Comparative Transcriptomic Analysis of Male and Female Gonads in the Zig-Zag Eel (Mastacembelus armatus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Handling
2.2. Paraffin Sectioning of M. armatus Gonads
2.3. Total RNA Extraction and Library Construction for Sequencing
2.4. Transcriptome Sequencing and Analysis
2.5. Real-Time Quantitative PCR (RT-qPCR) Validation
3. Results and Analysis
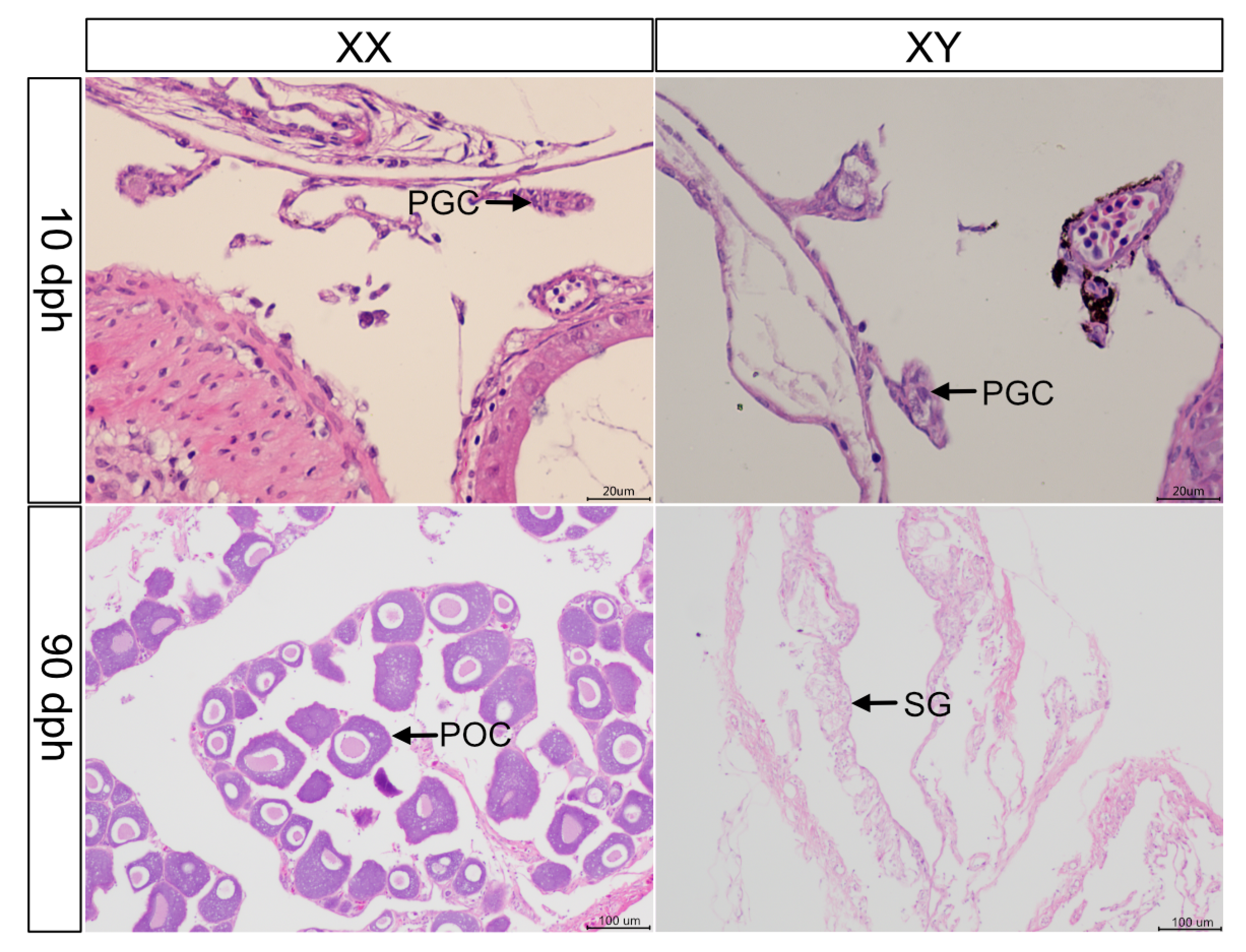
3.1. Histological Observation
3.2. Transcriptome Sequencing Results
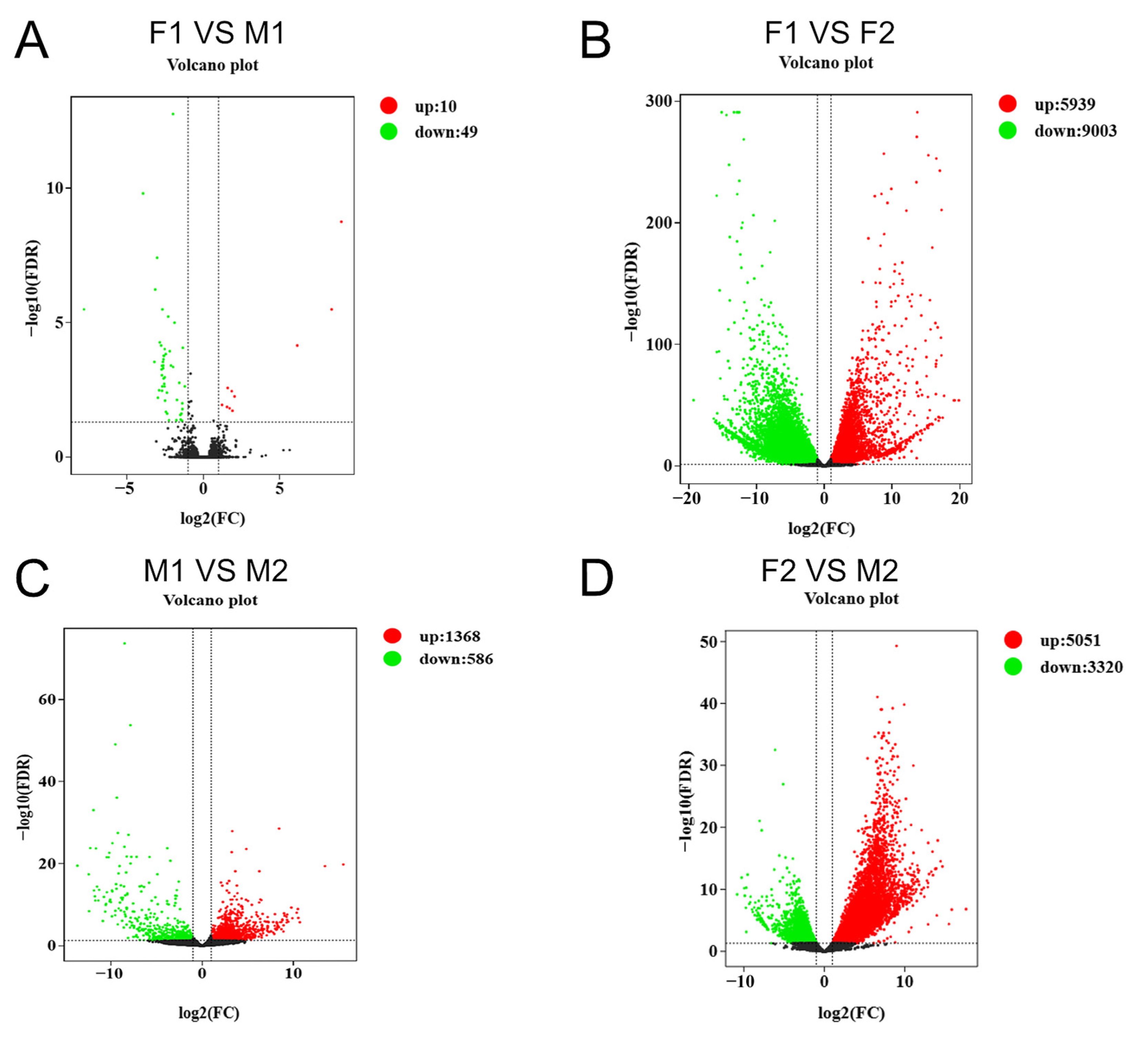
3.3. Differential Gene Expression Analysis
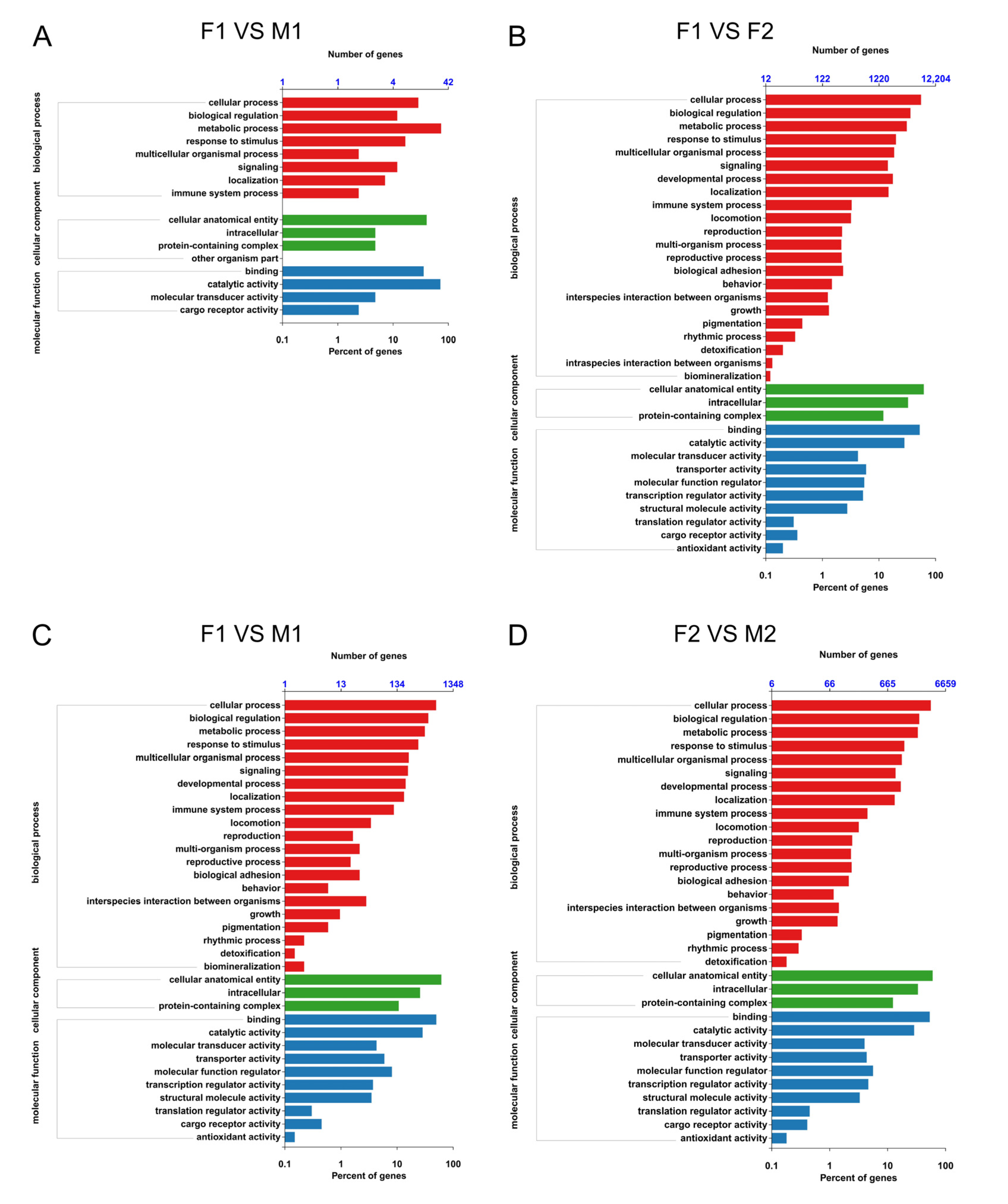
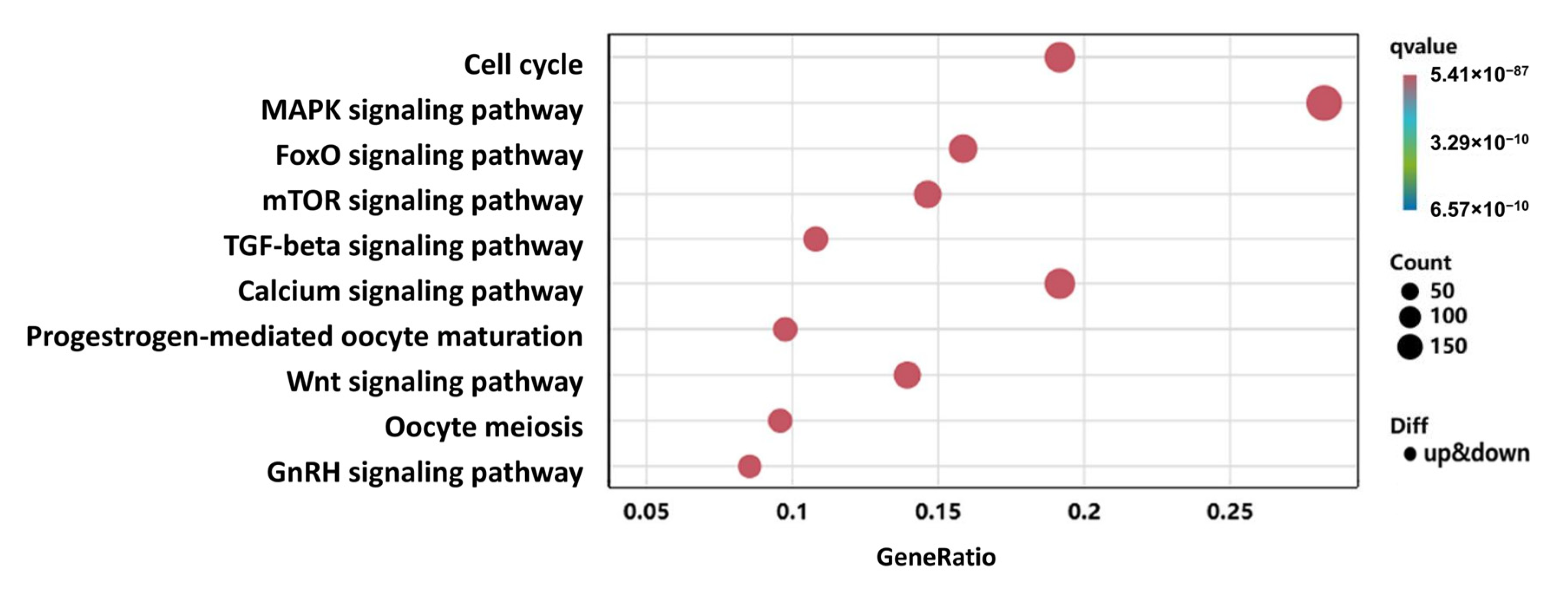
3.4. GO and KEGG Enrichment Analysis
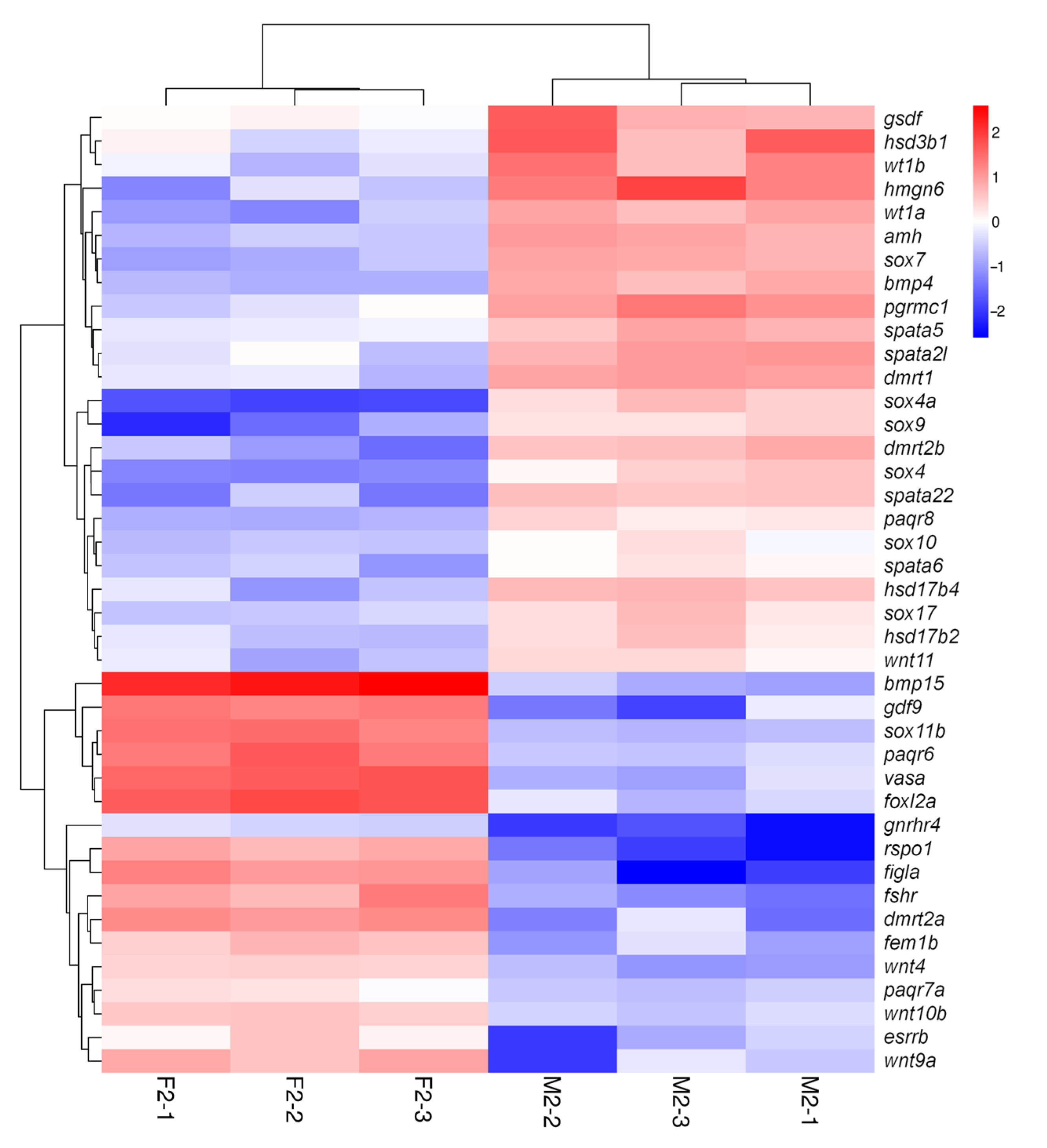
3.5. Screening and Analysis of Key Sex-Related DEGs
3.6. RT-qPCR Validation
4. Discussion
4.1. Sex-Related DEGs in M. armatus
4.2. Role and Interaction of Signaling Pathways in Gonadal Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mei, J.; Gui, J.F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef]
- Wang, H.P.; Gao, Z.X.; Rapp, D.; O’Bryant, P.; Yao, H.; Cao, X.J. Effects of temperature and genotype on sex determination and sexual size dimorphism of bluegill sunfish Lepomis macrochirus. Aquaculture 2014, 420, S64–S71. [Google Scholar] [CrossRef]
- Tao, W.J.; Zhu, X.; Cao, J.M.; Xiao, H.S.; Dong, J.J.; Kocher, T.D.; Lu, M.X.; Wang, D.S. Screening and characterization of sex-linked DNA markers in Mozambique tilapia (Oreochromis mossambicus). Aquaculture 2022, 557, 738331. [Google Scholar] [CrossRef]
- Wang, L.Y.; Qi, P.P.; Chen, M.; Yuan, Y.C.; Shen, Z.G.; Fan, Q.X. Effects of sex steroid hormones on sexual size dimorphism in yellow catfish (Tachysurus fulvidraco). Acta Hydrobiol. Sin. 2020, 44, 379–388. [Google Scholar] [CrossRef]
