DNA Barcoding of the Genus Discogobio (Teleostei, Cyprinidae) in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Identification
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Sequence Characterization, Genetic Distance, and Phylogenetic Analysis
2.4. Species Delimitation
3. Results
3.1. Sequence Characterization and Population Genetic Diversity
3.2. Species Boundary Discordance Across Analytical Approaches
3.3. Genetic Distance and Phylogenetic Analysis
4. Discussion
4.1. Applicability of DNA Barcode in the Identification of the Genus Discogobio
4.2. Different Methods Result in Different Species Delimitation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, J.L.; Dujsebayeva, T.N.; Chirikova, M.A.; Gong, X.; Li, D.J.; Guo, X.G. Does the Dzungarian racerunner (Eremias dzungarica Orlova, Poyarkov, Chirikova, Nazarov, Munkhbaatar, Munkhbayar & Terbish, 2017) occur in China? Species delimitation and identification with DNA barcoding and morphometric analyses. Zool. Res. 2021, 42, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.; Costa, F.O.; Walker, D.I.; North, A.W.; Hutchinson, W.F.; Carvalho, G.R. DNA barcodes of fish of the Scotia Sea, Antarctica indicate priority groups for taxonomic and systematics focus. Antarct. Sci. 2008, 20, 253–262. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Allison Jr, P.F.; Pickich, E.T.; Barnett, Z.C.; Garrick, R.C. DNA barcoding is currently unreliable for species identification in most crayfishes. Ecol. Evol. 2024, 14, e70050. [Google Scholar] [CrossRef]
- Zhou, Z.; Guo, H.; Han, L.; Chai, J.; Che, X.; Shi, F. Singleton molecular species delimitation based on COI-5P barcode sequences revealed high cryptic/undescribed diversity for Chinese katydids (Orthoptera: Tettigoniidae). BMC Evol. Biol. 2019, 19, 79. [Google Scholar] [CrossRef]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, B.L.; Baton, L.A.; Shimabukuro, P.H.F. Single-locus DNA barcoding and species delimitation of the sandfly subgenus Evandromyia (Aldamyia). Med. Vet. Entomol. 2020, 34, 420–431. [Google Scholar] [CrossRef]
- Yan, Y.B.; Cheng, Q.Q. Study on molecular taxonomy of genus Pampus based on complete mitochondrial genome structure. Mar. Fish. 2022, 44, 31–44. (In Chinese) [Google Scholar] [CrossRef]
- Lee, H.L.; Yu, H.J.; Kim, J.K. New descriptions of four larval lanternfish species and cryptic diversity of Benthosema pterotum (Pisces: Myctophidae) from the Northwest Pacific. J. Fish Biol. 2022, 101, 1474–1500. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, Y.P.; Ye, J.W.; Chen, H.Y.; Chen, Z.H.; Liang, R.S. DNA barcoding and molecular phylogeny of Pteroninae species in China Sea. Mar. Fish. 2024, 46, 257–265. [Google Scholar] [CrossRef]
- Baisvar, V.S.; Kumar, R.; Singh, M.; Kushwaha, B. Cytochrome-C oxidase I gene-based genetic divergence and molecular phylogeny among the species of fish genus Channa. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2019, 89, 1455–1463. [Google Scholar] [CrossRef]
- Bektas, Y.; Aksu, İ.; Kaya, C.; Bayçelebi, E.; Turan, D. DNA barcoding and species delimitation of the genus Oxynoemacheilus (Teleostei: Nemacheilidae) in Anatolia. J. Fish Biol. 2022, 101, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Yue, P.Q.; Chen, J.X. Labeoninae. In Fauna Sinica Osteichthyes Cypriniformes; Yue, P.Q., Ed.; Beijing Science Press: Beijing, China, 2000; pp. 171–174. (In Chinese) [Google Scholar]
- Tan, H.; Zhang, M.; Cheng, G.; Chen, X. The complete mitochondrial genome of Discogobio laticeps (Cyprinidae: Labeoninae). Mitochondrial DNA Part B 2019, 4, 1981–1982. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Catalog of Fishes: Genera, Species, References; California Academy of Sciences: San Francisco, CA, USA, 2024; Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 22 December 2024).
