Environmental DNA Was Utilized to Assess Fish Diversity and Community Structure in the Qingshui River
Abstract
1. Introduction
2. Materials and Methods
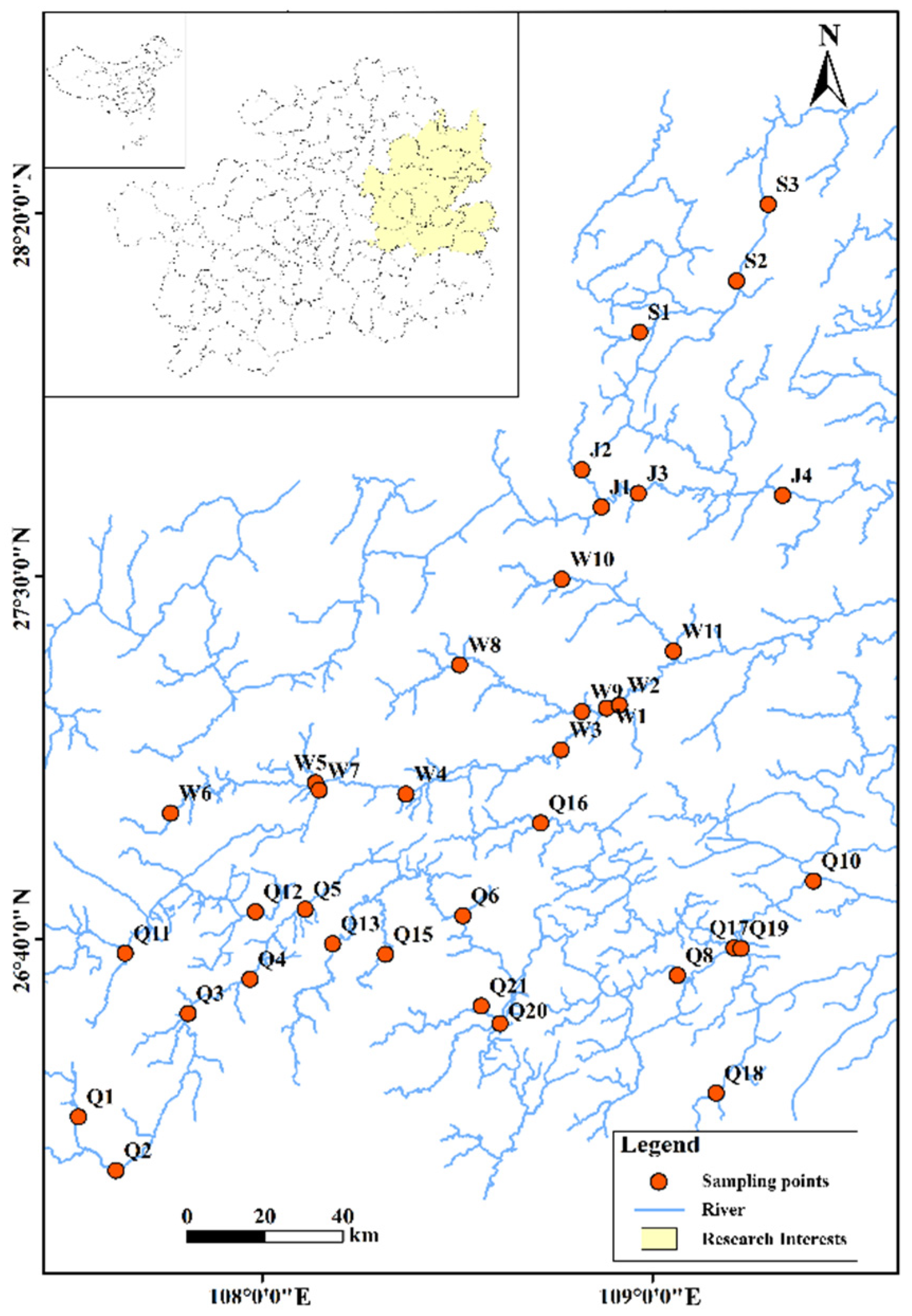
2.1. Sampling Site
Sampling Method
2.2. Data Analysis
2.2.1. DNA Analysis and Species Identification
2.2.2. Statistical Analysis
2.3. Alpha Diversity Analysis
2.4. Beta Diversity Analysis
2.5. Redundancy Analysis of Environmental Factors
3. Results
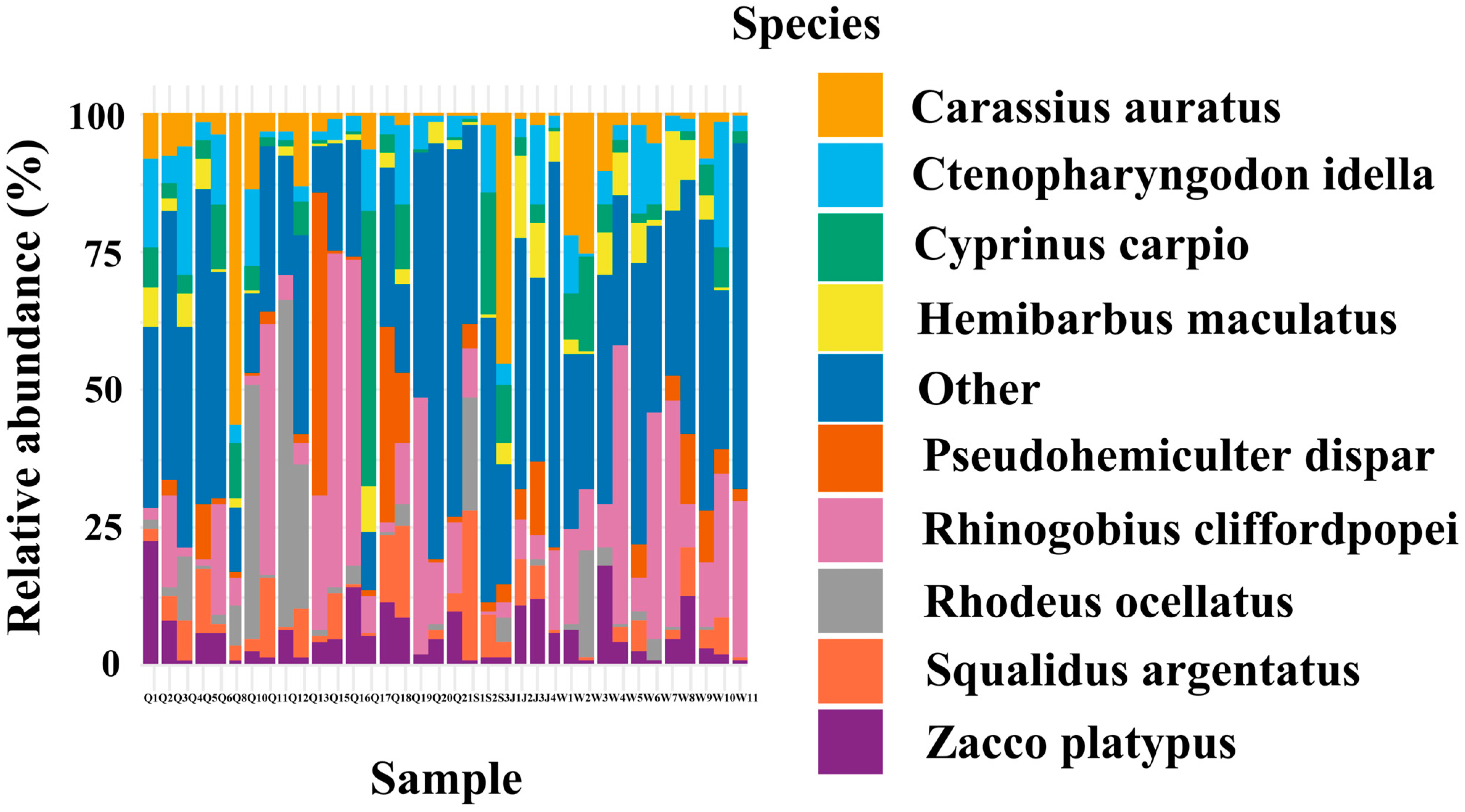
3.1. Composition of Fish Community
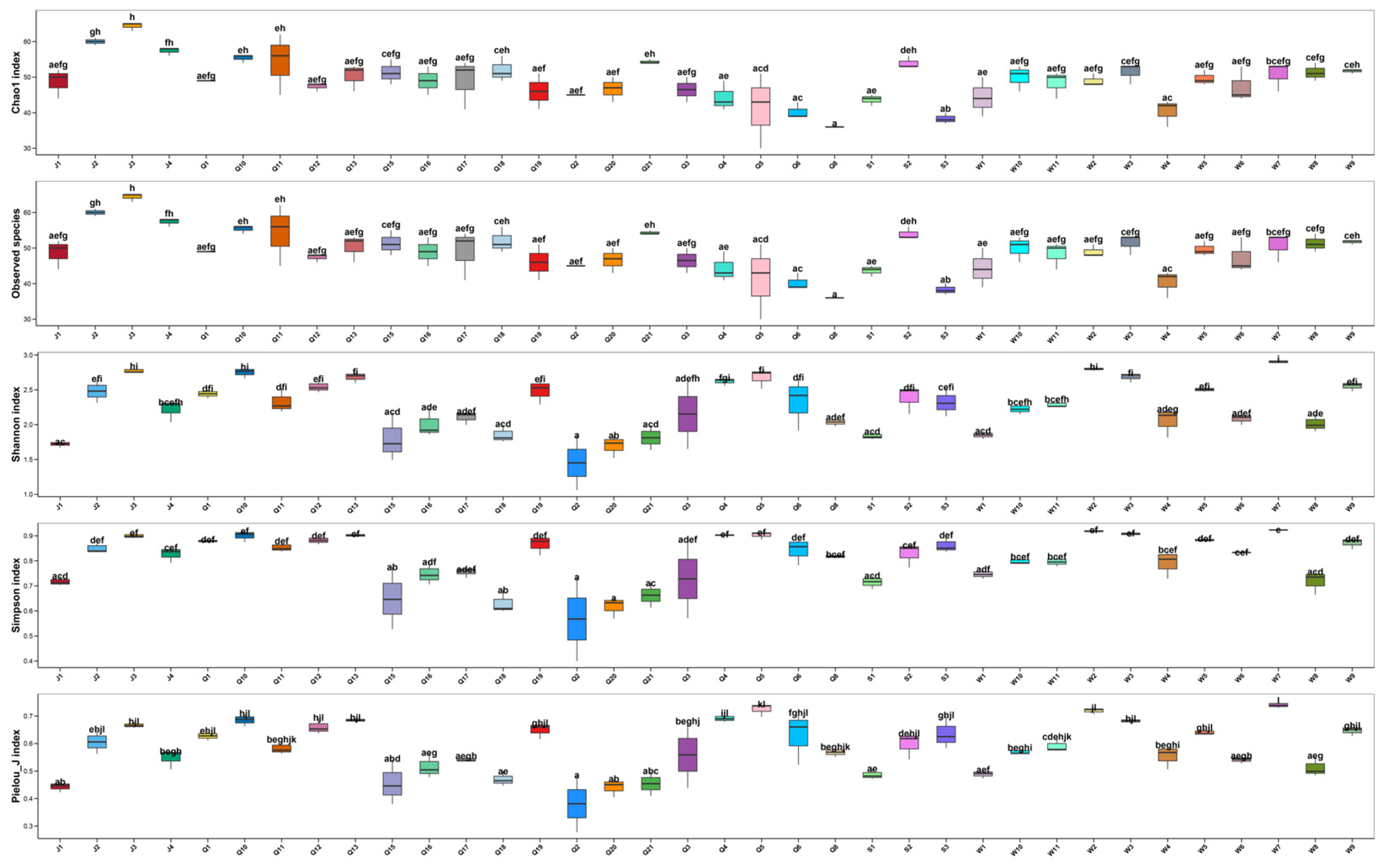
3.2. Alpha Diversity
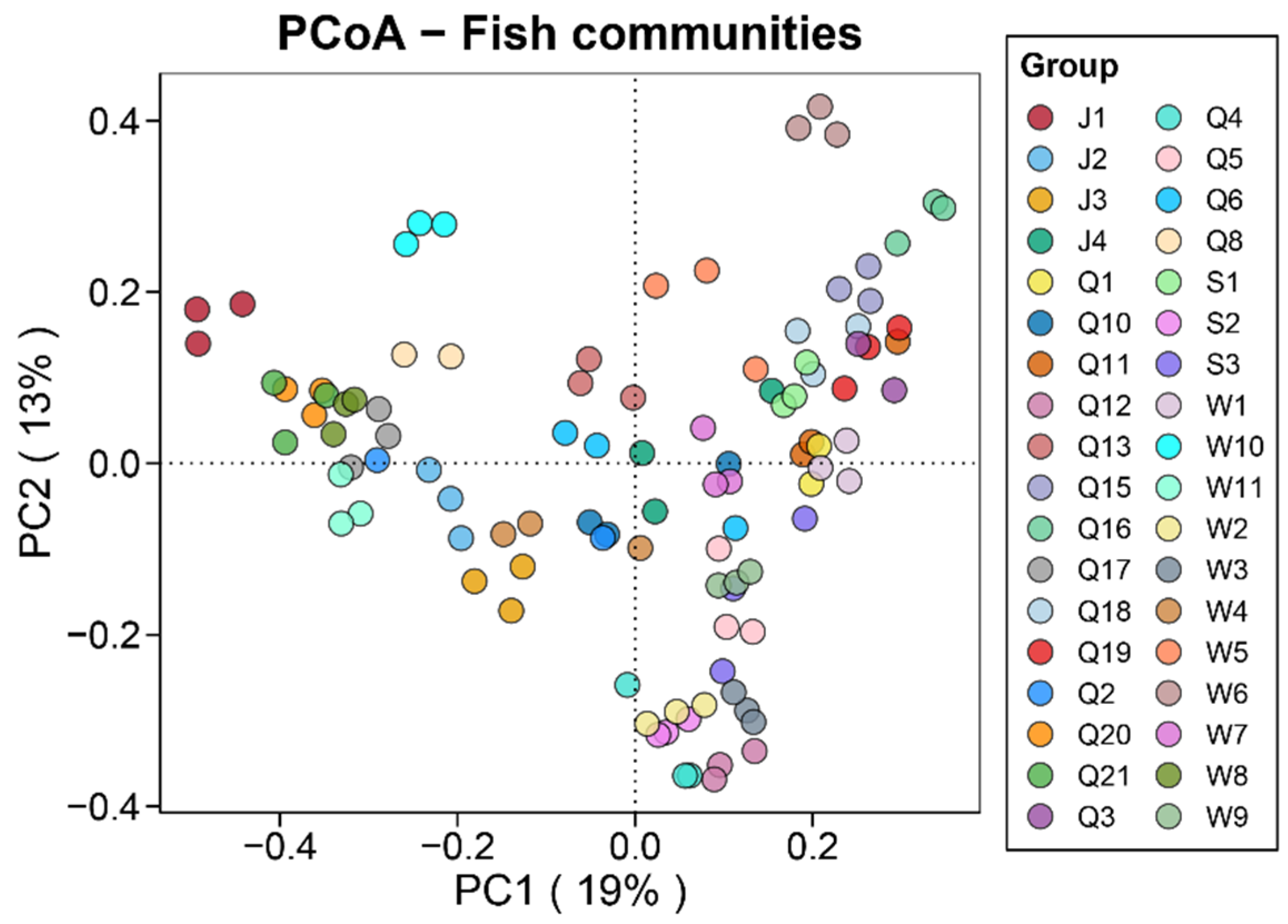
3.3. Beta Diversity
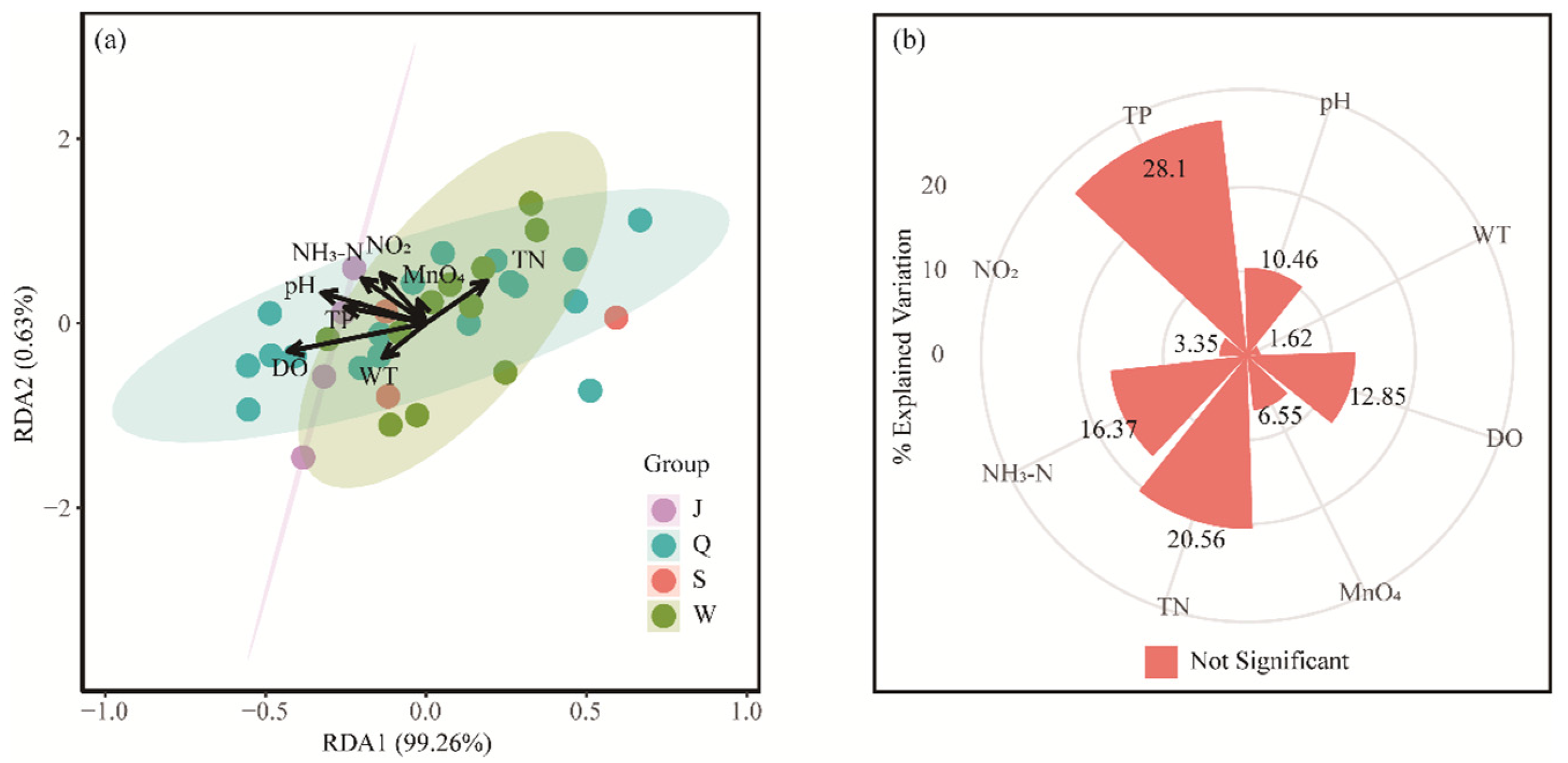
3.4. Redundancy Analysis
4. Discussion
4.1. eDNA Technology Enriches Historical Monitoring Data
4.2. Analysis of Differences in Fish Community Structure in the Qingshui River
4.3. Effects of Spatial Variations in Environmental Factors on Fish Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Z.N.; Tong, A.N.; Wu, Y.N.; Shui, B.N.; Hu, C.Y.; Wang, J. The impact of rare species loss on species diversity and functional diversity offish communities. Acta Ecol. Sin. 2025, 45, 252–264. [Google Scholar]