- Dai, S.; Chen, M.; Zheng, S.; Su, J.; Wu, J.; Han, L.; Zhou, C.; Zou, Y.; Wang, D.; Li, M. Sex-linked DNA marker screening and characterization in albino northern snakehead (Channa argus var.) via third-generation sequencing and pool resequencing. Aquaculture 2025, 594, 741449. [Google Scholar] [CrossRef]
- Zhai, G.; Shu, T.; Chen, K.; Lou, Q.; Jia, J.; Huang, J.; Shi, C.; Jin, X.; He, J.; Jiang, D. Successful production of an all-female common carp (Cyprinus carpio L.) population using cyp17a1-deficient neomale carp. Engineering 2021, 8, 181–189. [Google Scholar] [CrossRef]
- Li, X.Y.; Mei, J.; Ge, C.T.; Liu, X.L.; Gui, J.F. Sex determination mechanisms and sex control approaches in aquaculture animals. Sci. China Life Sci. 2022, 65, 1091–1122. [Google Scholar] [CrossRef]
- Wang, N.; Wang, R.; Wang, R.; Chen, S. Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integr. Genom. 2018, 18, 327–339. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef]
- Tao, W.; Chen, J.; Tan, D.; Yang, J.; Sun, L.; Wei, J.; Conte, M.A.; Kocher, T.D.; Wang, D. Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genom. 2018, 19, 363. [Google Scholar] [CrossRef]
- Ribas, L.; Robledo, D.; Gómez-Tato, A.; Viñas, A.; Martínez, P.; Piferrer, F. Comprehensive transcriptomic analysis of the process of gonadal sex differentiation in the turbot (Scophthalmus maximus). Mol. Cell. Endocrinol. 2016, 422, 132–149. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.H.; Ye, H.; Song, X.H.; Yue, H.M.; Huang, L.; Ruan, R.; Gao, W.H.; Li, C.J. Comparative transcriptomic analysis of male and female gonads of the Chinese longsnout catfish (Leiocassis longirostris). J. Fish. Sci. China 2024, 31, 129–143. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Talwar, P.K.; Jhingran, A.G. Inland Fishes of India and Adjacent Countries; CRC Press: Boca Raton, FL, USA, 1991. [Google Scholar]
- Anand, S.; Gedam, A.; Sonawane, S. Structures associated with feeding in Mastacembelus armatus (Lacepede, 1800) from Kaigaon Toka (MS). J. Exp. Sci 2012, 3, 17–20. [Google Scholar] [CrossRef]
- Xue, L. Observation on the embryonic development of Mastacembelue armatus. Freshw. Fish. 2016, 44, 101–104. [Google Scholar] [CrossRef]
- Xue, L.; Gao, Y.; Wu, M.; Tian, T.; Fan, H.; Huang, Y.; Huang, Z.; Li, D.; Xu, L. Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 2021, 22, 203. [Google Scholar] [CrossRef]
- Zhou, H.Q.; Li, F.; Shu, H.; Zhong, D.M.; He, P.Y.; Huang, X.Q.; Chen, Z.K. Analysis on morphological indexes and discrimination of male and female Mastacembelus armatus. J. Guangdong Ocean Univ. 2019, 39, 1–6. [Google Scholar] [CrossRef]
- Qin, W.; Han, C.; Yang, J.; Yu, Z.; Feng, Y.; Wu, Y.; Lu, B.; Cui, M.; Shu, H. Development of genetic sex markers of zig-zag eel (Mastacembelus armatus) by a NGS method. Aquaculture 2023, 571, 739498. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Niedziela, G.; Szabelska-Beręsewicz, A.; Zyprych-Walczak, J.; Graczyk, M. Application of edgeR and DESeq2 methods in plant experiments based on RNA-seq technology. Biom. Lett. 2022, 59, 127–139. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Guiguen, Y.; Fostier, A.; Herpin, A. Sex Determination and Differentiation in Fish: Genetic, Genomic, and Endocrine Aspects; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Matson, C.K.; Zarkower, D. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012, 13, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Lavery, R.; Chassot, A.-A.; Pauper, E.; Gregoire, E.P.; Klopfenstein, M.; de Rooij, D.G.; Mark, M.; Schedl, A.; Ghyselinck, N.B.; Chaboissier, M.-C. Testicular differentiation occurs in absence of R-spondin1 and Sox9 in mouse sex reversals. PLoS Genet. 2012, 8, e1003170. [Google Scholar] [CrossRef] [PubMed]
- Zarkower, D.; Murphy, M.W. DMRT1: An ancient sexual regulator required for human gonadogenesis. Sex. Dev. 2022, 16, 112–125. [Google Scholar] [CrossRef]
- Minkina, A.; Matson, C.K.; Lindeman, R.E.; Ghyselinck, N.B.; Bardwell, V.J.; Zarkower, D. DMRT1 protects male gonadal cells from retinoid-dependent sexual transdifferentiation. Dev. Cell 2014, 29, 511–520. [Google Scholar] [CrossRef]
- Wagner, S.; Whiteley, S.L.; Castelli, M.; Patel, H.R.; Deveson, I.W.; Blackburn, J.; Holleley, C.E.; Marshall Graves, J.A.; Georges, A. Gene expression of male pathway genes sox9 and amh during early sex differentiation in a reptile departs from the classical amniote model. BMC Genom. 2023, 24, 243. [Google Scholar] [CrossRef]
- Vining, B.; Ming, Z.; Bagheri-Fam, S.; Harley, V. Diverse regulation but conserved function: SOX9 in vertebrate sex determination. Genes 2021, 12, 486. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, W.; Cai, H.; Bao, H.; Zhang, Y.; Qian, G.; Ge, C. The role of anti-Müllerian hormone in testis differentiation reveals the significance of the TGF-β pathway in reptilian sex determination. Genetics 2019, 213, 1317–1327. [Google Scholar] [CrossRef]