- Lin, S.Y. Carps and Carp-Like Fishes of Kwangtung and Adjacent Inlands; Fishery Experimental Station, Bureau of Reconstruction: Shenzhen, China, 1931; p. 167. [Google Scholar]
- Wu, X.; Lin, R.; Chen, X.; He, M. The Cyprinid Fishes of China; Shanghai Scientific Technical Press: Shanghai, China, 1977; Volume 2, pp. 229–394. [Google Scholar]
- Huang, S.Y. Four new species of the genus Discogobio from Yunnan, China. Zool. Res. 1989, 10, 356–361. (In Chinese) [Google Scholar]
- Chu, X.L.; Cui, G.H.; Zhou, W. A taxonomic review of fishes of the genus Discogobio, with description of two new species (Cypriniformes: Cyprinidae). Acta Zootaxonomica Sin. 1993, 18, 237–246. (In Chinese) [Google Scholar]
- Cui, G.H.; Zhou, W.; Lan, J.H. Discogobio multilineatus, a new cyprinid species from China (Teleostei: Cyprinidae). Ichthyol. Explor. Freshw. 1993, 4, 155–160. [Google Scholar]
- Zheng, L.; Zhou, W. Revision of the cyprinid genus Discogobio Lin, 1931 (Pisces: Teleostei) from the upper Red River basin in Wenshan Prefecture, Yunnan, China, with descriptions of three new species. Environ. Biol. Fishes 2008, 81, 255–266. [Google Scholar] [CrossRef]
- Yang, Q.; Zhou, W.; Shu, S.S. Morphological and differentiation of Discogobio yunnanensis from different population. Acta Zoo Taxon. Ica Sin. 2011, 36, 117–124. [Google Scholar]