- Yu, Y.; Lu, J.; Xie, B.; Xie, B.; Deng, Y.Q.; Chen, S.Z.; Liu, Y.D.; Qu, X.D. Fish Community Structure and Species Biodiversity in 13 Lakes of Nanchang City. J. Hydroecol. 2021, 42, 110–118. [Google Scholar]
- Chen, H.D.; Zhang, H.; Fu, S.; Lin, Z.M.; Liu, C.; Chen, K.N.; Zhang, Y.G.; Mao, Z.G.; Mei, X.Y.; Liu, Z.W.; et al. Spatiotemporal patterns of fish communities and diversity in Lake Hengshui and their relationships with water environmental factors. J. Lake Sci. 2024, 36, 1447–1458. [Google Scholar]
- Lin, T.; Cui, F.Y.; Liu, D.M.; Ma, F.; Zhang, L.Q. Effect of food habits of some fishes on the biomanipulation in drinking water source management. J. Harbin Inst. Technol. 2006, 38, 35–37+148. [Google Scholar]
- Chen, B.H.; Liu, Z.W. Effect of filter-feeding omnivorous fish on zooplankton community. Ecol. Sci. 2012, 31, 161–166. [Google Scholar]
- Yu, J.; Liu, Z.; Li, K.; Chen, F.; Guan, B.; Hu, Y.; Zhong, P.; Tang, Y.; Zhao, X.; He, H.; et al. Restoration of shallow lakes in subtropical and tropical China:Response of nutrients and water clarity to biomanipulation by fish removal and submerged plant transplantation. Water 2016, 8, 438. [Google Scholar] [CrossRef]
- Joy, M.K.; Death, R.G. Predictive modelling of freshwater fish as a biomonitoring tool in New Zealand. Freshw. Biol. 2002, 47, 2261–2275. [Google Scholar] [CrossRef]
