyses. Ecol. Evol. 2020, 10, 9721–9739. [Google Scholar] [CrossRef]
- Nosova, A.Y.; Kipen, V.N.; Tsar, A.I.; Lemesh, V.A. Differentiation of Hybrid Progeny of Silver Carp (Hypophthalmichthys molitrix Val.) and Bighead Carp (H. nobilis Rich.) Based on Microsatellite Polymorphism. Russ. J. Genet. 2020, 56, 317–323. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Schabacker, J.; Smith, S.; Al-Chokhachy, R.; Luikart, G.; Amish, S.J. Improved Detection of Rare, Endangered and Invasive Trout in Using a New Large-volume Sampling Method for eDNA Capture. Environ. DNA 2019, 1, 227–237. [Google Scholar] [CrossRef]
- Eick, D. Habitat Preferences of the Burbot (Lota lota) from the River Elbe: An Experimental Approach. J. Appl. Ichthyol. 2013, 29, 541–548. [Google Scholar] [CrossRef]
- Blabolil, P.; Duras, J.; Jůza, T.; Kočvara, L.; Matěna, J.; Muška, M.; Říha, M.; Vejřík, L.; Holubová, M.; Peterka, J. Assessment of Burbot Lota lota (L. 1758) Population Sustainability in Central European Reservoirs. J. Fish Biol. 2018, 92, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Mehner, T.; Diekmann, M.; Brämick, U.; Lemcke, R. Composition of Fish Communities in German Lakes as Related to Lake Morphology, Trophic State, Shore Structure and Human-use Intensity. Freshw. Biol. 2005, 50, 70–85. [Google Scholar] [CrossRef]
- Willemsen, J. Fishery-Aspects of Eutrophication. Hydrobiol. Bull. 1980, 14, 12–21. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of Temperature and Trophic State on Degradation of Environmental DNA in Lake Water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Leuven, R.S.E.W.; Hendriks, A.J.; Huijbregts, M.A.J.; Lenders, H.J.R.; Matthews, J.; Velde, G.V.D. Differences in Sensitivity of Native and Exotic Fish Species to Changes in River Temperature. Curr. Zool. 2011, 57, 852–862. [Google Scholar] [CrossRef]