- Mustapha, U.F.; Peng, Y.-X.; Huang, Y.-Q.; Assan, D.; Zhi, F.; Shi, G.; Huang, Y.; Li, G.-L.; Jiang, D.-N. Comparative transcriptome analysis of the differentiating gonads in Scatophagus argus. Front. Mar. Sci. 2022, 9, 962534. [Google Scholar] [CrossRef]
- Wang, M.; Chen, L.; Zhou, Z.; Xiao, J.; Chen, B.; Huang, P.; Li, C.; Xue, Y.; Liu, R.; Bai, Y. Comparative transcriptome analysis of early sexual differentiation in the male and female gonads of common carp (Cyprinus carpio). Aquaculture 2023, 563, 738984. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Jakob, S.; Anlag, K.; Eisenberger, T.; Sekido, R.; Kress, J.; Treier, A.-C.; Klugmann, C.; Klasen, C.; Holter, N.I. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 2009, 139, 1130–1142. [Google Scholar] [CrossRef]
- Georges, A.; Auguste, A.; Bessiere, L.; Vanet, A.; Todeschini, A.-L.; Veitia, R.A. FOXL2: A central transcription factor of the ovary. J. Mol. Endocrinol. 2014, 52, R17–R33. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, H.; Xiang, Y.; Jia, K.; Luo, M.; Yi, M. A novel germline and somatic cell expression of two sexual differentiation genes, Dmrt1 and Foxl2 in marbled goby (Oxyeleotris marmorata). Aquaculture 2020, 516, 734619. [Google Scholar] [CrossRef]
- Zhou, L.; Charkraborty, T.; Yu, X.; Wu, L.; Liu, G.; Mohapatra, S.; Wang, D.; Nagahama, Y. R-spondins are involved in the ovarian differentiation in a teleost, medaka (Oryzias latipes). BMC Dev. Biol. 2012, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Tomizuka, K.; Horikoshi, K.; Kitada, R.; Sugawara, Y.; Iba, Y.; Kojima, A.; Yoshitome, A.; Yamawaki, K.; Amagai, M.; Inoue, A. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum. Mol. Genet. 2008, 17, 1278–1291. [Google Scholar] [CrossRef]