- Xue, D. Guide to Collection, Preservation, Identification and Information Share of Animal Specimens; Standard Press of China: Beijing, China, 2010; pp. 103–125. [Google Scholar]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Moberly, J.G.; Bernards, M.T.; Waynant, K.V. Key features and updates for origin 2018. J. Cheminform. 2018, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- McLean, D. Adobe photoshop and illustrator techniques. J. Audiov. Media Med. 2002, 25, 79–81. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G.J. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, M.T.; Wild, R.; Elliot, M.; Fujisawa, T.; Balke, M.; Inward, D.J.; Lees, D.C.; Ranaivosolo, R.; Eggleton, P.; Barraclough, T.G.; et al. Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 2009, 58, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Barraclough, T. Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Zheng, L.P.; Geng, Y.M. Complete mitochondrial genome of Guigarra cailaoensis Wang, Chen & Zheng, 2022 (Cypriniformes, Cyprinidae) and its phylogenetic implications. ZooKeys 2024, 1190, 75–89. [Google Scholar] [CrossRef]
- Miralles, A.; Puillandre, N.; Vences, M. DNA barcoding in species delimitation: From genetic distances to integrative taxonomy. Methods Mol. Biol. 2024, 2744, 77–104. [Google Scholar] [CrossRef]
- Lara, A.; Ponce de León, J.L.; Rodriguez, R.; Casane, D.; Cote, G.; Bernatchez, L.; García-Machado, E.R. DNA barcoding of Cuban freshwater fishes: Evidence for cryptic species and taxonomic conflicts. Mol. Ecol. Resour. 2010, 10, 421–430. [Google Scholar] [CrossRef]