- Van De Pol, I.; Flik, G.; Gorissen, M. Comparative Physiology of Energy Metabolism: Fishing for Endocrine Signals in the Early Vertebrate Pool. Front. Endocrinol. 2017, 8, 36. [Google Scholar] [CrossRef]
- De Souza, L.S.; Godwin, J.C.; Renshaw, M.A.; Larson, E. Environmental DNA (eDNA) Detection Probability Is Influenced by Seasonal Activity of Organisms. PLoS ONE 2016, 11, e0165273. [Google Scholar] [CrossRef]
- Cherry, D.S.; Dickson, K.L.; Cairns, J., Jr.; Stauffer, J.R. Preferred, Avoided, and Lethal Temperatures of Fish During Rising Temperature Conditions. J. Fish. Res. Board Can. 1977, 34, 239–246. [Google Scholar] [CrossRef]
- Cherry, D.S.; Cairns, J. Biological Monitoring Part V—Preference and Avoidance Studies. Water Res. 1982, 16, 263–301. [Google Scholar] [CrossRef]
- Schenekar, T.; Schletterer, M.; Lecaudey, L.A.; Weiss, S.J. Reference Databases, Primer Choice, and Assay Sensitivity for Environmental Metabarcoding: Lessons Learnt from a Re-evaluation of an eDNA Fish Assessment in the Volga Headwaters. River Res. Appl. 2020, 36, 1004–1013. [Google Scholar] [CrossRef]
- Lladó Fernández, S.; Větrovský, T.; Baldrian, P. The Concept of Operational Taxonomic Units Revisited: Genomes of Bacteria That Are Regarded as Closely Related Are Often Highly Dissimilar. Folia Microbiol. 2019, 64, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Nearing, J.T.; Douglas, G.M.; Comeau, A.M.; Langille, M.G.I. Denoising the Denoisers: An Independent Evaluation of Microbiome Sequence Error-Correction Approaches. PeerJ 2018, 6, e5364. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.J.; Rasmussen, L.; Asplund, M.; Knudsen, S.W.; Clausen, M.-L.; Agner, T.; Hansen, A.J. Comparing DADA2 and OTU Clustering Approaches in Studying the Bacterial Communities of Atopic Dermatitis. J. Med. Microbiol. 2020, 69, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Pauvert, C.; Buée, M.; Laval, V.; Edel-Hermann, V.; Fauchery, L.; Gautier, A.; Lesur, I.; Vallance, J.; Vacher, C. Bioinformatics Matters: The Accuracy of Plant and Soil Fungal Community Data Is Highly Dependent on the Metabarcoding Pipeline. Fungal Ecol. 2019, 41, 23–33. [Google Scholar] [CrossRef]
- Antich, A.; Palacin, C.; Wangensteen, O.S.; Turon, X. To Denoise or to Cluster, That Is Not the Question: Optimizing Pipelines for COI Metabarcoding and Metaphylogeography. BMC Bioinform. 2021, 22, 177. [Google Scholar] [CrossRef]
- Lamb, P.D.; Hunter, E.; Pinnegar, J.K.; Creer, S.; Davies, R.G.; Taylor, M.I. How Quantitative Is Metabarcoding: A Meta-analytical Approach. Mol. Ecol. 2019, 28, 420–430. [Google Scholar] [CrossRef]
- Zielezinski, A.; Vinga, S.; Almeida, J.; Karlowski, W.M. Alignment-Free Sequence Comparison: Benefits, Applications, and Tools. Genome Biol. 2017, 18, 186. [Google Scholar] [CrossRef]
- Hakimzadeh, A.; Abdala Asbun, A.; Albanese, D.; Bernard, M.; Buchner, D.; Callahan, B.; Caporaso, J.G.; Curd, E.; Djemiel, C.; Brandström Durling, M.; et al. Pile of Pipelines: An Overview of the Bioinformatics Software for Metabarcoding Data Analyses. Mol. Ecol. Resour. 2024, 24, e13847. [Google Scholar] [CrossRef]