- Zhou, B.Z.; Wang, H.X.; Shen, P. Ecological Environmental Vicissitudes of Qingshui River Basins and Sustainable Development. Chongqing Environ. Sci. 2000, 14–16. [Google Scholar]
- Hu, S.R.; Mo, R.B.; Ye, G.K.; Zhou, C.H.; Yang, Z.H. Thoughts on the Sustainable Development of Ecological Fisheries in Qingshuijiang of Qiandongnan. Guizhou J. Anim. Husb. Vet. Med. 2019, 43, 23–27. [Google Scholar]
- Li, S. Genetic Structure and Genetic Diversity of Sinipercascherzeri Population in Qingshuijiang River. Master’s Thesis, Guizhou University, Guiyang, China, 2018. [Google Scholar]
- Zeng, S.; Liu, W.; Du, Q.; Wang, Y.Y.; Zhou, Z.; Hu, S.R. Early Development of Gonad and Preliminary Analysis of Foxl2 Gene Function in Qingshuijiang Cyprinus carpio. Guizhou Agric. Sci. 2020, 48, 82–86+173. [Google Scholar]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 2019, 286, 20191409. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, H.D.; Misutka, M.D.; Glover, C.N. Environmental DNA and environmental RNA: Current and prospective applications for biological monitoring. Sci. Total Environ. 2021, 782, 146891. [Google Scholar] [PubMed]