- Shen, Y.J.; Hubert, N.; Huang, Y.; Wang, X.Z.; Gan, X.N.; Peng, Z.G. DNA barcoding the ichthyofauna of the Yangtze River: Insights from the molecular inventory of a mega-diverse temperate fauna. Mol. Ecol. Resour. 2019, 19, 1278–1291. [Google Scholar] [CrossRef]
- Tang, Q.; Deng, L.; Luo, Q.; Duan, Q.; Wang, X.; Zhang, R. DNA barcoding of fish species diversity in Guizhou, China. Diversity 2023, 15, 203. [Google Scholar] [CrossRef]
- Greenstone, M.H.; Vandenberg, N.J.; Hu, J. Barcode haplotype variation in North American agroecosystem lady beetles (Coleoptera: Coccinellidae). Mol. Ecol. Resour. 2011, 11, 629–637. [Google Scholar] [CrossRef]
- Li, Q.; Liu, Q.; Yu, Y.; Lin, X.; He, X.; Huang, X. Revealing cryptic diversity and population divergence in subtropical aphids through DNA barcoding. Zool. Scr. 2023, 52, 517–530. [Google Scholar] [CrossRef]
- Ren, J.L.; Ren, L.; Zhang, R.Z. Delimiting species, revealing cryptic diversity, and population divergence in Qinghai-Tibet Plateau weevils through DNA barcoding. Ecol. Evol. 2024, 14, e11592. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Y.P.; Yang, Z.Y.; Liu, Z.; Du, Y.Y. DNA barcoding reveals cryptic diversity in the underestimated genus Triplophysa (Cypriniformes: Cobitidae, Nemacheilinae) from the northeastern Qinghai-Tibet Plateau. BMC Evol. Biol. 2020, 20, 151. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Qin, G.; Zhang, H.X.; Wang, X.; Lin, Q. DNA barcoding reflects the diversity and variety of brooding traits of fish species in the family Syngnathidae along China’s coast. Fish. Res. 2017, 185, 137–144. [Google Scholar] [CrossRef]