- Mathon, L.; Valentini, A.; Guérin, P.; Normandeau, E.; Noel, C.; Lionnet, C.; Boulanger, E.; Thuiller, W.; Bernatchez, L.; Mouillot, D.; et al. Benchmarking Bioinformatic Tools for Fast and Accurate eDNA Metabarcoding Species Identification. Mol. Ecol. Resour. 2021, 21, 2565–2579. [Google Scholar] [CrossRef]
- Dufresne, Y.; Lejzerowicz, F.; Perret-Gentil, L.A.; Pawlowski, J.; Cordier, T. SLIM: A Flexible Web Application for the Reproducible Processing of Environmental DNA Metabarcoding Data. BMC Bioinform. 2019, 20, 88. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA Barcode Reference Libraries for the Monitoring of Aquatic Biota in Europe: Gap-Analysis and Recommendations for Future Work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef] [PubMed]
- Günther, B.; Knebelsberger, T.; Neumann, H.; Laakmann, S.; Martínez Arbizu, P. Metabarcoding of Marine Environmental DNA Based on Mitochondrial and Nuclear Genes. Sci. Rep. 2018, 8, 14822. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, M.; Lu, S.; Zhang, X.; Fang, C.; Tan, L.; Xiong, F.; Zeng, H.; He, S. A Comparative Evaluation of eDNA Metabarcoding Primers in Fish Community Monitoring in the East Lake. Water 2024, 16, 631. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for Methods Utilizing Environmental DNA for Detection of Fish Species. Genes 2020, 11, 296. [Google Scholar] [CrossRef]
- Brys, R.; Everts, T.; Halfmaerten, D.; Neyrinck, S.; Mauvisseau, Q. The Use of Multiple Markers and Internal Positive Controls Significantly Improves Species eDNA Detection Rates and Data Reliability. ARPHA Conf. Abstr. 2021, 4, e65064. [Google Scholar] [CrossRef]