- Nicol, B.; Estermann, M.A.; Yao, H.H.; Mellouk, N. Becoming female: Ovarian differentiation from an evolutionary perspective. Front. Cell Dev. Biol. 2022, 10, 944776. [Google Scholar] [CrossRef]
- Kim, Y.; Kobayashi, A.; Sekido, R.; DiNapoli, L.; Brennan, J.; Chaboissier, M.-C.; Poulat, F.; Behringer, R.R.; Lovell-Badge, R.; Capel, B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006, 4, e187. [Google Scholar] [CrossRef]
- Kocer, A.; Pinheiro, I.; Pannetier, M.; Renault, L.; Parma, P.; Radi, O.; Kim, K.-A.; Camerino, G.; Pailhoux, E. R-spondin1 and FOXL2 act into two distinct cellular types during goat ovarian differentiation. BMC Dev. Biol. 2008, 8, 36. [Google Scholar] [CrossRef]
- Cederroth, C.R.; Pitetti, J.L.; Papaioannou, M.D.; Nef, S. Genetic programs that regulate testicular and ovarian development. Mol. Cell. Endocrinol. 2007, 265, 3–9. [Google Scholar] [CrossRef]
- Chen, W.; Liu, L.; Ge, W. Expression analysis of growth differentiation factor 9 (Gdf9/gdf9), anti-Müllerian hormone (Amh/amh) and aromatase (Cyp19a1a/cyp19a1a) during gonadal differentiation of the zebrafish, Danio rerio. Biol. Reprod. 2017, 96, 401–413. [Google Scholar] [CrossRef]
- Halm, S.; Ibáñez, A.J.; Tyler, C.R.; Prat, F. Molecular characterisation of growth differentiation factor 9 (gdf9) and bone morphogenetic protein 15 (bmp15) and their patterns of gene expression during the ovarian reproductive cycle in the European sea bass. Mol. Cell. Endocrinol. 2008, 291, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Lu, C. Molecular Regulation Mechanism of MKPs Toward MAPKs in the MAPK Pathway; Tsinghua University: Beijing, China, 2018. [Google Scholar]