- Hsiao, Y.; Oberprieler, R.G. An integrative taxonomic and phylogenetic approach reveals a new genus of Australasian Cycas-pollinating weevils (Coleoptera: Curculionidae: Cossoninae). Zool. J. Linn. Soc. 2024, 202, zlad190. [Google Scholar] [CrossRef]
- Dupuis, J.R.; Roe, A.D.; Sperling, F.A. Multi-locus species delimitation in closely related animals and fungi: One marker is not enough. Mol. Ecol. 2012, 21, 4422–4436. [Google Scholar] [CrossRef]
- Knowles, L.L.; Carstens, B.C. Delimiting species without monophyletic gene trees. Syst. Biol. 2007, 56, 887–895. [Google Scholar] [CrossRef]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Li, T.C.; Wang, Y.K.; Sui, Z.X.; Wang, T.; Nian, J.Z.; Jiang, J.Z.; Tang, T.; Liu, F.S.; Xiao, J.H.; Huang, D.W. Multiple mitochondrial haplotypes within individual specimens may interfere with species identification and biodiversity estimation by DNA barcoding and metabarcoding in fig wasps. Syst. Entomol. 2021, 46, 887–899. [Google Scholar] [CrossRef]
- Li, J.J.; Li, Q.; Wu, Y.X.; Ye, L.Q.; Liu, H.H.; Wei, J.F.; Huang, X.L. Mountains act as museums and cradles for hemipteran insects in China: Evidence from patterns of richness and phylogenetic structure. Glob. Ecol. Biogeogr. 2021, 30, 1070–1085. [Google Scholar] [CrossRef]