- Ruan, H.-T.; Wang, R.-L.; Li, H.-T.; Liu, L.; Kuang, T.-X.; Li, M.; Zou, K.-S. Effects of Sampling Strategies and DNA Extraction Methods on eDNA Metabarcoding: A Case Study of Estuarine Fish Diversity Monitoring. Zool. Res. 2022, 43, 192–204. [Google Scholar] [CrossRef]
- Schabacker, J.C.; Amish, S.J.; Ellis, B.K.; Gardner, B.; Miller, D.L.; Rutledge, E.A.; Sepulveda, A.J.; Luikart, G. Increased eDNA Detection Sensitivity Using a Novel High-volume Water Sampling Method. Environ. DNA 2020, 2, 244–251. [Google Scholar] [CrossRef]
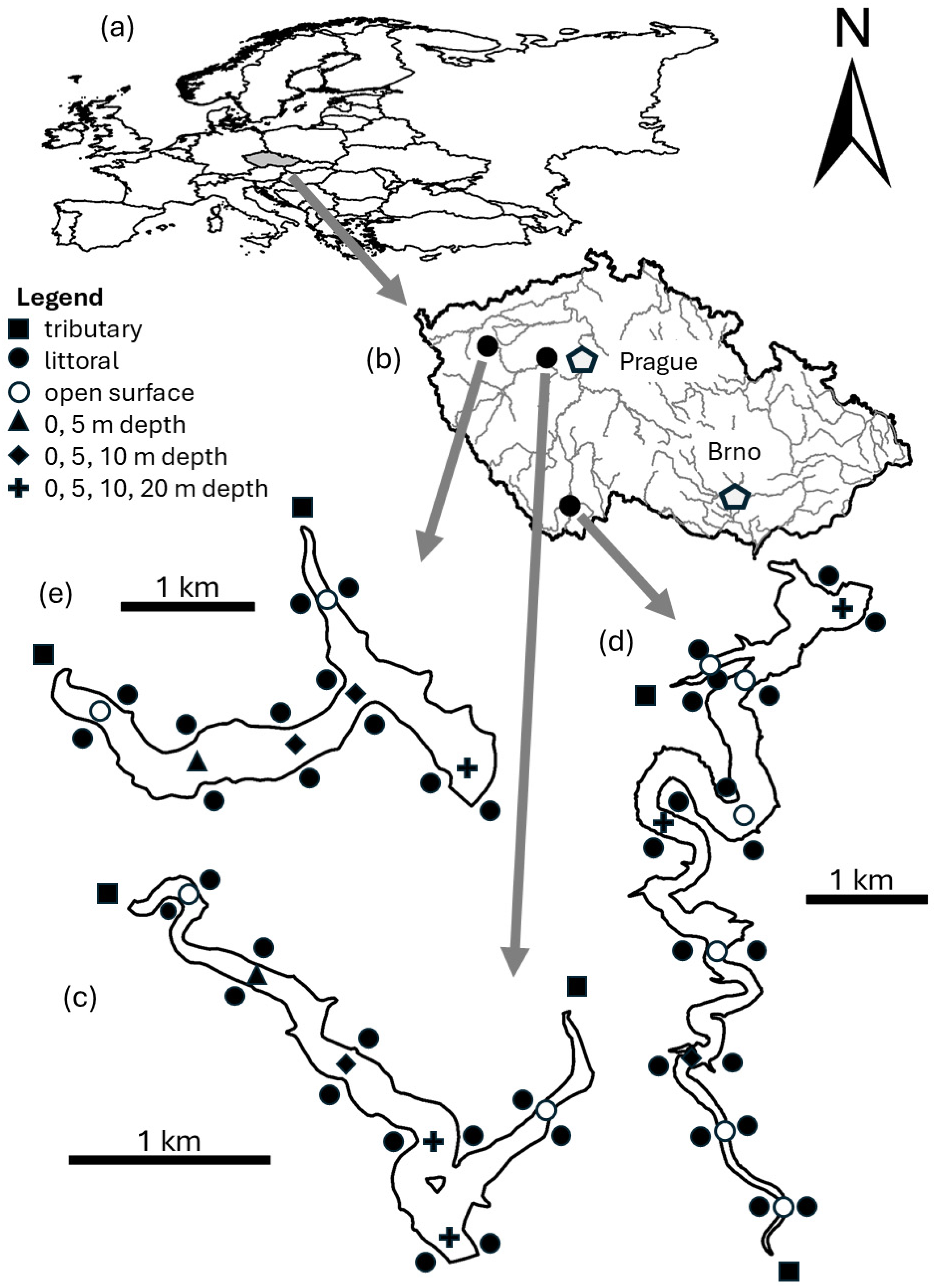
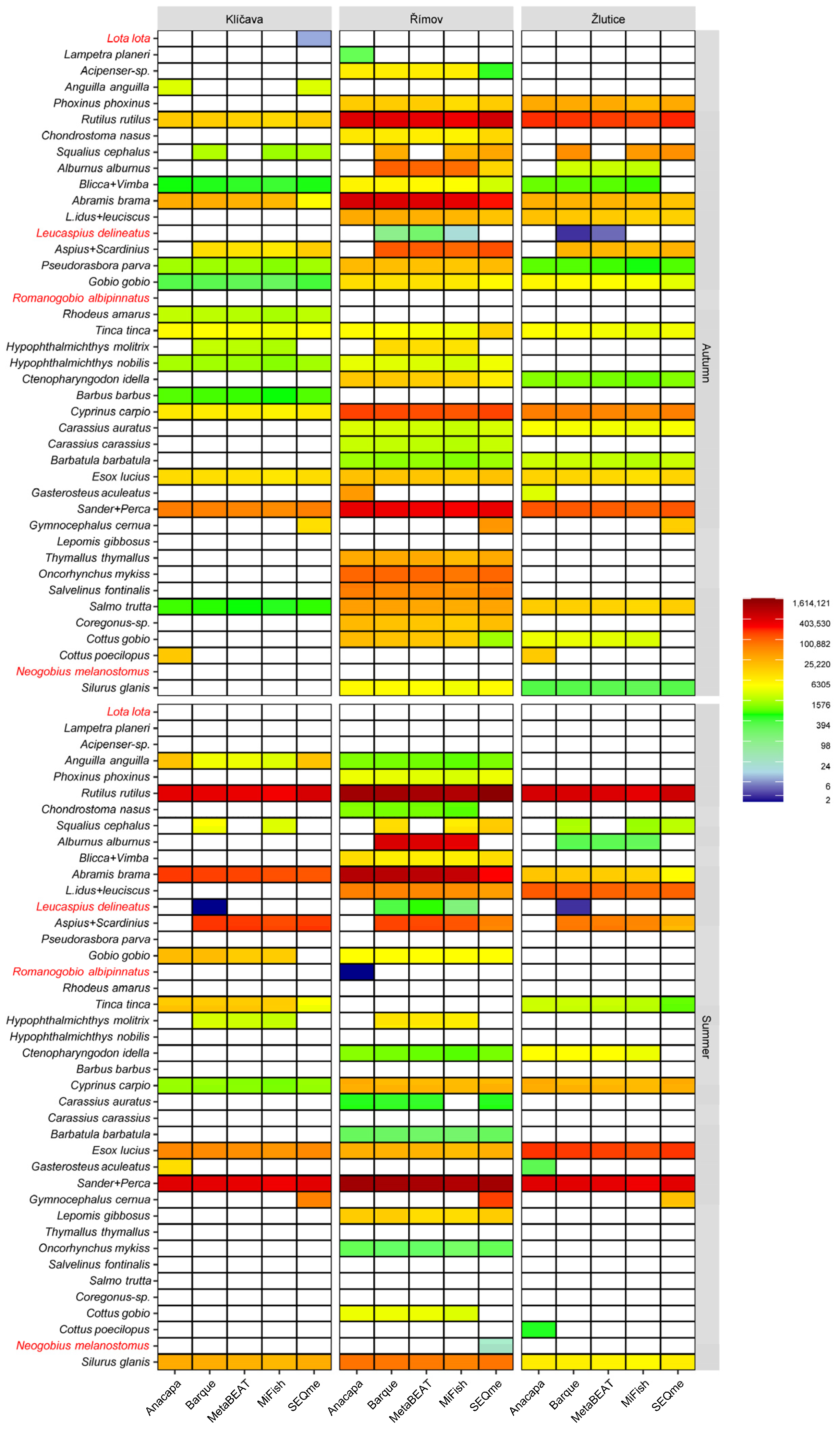
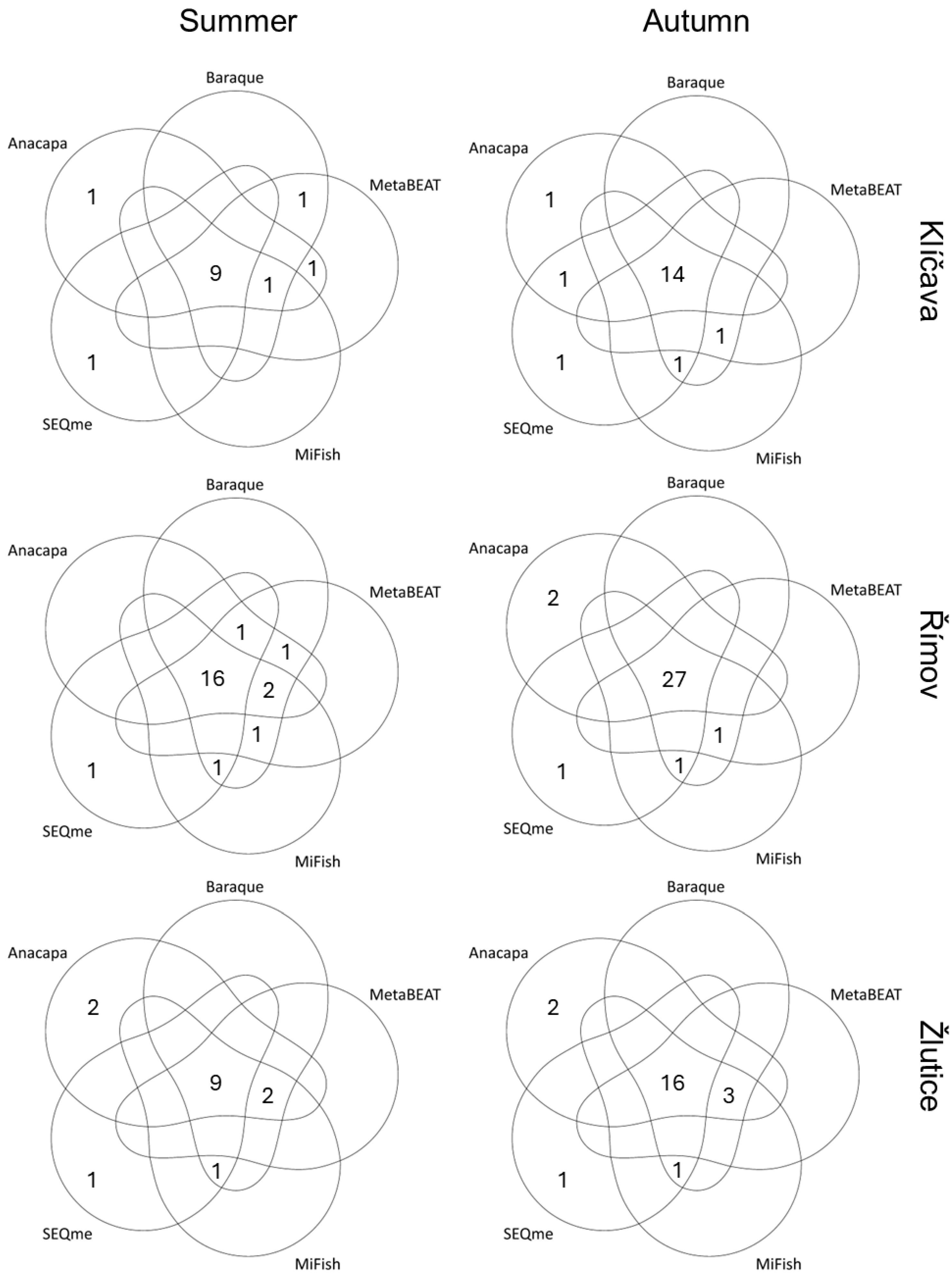
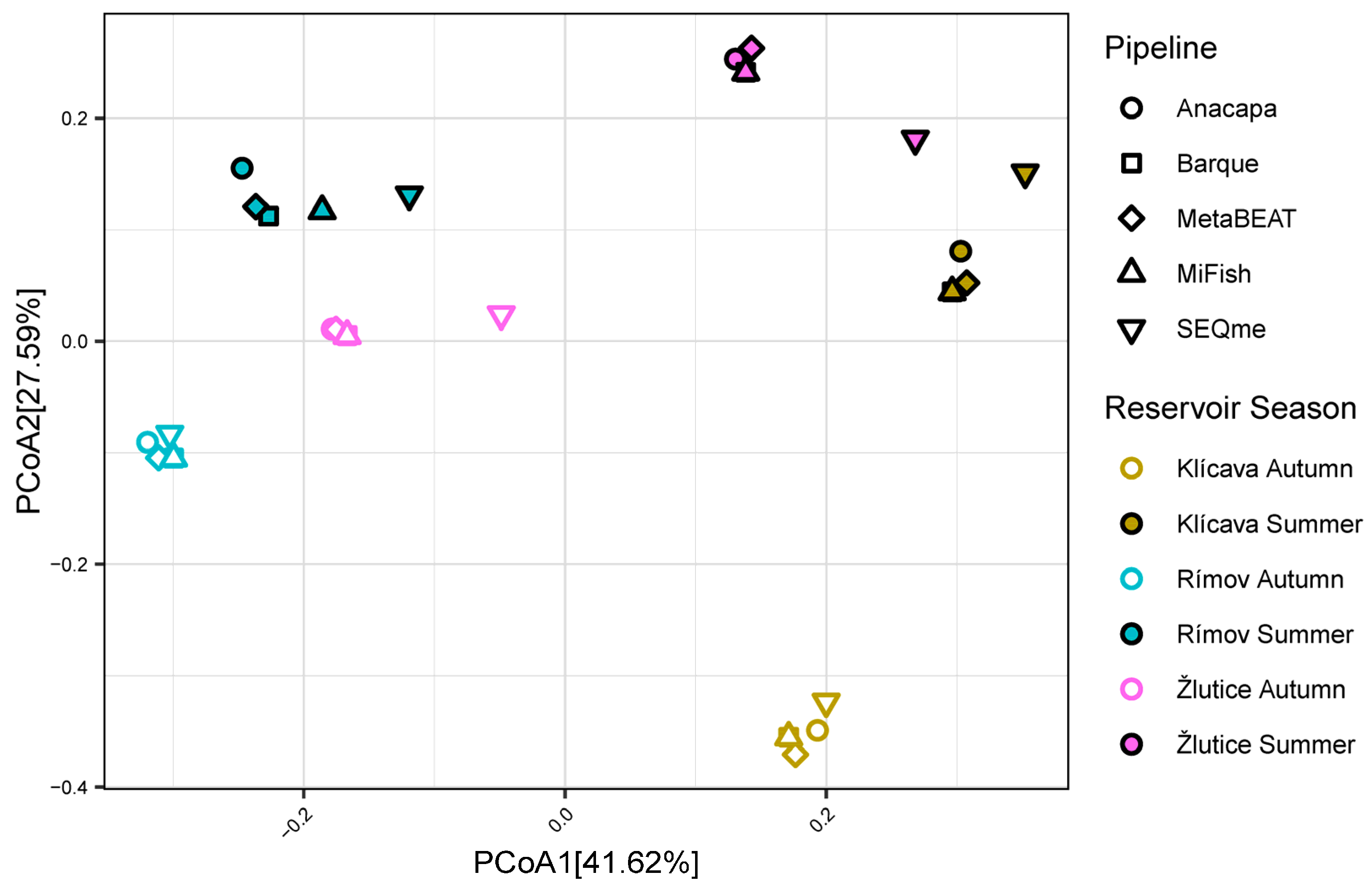
| Parameter | Klíčava | Římov | Žlutice |
|---|---|---|---|
| Trophic state | mesotrophic | eutrophic | eutrophic |
| Elevation above sea level (m) | 294 | 470 | 509 |
| Volume (mil. m3) | 8.3 | 34 | 14 |
| Flooded area (km2) | 0.62 | 2.1 | 1.6 |
| Maximum depth (m) | 34 | 42 | 23 |
| Average depth (m) | 13 | 16 | 9 |
| Boat electrofishing | 2018–2020 | 2018-2020 | 2018–2020 |
| Shore seining | 2019–2020 | 2018–2020 | 2019 |
| Pelagic trawling | 2018–2020 | 2018–2020 | 2018–2020 |
| Hook lines | 2018–2020 | 2018–2019 | 2018–2019 |
| Benthic and pelagic gillnets | 2018–2020 | 2018–2020 | 2018–2020 |
| Species | Klíčava | Římov | Žlutice |
|---|---|---|---|
| Lampetra planeri | X | ||
| Acipenser baerii | X | ||
| Anguilla anguilla | X | X | X |
| Rutilus rutilus | X | X | X |
| Chondrostoma nasus | X | ||
| Squalius cephalus | X | X | X |
| Alburnus alburnus | X | X | X |