- Huang, T.; Gu, W.; Liu, E.; Zhang, L.; Dong, F.; He, X.; Jiao, W.; Li, C.; Wang, B.; Xu, G. Screening and validation of p38 MAPK involved in ovarian development of Brachymystax lenok. Front. Vet. Sci. 2022, 9, 752521. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Luo, Y.R.; Luo, Z.W.; Wang, J.; Xiao, Y.M. Expression of MAPK in the Gonadal Tissue of Different Reproductive Characteristics of the Hybrid Crucian Carp. Life Sci. Res. 2016, 20, 95–101. [Google Scholar]
- Ten Dijke, P.; Goumans, M.J.; Itoh, F.; Itoh, S. Regulation of cell proliferation by Smad proteins. J. Cell. Physiol. 2002, 191, 1–16. [Google Scholar] [CrossRef]
- Shi, Y.; Massagué, J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Sower, S.A. Landmark discoveries in elucidating the origins of the hypothalamic-pituitary system from the perspective of a basal vertebrate, sea lamprey. Gen. Comp. Endocrinol. 2018, 264, 3–15. [Google Scholar] [CrossRef]
- Stamatiades, G.A.; Carroll, R.S.; Kaiser, U.B. GnRH—A key regulator of FSH. Endocrinology 2019, 160, 57–67. [Google Scholar] [CrossRef]
- Hu, K.L.; Chen, Z.M.; Li, X.X.; Cai, E.; Yang, H.Y.; Chen, Y.; Wang, C.Y.; Ju, L.P.; Deng, W.H.; Mu, L.S. Advances in clinical applications of kisspeptin-GnRH pathway in female reproduction. Reprod. Biol. Endocrinol. 2022, 20, 81. [Google Scholar] [CrossRef]


| Gene | Sequences From (5′-3′) | Gene | Sequences From (5′-3′) |
|---|---|---|---|
| dmrt1 | F:CGGCCCAGGTTGCCTTGAG | foxl2 | F:TCCGTCCCAGAAACCACCGTAT |
| R:CCAGCTTCATTCTTCACCATCA | R:CCTGATGCTGTTCTGCCAACCT | ||
| sox9 | F:GAAGGACGAGGACGATAAGTT | rspo1 | F:AAAGGCTCACAATCTCGG |
| R:TGGCATAGGCACGAGGGT | R:CCTCCTCTACTGCCATCC | ||
| amh | F:TCTGGCACTCAGCTTATCC | wnt4 | F:TGGGCAACATCATCAAGG |
| R:CCATCTCCTCCTCCCTTA | R:TGGATCATAGTCGCAGAAA | ||