- Rubinoff, D.; Cameron, S.; Will, K. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J. Hered. 2006, 97, 581–594. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Moulton, M.J.; Whiting, M.F. Rampant nuclear insertion of mtDNA across diverse lineages within Orthoptera (Insecta). PLoS ONE 2014, 9, e110508. [Google Scholar] [CrossRef]
- Scribner, K.T.; Page, K.S.; Bartron, M.L. Hybridization in freshwater fishes: A review of case studies and cytonuclear methods of biological inference. Rev. Fish Biol. Fish. 2000, 10, 293–323. [Google Scholar] [CrossRef]
- Mastrantonio, V.; Porretta, D.; Urbanelli, S.; Crasta, G.; Nascetti, G. Dynamics of mtDNA introgression during species range expansion: Insights from an experimental longitudinal study. Sci. Rep. 2016, 6, 30355. [Google Scholar] [CrossRef]
- Carstens, B.C.; Pelletier, T.A.; Reid, N.M.; Satler, J.D. How to fail at species delimitation. Mol. Ecol. 2013, 22, 4369–4383. [Google Scholar] [CrossRef]
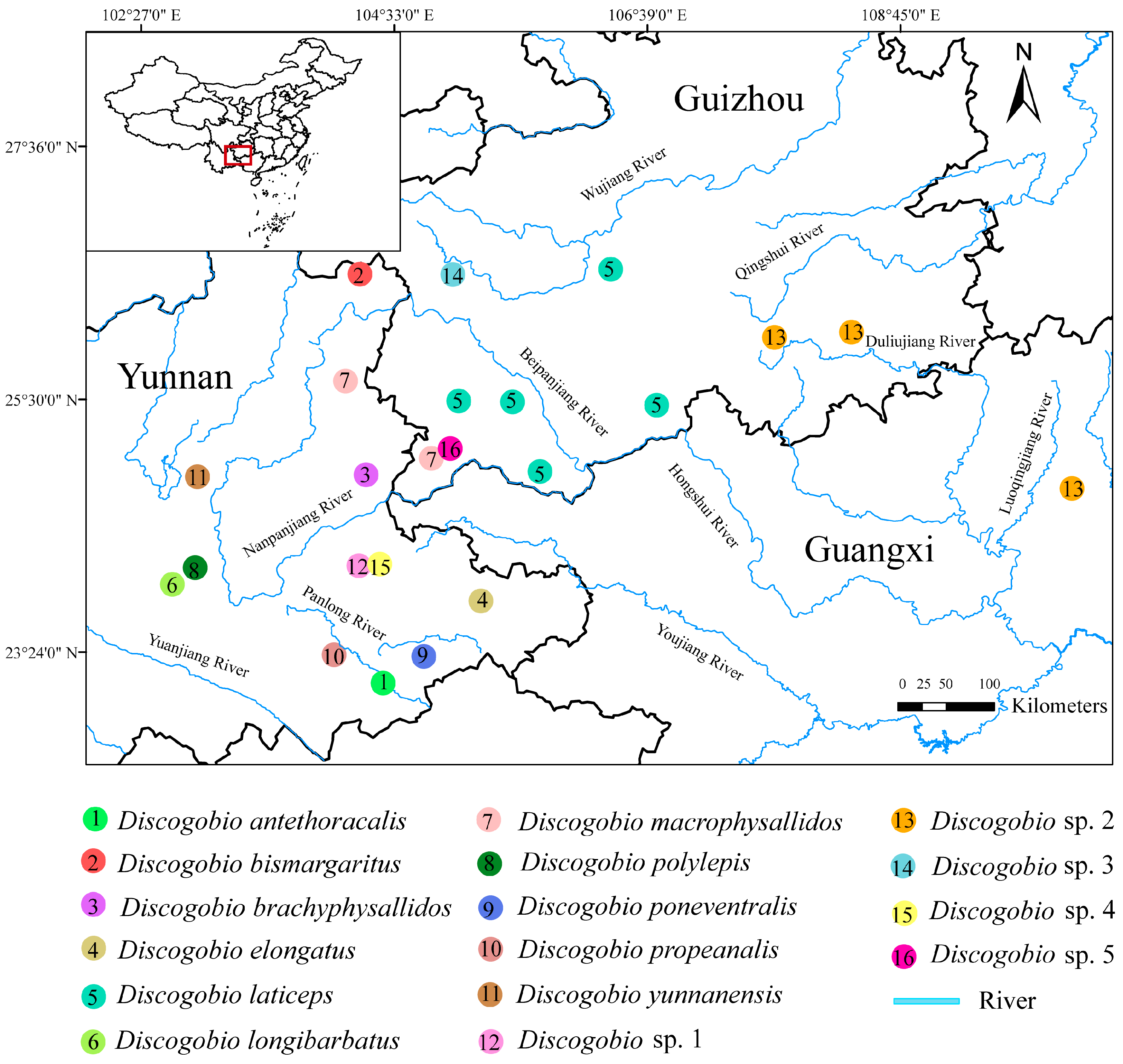
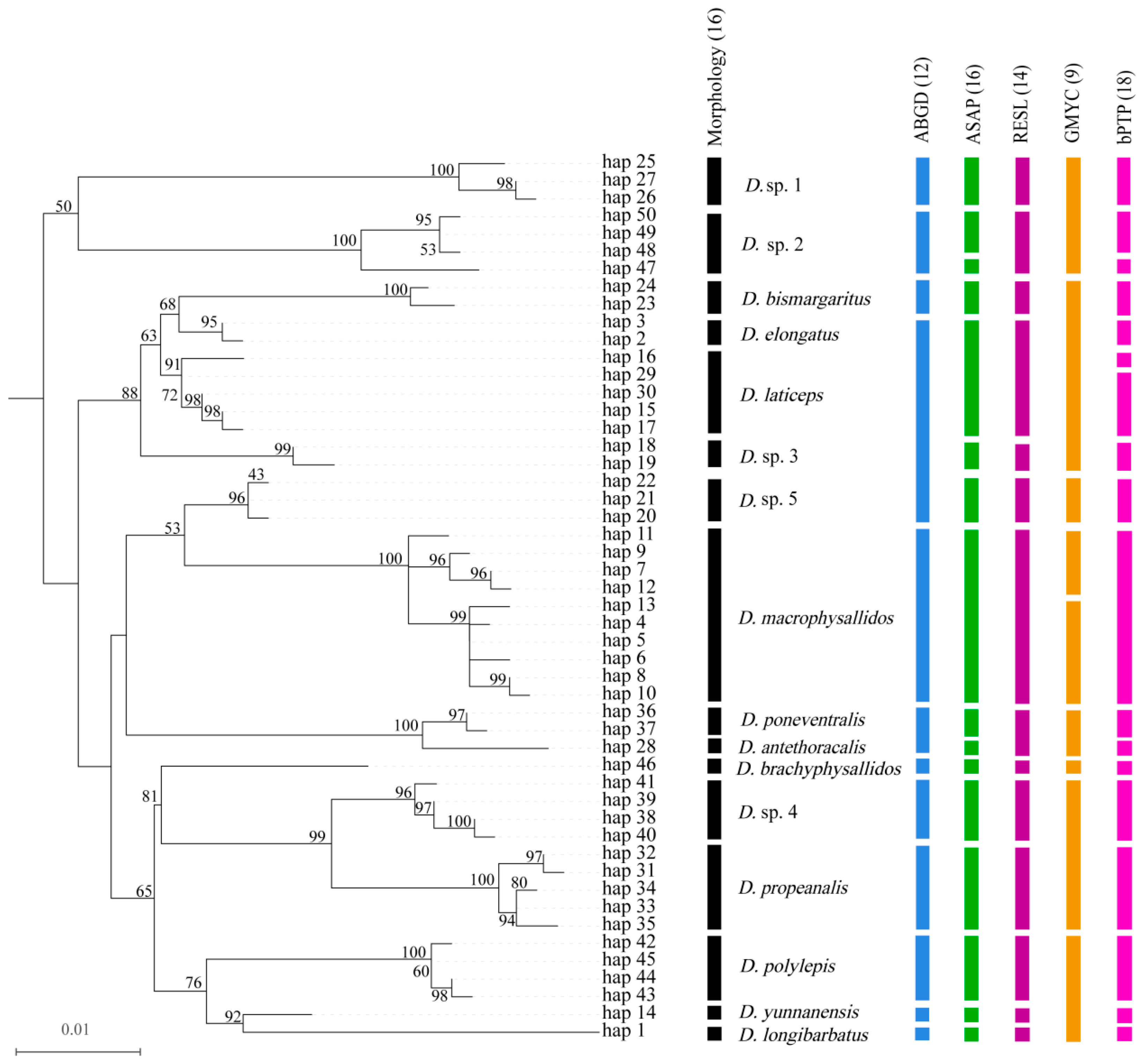
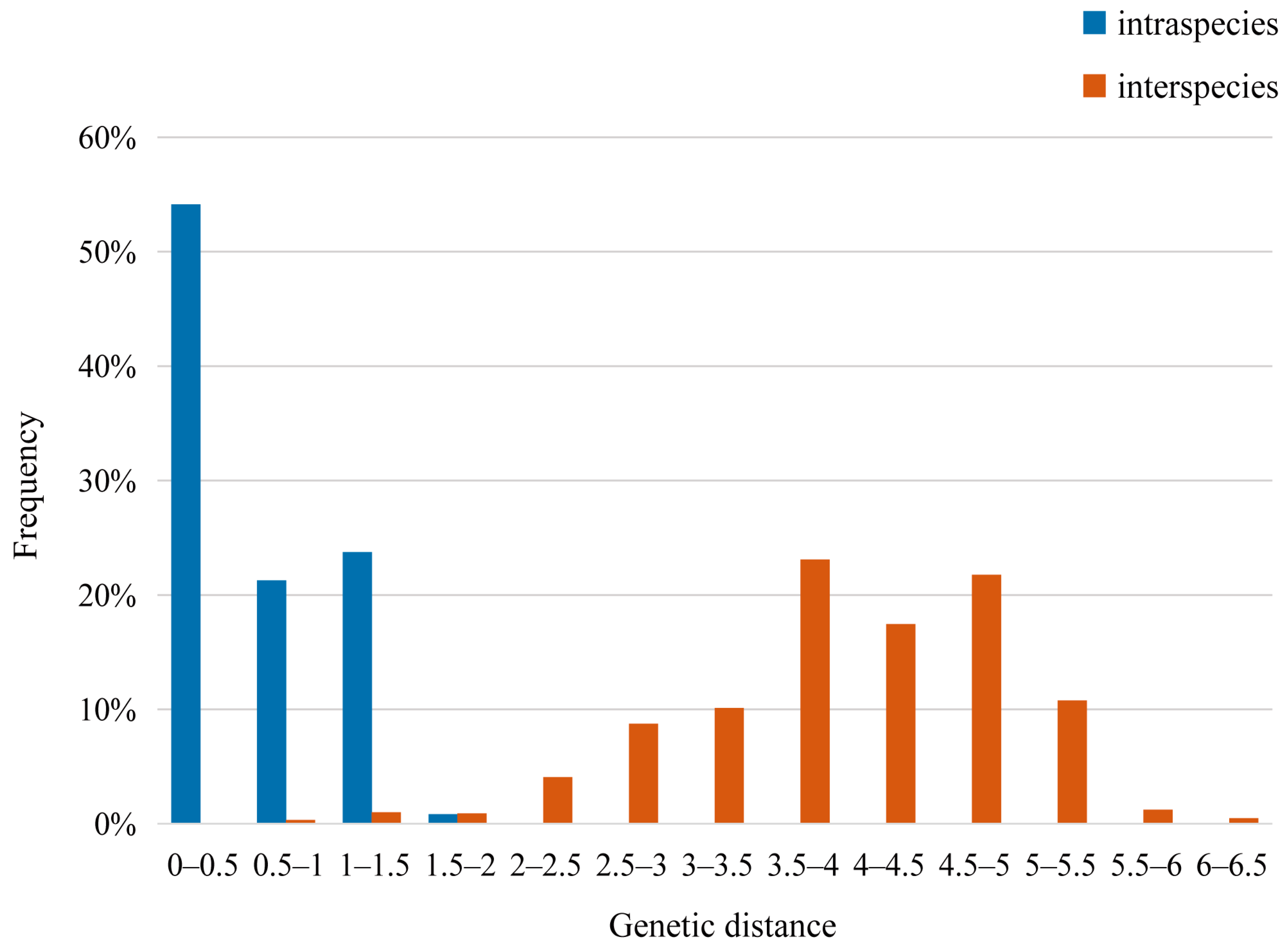
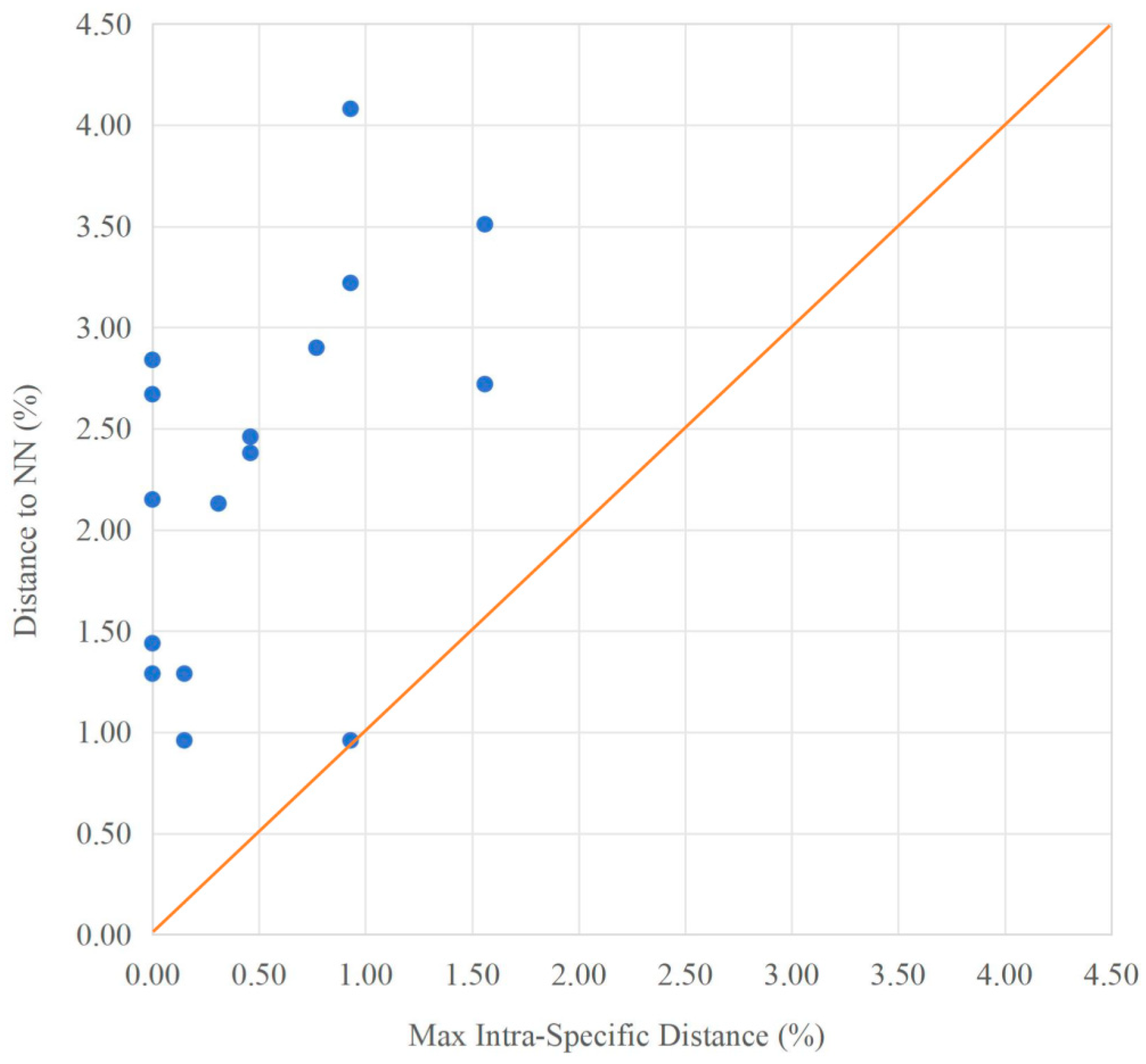
| Species | Collection Site | Haplotype | Number of Specimens (N) | Number of Haplotypes (Nh) | Haplotype Diversity (h) | Nucleotide Diversity (π) |
|---|---|---|---|---|---|---|
| D. antethoracalis | Wenshan, Yunnan, China | Hap 28 | 12 | 1 | / | / |
| D. bismargaritus | Guangnan, Yunnan, China | Hap 23–24 | 4 | 2 | 0.500 ± 0.265 | 0.00230 ± 0.00122 |
| D. brachyphysallidos | Yiliang, Yunnan, China | Hap 46 | 3 | 1 | / | / |
| D. elongatus | Xuanwei, Yunnan, China | Hap 2–3 | 4 | 2 | 0.500 ± 0.265 | 0.00077 ± 0.00041 |