- Yao, M.; Zhang, S.; Lu, Q.; Chen, X.; Zhang, S.Y.; Kong, Y.; Zhao, J. Fishing for fish environmental DNA: Ecological applications, methodological considerations, surveying designs, and ways forward. Mol. Ecol. 2022, 31, 5132–5164. [Google Scholar] [PubMed]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating Occupancy and Abundance of Stream Amphibians Using Environmental DNA from Filtered Water Samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar]
- Hou, T.; Lu, S.; Shao, J.; Dong, X.; Yang, Z.; Yang, Y.; Yao, D.; Lin, Y. Assessment of planktonic community diversity and stability in lakes and reservoirs based on eDNA metabarcoding—A case study of Minghu National Wetland Park, China. Environ. Res. 2025, 271, 121025. [Google Scholar]
- Zhang, S.; Lu, Q.; Wang, Y.; Wang, X.; Zhao, J.; Yao, M. Assessment of fish communities using environmental DNA: Effect of spatial sampling design in lentic systems of different sizes. Mol. Ecol. Resour. 2020, 20, 242–255. [Google Scholar] [CrossRef]
- Chen, W.; Cheng, Y.; Zhang, S.; Pan, Q. Heuristic Clustering Method Based on Neighbor-Seeds for 454 Sequencing Data. J. Softw. 2014, 25, 929–938. [Google Scholar]
- Li, B.; Zhang, X.; Guo, F.; Wu, W.; Zhang, T. Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res. 2013, 47, 4207–4216. [Google Scholar]