| wt1 | F:TGCGTTCACCGTCCACTT | gdf9 | F:ATCTACACCTGCTCATCAA |
| R:CGACCGTGCTGTAACCTG | R:ACTGACTACTGAACCCTGAT | ||
| bmp4 | F:GTATCGGCTACAGTCAGGG | bmp15 | F:GCAGAAAGCGGACCAGAA |
| R:ATCTTCGGGAATGGTGCT | R:CGAGGGAAGAGTGTCAAGC | ||
| sox4 | F:GGGACTTGGATTTGAACTTTG | dmrt2a | F:CGGGAATACAAAGAACGAGA |
| R:TCGCTCACTTCGGGCGTA | R:CGCTGACATTGGAGGAGAT | ||
| dmrt2b | F:AACCAGGGAGGATAAGGA | β-actin | F:TCATGAGGTAGTCTGTGAGGTCCC |
| R:GCTGACGTGCTATTTGAGT | R:GCCTCTGGTCGTACCACTGGTATT | ||
| gsdf | F:TCCAAGGAAGAACCTGCAACCT | ||
| R:CAGGCATCCATGGCTCAGACTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, F.; Wang, Y.; Liang, H.; Yang, Y.; Jiang, Z.; Song, J.; Liu, C.; Wu, Y.; Mu, X.; Liu, Y. Comparative Transcriptomic Analysis of Male and Female Gonads in the Zig-Zag Eel (Mastacembelus armatus). Fishes 2025, 10, 117. https://doi.org/10.3390/fishes10030117
Cui F, Wang Y, Liang H, Yang Y, Jiang Z, Song J, Liu C, Wu Y, Mu X, Liu Y. Comparative Transcriptomic Analysis of Male and Female Gonads in the Zig-Zag Eel (Mastacembelus armatus). Fishes. 2025; 10(3):117. https://doi.org/10.3390/fishes10030117
Chicago/Turabian StyleCui, Fangyu, Yuanyuan Wang, Haiyan Liang, Yexin Yang, Zhiyong Jiang, Jiahuan Song, Chao Liu, Yuli Wu, Xidong Mu, and Yi Liu. 2025. "Comparative Transcriptomic Analysis of Male and Female Gonads in the Zig-Zag Eel (Mastacembelus armatus)" Fishes 10, no. 3: 117. https://doi.org/10.3390/fishes10030117
APA StyleCui, F., Wang, Y., Liang, H., Yang, Y., Jiang, Z., Song, J., Liu, C., Wu, Y., Mu, X., & Liu, Y. (2025). Comparative Transcriptomic Analysis of Male and Female Gonads in the Zig-Zag Eel (Mastacembelus armatus). Fishes, 10(3), 117. https://doi.org/10.3390/fishes10030117