| Blicca bjoerkna | X | ||
| Abramis brama | X | X | X |
| Leuciscus idus | X | X | |
| Leuciscus leuciscus | X | X | X |
| Leuciscus aspius | X | X | X |
| Scardinius erythrophthalmus | X | X | X |
| Pseudorasbora parva | X | X | X |
| Gobio gobio | X | X | |
| Tinca tinca | X | X | X |
| Hypophthalmichthys molitrix | X | ||
| Hypophthalmichthys nobilis | X | ||
| Ctenopharyngodon idella | X | X | |
| Cyprinus carpio | X | X | X |
| Carassius auratus | X | X | X |
| Barbatula barbatula | X | ||
| Esox Lucius | X | X | X |
| Sander lucioperca | X | X | X |
| Perca fluviatilis | X | X | X |
| Gymnocephalus cernua | X | X | X |
| Lepomis gibbosus | X | ||
| Oncorhynchus mykiss | X | ||
| Salmo trutta | X | ||
| Coregonus maraena | X | ||
| Silurus glanis | X | X | X |
| Lota lota | X | X |
| Data Processing Steps | Anacapa | Barque | MetaBEAT | MiFish | SEQme |
|---|---|---|---|---|---|
| Total from original data | 22,464,147 | 22,464,147 | 22,464,147 | 22,464,147 | 22,464,147 |
| Total after demultiplexing | 20,910,517 | 20,910,517 | 20,910,517 | 20,910,517 | 20,910,517 |
| Trimmed and filtered | 19,095,153 | 18,632,248 | 18,513,853 | 20,910,517 | 19,384,077 |
| Merged | 18,944,446 | 18,619,523 | 17,792,519 | 17,145,436 | 19,384,077 |
| Filtered and chimera removed | 18,271,442 | 17,889,794 | 17,782,513 | 15,876,672 | 17,454,382 |
| Assigned | 11,886,532 | 11,112,721 | 10,347,227 | 8,940,467 | 10,650,189 |
| Unassigned (original data) | 10,577,615 | 11,351,412 | 12,114,223 | 13,523,761 | 10,794,328 |
| Unassigned (demultiplexed data) | 9,023,985 | 9,797,796 | 10,563,290 | 11,970,037 | 9,950,328 |