- Qu, J.Q.; Hao, T.F.; Li, Y.G.; Liu, L.M.; Hao, D.S.; Jia, C.X.; Zhang, Q.J. Analysis of fish community structure and ecological niche changes in Miyun Reservoir based on environmental DNA technology. J. Dalian Ocean Univ. 2024, 39, 298–307. [Google Scholar]
- Liu, W.Y.; Huang, J.Q.; Dai, G.X.; Liu, G.Q.; Lei, X.T.; Zhou, Y.D.; Ye, Y.C. RDA analysis between fish community structure and environmental factors in the Lingdingyang Estuary. Fish. Inf. Strategy 2023, 38, 32–41. [Google Scholar]
- Nakagawa, H.; Yamamoto, S.; Sato, Y.; Sado, T.; Minamoto, T.; Miya, M. Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshw. Biol. 2018, 6, 505–612. [Google Scholar] [CrossRef]
- Dai, Y.G.; Chen, Y.F. Status of fish resources in Qingshuijiang River and protection measures. J. Hydroecol. 2007, 75–78. [Google Scholar]
- Wang, M.; Jin, X.W.; Lin, X.L.; Du, L.N.; Cui, Y.D.; Wu, X.P.; Sun, H.Y.; Xie, Z.C.; Wang, X.H.; Wang, B.X. Advances in the macrozoobenthos biodiversity monitoring and ecosystem assessment using environmental DNA metabarcoding. Acta Ecol. Sin. 2021, 41, 7440–7453. [Google Scholar]
- Norshida, I.; Mohd Nasir, M.S.A.; Khaleel, A.G.; Sallehuddin, A.S.; Syed Idrus, S.N.; Istiqomah, I.; Venmathi, M.B.; Ahmad, S. First wild record of Australian redclaw crayfish Cherax quadricarinatus (von Martens, 1868) in the East Coast of Peninsular Malaysia. BioInvasions Rec. 2021, 10, 360–368. [Google Scholar]