| D. laticeps | Zhenfeng, Xingren, Xiuwen, Ceheng, Luodian, Guizhou, China | Hap 15–17 Hap 29–30 | 26 | 5 | 0.732 ± 0.054 | 0.00371 ± 0.00041 |
| D. longibarbatus | Yuxi, Yunnan, China | Hap 1 | 3 | 1 | / | / |
| D. macrophysallidos | Fuyuan, Yunnan; Xingyi, Guizhou, China | Hap 4–13 | 67 | 10 | 0.698 ± 0.055 | 0.00605 ± 0.00063 |
| D. polylepis | Yuxi, Yunnan, China | Hap 42–45 | 10 | 4 | 0.733 ± 0.120 | 0.00157 ± 0.00040 |
| D. poneventralis | Wenshan, Yunnan, China | Hap 36–37 | 9 | 2 | 0.500 ± 0.128 | 0.00077 ± 0.00020 |
| D. propeanalis | Wenshan, Yunnan, China | Hap 31–35 | 15 | 5 | 0.705 ± 0.088 | 0.00249 ± 0.00075 |
| D. yunnanensis | Yiliang, Yuannan, China | Hap 14 | 1 | 1 | / | / |
| D. sp. 1 | Wenshan, Yunnan, China | Hap 25–27 | 5 | 3 | 0.800 ± 0.164 | 0.00553 ± 0.00142 |
| D. sp. 2 | Rongjiang, Sandu, Guizhou; Gongcheng, Guangxi, China | Hap 47–50 | 13 | 4 | 0.615 ± 0.136 | 0.00492 ± 0.00201 |
| D. sp. 3 | Shuicheng, Guizhou, China | Hap 18–19 | 4 | 2 | 0.500 ± 0.265/ | 0.00154 ± 0.00081/ |
| D. sp. 4 | Wenshan, Yunnan, China | Hap 38–41 | 13 | 4 | 0.603 ± 0.131 | 0.00213 ± 0.00061 |
| D. sp. 5 | Xingyi, Guizhou, China | Hap 20–22 | 17 | 3 | 0.404 ± 0.130 | 0.00077 ± 0.00028 |
| Species | Mean Intra | Max Intra | NN Dist | NN |
|---|---|---|---|---|
| D. antethoracalis | 0.00% | 0.00% | 1.29% | D. poneventralis |
| D. bismargaritus | 0.23% | 0.46% | 2.38% | D. laticeps |
| D. brachyphysallidos | 0.00% | 0.00% | 2.67% | D. yunnanensis |
| D. elongatus | 0.08% | 0.15% | 0.96% | D. laticeps |