| Bioinformatic Pipeline | Reservoir | Klíčava | Římov | Žlutice | Klíčava | Římov | Žlutice |
|---|---|---|---|---|---|---|---|
| Season | Summer | Autumn | |||||
| Anacapa | Taxa | 12 | 20 | 13 | 17 | 30 | 21 |
| Reads | 1,027,223 | 3,207,005 | 1,171,369 | 152,115 | 1,748,946 | 509,967 | |
| Barque | Taxa | 12 | 22 | 12 | 16 | 29 | 20 |
| Reads | 1,125,257 | 3,610,686 | 1,171,767 | 139,839 | 1,826,975 | 535,515 | |
| MetaBEAT | Taxa | 11 | 21 | 11 | 15 | 28 | 19 |
| Reads | 1,060,167 | 3,417,605 | 1,102,758 | 129,156 | 1,697,086 | 452,972 | |
| MiFish | Taxa | 12 | 21 | 12 | 16 | 29 | 20 |
| Reads | 916,496 | 2,927,711 | 939,469 | 113,122 | 1,490,978 | 432,617 | |
| SEQme | Taxa | 10 | 19 | 11 | 17 | 29 | 18 |
| Reads | 1,230,336 | 3,316,388 | 1,228,640 | 135,439 | 1,729,903 | 559,636 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, R.A.; Blabolil, P. Comparison of Bioinformatic Pipelines for eDNA Metabarcoding Data Analysis of Fish Populations. Fishes 2025, 10, 214. https://doi.org/10.3390/fishes10050214
dos Santos RA, Blabolil P. Comparison of Bioinformatic Pipelines for eDNA Metabarcoding Data Analysis of Fish Populations. Fishes. 2025; 10(5):214. https://doi.org/10.3390/fishes10050214
Chicago/Turabian Styledos Santos, Romulo A., and Petr Blabolil. 2025. "Comparison of Bioinformatic Pipelines for eDNA Metabarcoding Data Analysis of Fish Populations" Fishes 10, no. 5: 214. https://doi.org/10.3390/fishes10050214
APA Styledos Santos, R. A., & Blabolil, P. (2025). Comparison of Bioinformatic Pipelines for eDNA Metabarcoding Data Analysis of Fish Populations. Fishes, 10(5), 214. https://doi.org/10.3390/fishes10050214