- Ma, X.; Yang, H.; Zhong, X.; Zeng, P.; Zhou, X.; Zeng, S.; Dong, X.; Min, W.; Huang, F. eDNA metabarcoding analysis of the composition and spatial patterns of fish communities in the Sanbanxi Reservoir, China. Sustainability 2022, 14, 12966. [Google Scholar] [CrossRef]
- Yu, X.D.; Luo, T.H.; Zhou, H.Z. Large-scale patterns in species diversity of fishes in the Yangtze River Basin. Biodivers. Sci. 2005, 13, 473. [Google Scholar] [CrossRef]
- Wang, S.; Xie, Y. China Species Red List: Vol. 1, Red List; Higher Education Press: Beijing, China, 2004. [Google Scholar]
- Dai, Y.G.; Chen, Y.F. Fish fauna and its ecological types of Qingshui River in Guizhou Province, Southwest China. Chin. J. Ecol. 2007, 26, 682–687. [Google Scholar]
- Sankian, Z.; Khosravi, S.; Kim, Y.; Lee, S. Dietary protein requirement for juvenile mandarin fish, Siniperca scherzeri. J. World Aquac. 2019, 50, 34–41. [Google Scholar] [CrossRef]
- Wolter, C. Temperature influence on the fish assemblage structure in a large lowland river, the lower Oder River, Germany. Ecol. Freshw. Fish 2007, 16, 493–503. [Google Scholar] [CrossRef]
- G8 3838-2002; Environmental Quality Standard for Surface Water. China Environmental Science Press: Beijing, China, 2002.