| D. laticeps | 0.37% | 0.93% | 0.96% | D. elongatus |
| D. longibarbatus | 0.00% | 0.00% | 2.84% | D. yunnanensis |
| D. macrophysallidos | 0.61% | 1.56% | 2.72% | D. sp. 4 |
| D. polylepis | 0.16% | 0.46% | 2.46% | D. yunnanensis |
| D. poneventralis | 0.08% | 0.15% | 1.29% | D. antethoracalis |
| D. propeanalis | 0.25% | 0.93% | 3.22% | D. sp. 4 |
| D. yunnanensis | / | / | 2.15% | D. sp. 4 |
| D. sp. 1 | 0.56% | 0.93% | 4.08% | D. laticeps |
| D. sp. 2 | 0.21% | 0.77% | 2.90% | D. yunnanensis |
| D. sp. 3 | 0.50% | 1.56% | 3.51% | D. elongatus |
| D. sp. 4 | 0.07% | 0.31% | 2.13% | D. laticeps |
| D. sp. 5 | 0.00% | 0.00% | 1.44% | D. elongatus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Cheng, H.; Huang, R.; Qiu, Z.; Zhang, R. DNA Barcoding of the Genus Discogobio (Teleostei, Cyprinidae) in China. Fishes 2025, 10, 157. https://doi.org/10.3390/fishes10040157
Li H, Cheng H, Huang R, Qiu Z, Zhang R. DNA Barcoding of the Genus Discogobio (Teleostei, Cyprinidae) in China. Fishes. 2025; 10(4):157. https://doi.org/10.3390/fishes10040157
Chicago/Turabian StyleLi, Hongmei, Huan Cheng, Renrong Huang, Zhenya Qiu, and Renyi Zhang. 2025. "DNA Barcoding of the Genus Discogobio (Teleostei, Cyprinidae) in China" Fishes 10, no. 4: 157. https://doi.org/10.3390/fishes10040157
APA StyleLi, H., Cheng, H., Huang, R., Qiu, Z., & Zhang, R. (2025). DNA Barcoding of the Genus Discogobio (Teleostei, Cyprinidae) in China. Fishes, 10(4), 157. https://doi.org/10.3390/fishes10040157