- Zhu, S.; Chen, W.; Li, X.; Li, J.; Li, Y. Pattern of fish assemblage structure and diversity in Liujiang River. Acta Hydrobiol. Sin. 2022, 46, 375–384. [Google Scholar]
- Li, J.; Qin, H.; Pei, S.; Yao, L.; Wen, W.; Yi, L.; Zhou, J.; Tang, L. Analysis of an Ecological Flow Regime during the Ctenopharyngodon Idella Spawning Period Based on Reservoir Operations. Water 2019, 11, 2034. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, Z.G.; Hu, H.Y.; Jing, Y.; Cui, H.W.; Ding, D.S.; Gao, Y. Variation of nitrogen, phosphorus and COD during the cultivation of Ruditapes philippinarum. J. Agro-Environ. Sci. 2025, 1–12. Available online: http://kns.cnki.net/kcms/detail/12.1347.S.20240716.1757.006.html (accessed on 20 March 2025).
- Sun, C.C. Diatoms Biological Diversity and Relationship Among Diatoms with Heavy Metals and TN &TP in Wuyang River. Master’s Thesis, Guizhou University, Guiyang, China, 2015. [Google Scholar]
- Du, H.; Chen, Z.; Mao, G.; Chen, L.; Crittenden, J.; Li, R.Y.M.; Chai, L. Evaluation of eutrophication in freshwater lakes: A new non-equilibrium statistical approach. Ecol. Indic. 2019, 102, 686–692. [Google Scholar]
- Li, Q.; Liu, Y.; Li, S.; Guo, X.; Fu, Y.; He, N.; Ruan, G.; Wang, Q.; Gao, W.; Fang, L. Impact of nitrite exposure on oxidative stress and antioxidative-related genes responses in the gills of Procambarus clarkii. Fish Shellfish Immunol. 2022, 131, 624–630. [Google Scholar] [CrossRef]
- Ciji, A.; Akhtar, M.S. Nitrite implications and its management strategies in aquaculture: A review. Rev. Aquac. 2020, 12, 878–908. [Google Scholar]
- Sercu, B.; Jones, A.D.G.; Wu, C.H.; Escobar, M.H.; Serlin, C.L.; Knapp, T.A.; Andersen, G.L.; Holden, P.A. The Influence of In Situ Chemical Oxidation on Microbial Community Composition in Groundwater Contaminated with Chlorinated Solvents. Microb. Ecol. 2013, 65, 39–49. [Google Scholar] [CrossRef]
- Song, Y.Q. Insight into the Kinetics and Mechanism of Refractory Organic Pollutants Degradation in the Electro-Permanganate-Ozone Process. Master’s Thesis, Chongqing University, Chongqing, China, 2021. [Google Scholar]
- Liu, S.; Cao, X.X.; Wu, P.; Yang, S.D.; Zhang, T.; Liao, L. Relationship between bacterial diversity and environmental factors in reservoirs and their inflow basins under the influence of acid mine drainage. Environ. Chem. 2022, 41, 2121–2132. [Google Scholar]
- Cao, X.J.; Long, X.M. From Local Division to Co-Governance: Experiences and Reflections on Watershed Governance—Taking the Water Pollution Control of Qingshui River in the Border Area of Hunan, Chongqing and Guizhou as an Example. Soc. Sci. Guangxi 2019, 294, 54–58. [Google Scholar]
| Species | Documented | Endemic Fishes of Yangtze River | Exotic Species |
|---|---|---|---|
| Cypriniformes | |||
| Cyprinidae | |||
| Acrossocheilus jishouensis | + | + | |
| Acrossocheilus monticola | + | + | |
| Acrossocheilus parallens | + | ||
| Acrossocheilus yunnanensis | + | ||
| Bangana rendahli | + | + | |
| Carassius auratus | + | ||
| Cyprinus carpio | + | ||
| Discogobio yunnanensis | + | ||
| Folifer brevifilis | + | ||
| Onychostoma barbatum | + | ||
| Onychostoma lini | + | ||
| Onychostoma rara | + | ||
| Onychostoma simum | + | ||
| Pseudogyrinocheilus prochilus | + | ||
| Spinibarbus hollandi | + | ||
| Spinibarbus sinensis | + | + | |
| Xenocyprididae | |||
| Aphyocypris chinensis | + | ||
| Aphyocypris normalis | + | ||
| Chanodichthys ilishaeformis | + | ||
| Ctenopharyngodon idella | + | ||
| Culter alburnus | + | ||
| Elopichthys bambusa | + | ||
| Hemiculter bleekeri | + | ||
| Hemiculter leucisculus | + | ||
| Hemiculterella sauvagei | + | + | |
| Hypophthalmichthys molitrix | + | ||
| Hypophthalmichthys nobilis | + | ||
| Mylopharyngodon piceus | + | ||
| Opsariichthys bidens | + | ||
| Parabramis pekinensis | + | ||
| Pseudohemiculter dispar | + | ||
| Pseudolaubuca sinensis | + | ||
| Sinibrama macrops | + | + | |
| Squaliobarbus curriculus | + | ||
| Xenocypris davidi | + | ||
| Zacco platypus | + | ||
| Gobionidae | |||
| Abbottina binhi | + | ||
| Abbottina rivularis | + | ||
| Gnathopogon tsinanensis | + | ||
| Hemibarbus labeo | + | ||
| Hemibarbus maculatus | + | ||
| Microphysogobio fukiensis | + | ||
| Microphysogobio tungtingensis | + | + | |
| Platysmacheilus exiguus | + | ||
| Pseudorasbora parva | + | ||
| Rhinogobio cylindricus | + | + | |
| Sarcocheilichthys nigripinnis | + | ||
| Sarcocheilichthys sinensis | + | ||
| Saurogobio dabryi | + | ||
| Saurogobio punctatus | + | + | |
| Squalidus argentatus | + | ||
| Squalidus wolterstorffi | + | + | |
| Nemacheilidae | |||
| Homatula potanini | + | + | |
| Schistura fasciolata | + | ||
| Cobitidae | |||
| Cobitis macrostigma | + | + | |
| Leptobotia elongata | + | + | |
| Leptobotia guilinensis | + | ||
| Leptobotia pellegrini | + | ||
| Misgurnus anguillicaudatus | + | ||
| Parabotia banarescui | + | ||
| Parabotia kiangsiensis | + | ||
| Parabotia lijiangensis | + | ||
| Balitoridae | |||
| Sinogastromyzon hsiashiensis | + | + | |
| Acheilognathidae | |||
| Acheilognathus macropterus | + | ||
| Acheilognathus rhombeus | + | ||
| Rhodeus ocellatus | + | ||
| Synbranchiformes | |||
| Mastacembelidae | |||
| Macrognathus aculeatus | + | ||
| Synbranchidae | |||
| Monopterus albus | + | ||
| Beloniformes | |||
| Adrianichthyidae | |||
| Oryzias sinensis | + | ||
| Cyprinodontiformes | |||
| Poeciliidae | |||
| Gambusia affinis | + | + | |
| Siluriformes | |||
| Bagridae | |||
| Hemibagrus macropterus | + | ||
| Pseudobagrus brachyrhabdion | + | ||
| Pseudobagrus truncatus | + | ||
| Tachysurus eupogon | + | ||
| Tachysurus fulvidraco | + | ||
| Tachysurus intermedius | + | ||
| Tachysurus nitidus | + | ||
| Tachysurus vachellii | + | ||
| Siluridae | |||
| Silurus asotus | + | ||
| Silurus meridionalis | + | ||
| Sisoridae | |||
| Glyptothorax sinensis | + | ||
| Odontobutidae | |||
| Micropercops swinhonis | + | ||
| Odontobutis sinensis | + | ||
| Gobiidae | |||
| Rhinogobius cliffordpopei | + | ||
| Rhinogobius giurinus | + | ||
| Anabantiformes | |||
| Channidae | |||
| Channa asiatica | + | ||
| Centrarchiformes | |||
| Sinipercidae | |||
| Siniperca knerii | + | ||
| Siniperca loona | + | ||
| Siniperca roulei | + | ||
| Siniperca scherzeri | + | ||
| Siniperca undulata | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, F.; Zhang, R.; Lv, Z.; Xiang, Y.; Min, W.; Wang, X.; Liu, W.; Wang, W.; Zeng, S. Environmental DNA Was Utilized to Assess Fish Diversity and Community Structure in the Qingshui River. Fishes 2025, 10, 165. https://doi.org/10.3390/fishes10040165
Huang F, Zhang R, Lv Z, Xiang Y, Min W, Wang X, Liu W, Wang W, Zeng S. Environmental DNA Was Utilized to Assess Fish Diversity and Community Structure in the Qingshui River. Fishes. 2025; 10(4):165. https://doi.org/10.3390/fishes10040165
Chicago/Turabian StyleHuang, Fujiang, Ruiyuan Zhang, Zhengyu Lv, Yan Xiang, Wenwu Min, Xue Wang, Wei Liu, Wei Wang, and Sheng Zeng. 2025. "Environmental DNA Was Utilized to Assess Fish Diversity and Community Structure in the Qingshui River" Fishes 10, no. 4: 165. https://doi.org/10.3390/fishes10040165
APA StyleHuang, F., Zhang, R., Lv, Z., Xiang, Y., Min, W., Wang, X., Liu, W., Wang, W., & Zeng, S. (2025). Environmental DNA Was Utilized to Assess Fish Diversity and Community Structure in the Qingshui River. Fishes, 10(4), 165. https://doi.org/10.3390/fishes10040165






