IsoSearch: An Untargeted and Unbiased Metabolite and Lipid Isotopomer Tracing Strategy from HR-LC-MS/MS Datasets
Abstract
:1. Introduction
2. Results
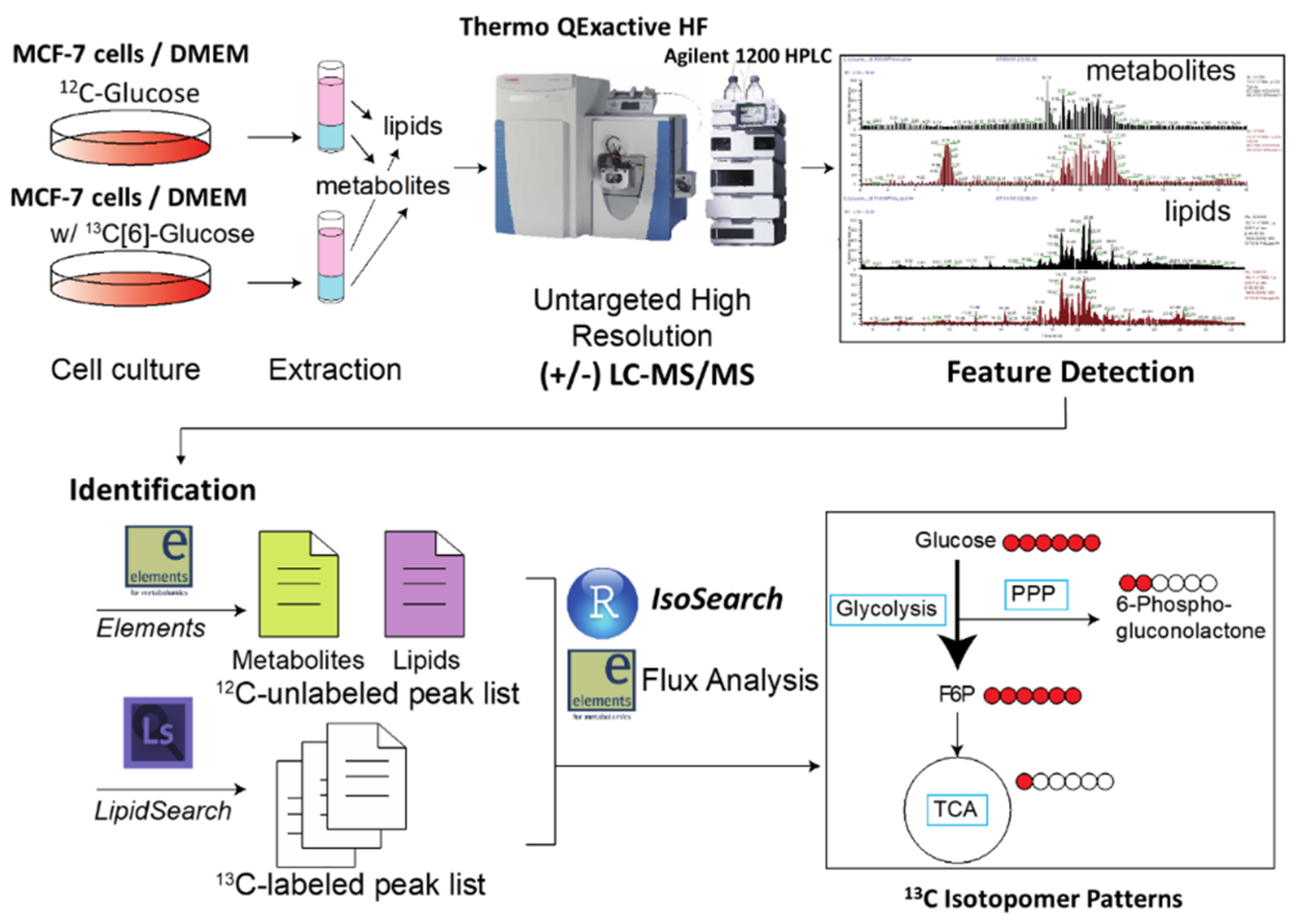
2.1. Development of IsoSearch Strategy-Identification and Peak Picking
2.2. IsoSearch Analysis
2.3. LC-MS/MS Data Preprocessing
3. Discussion
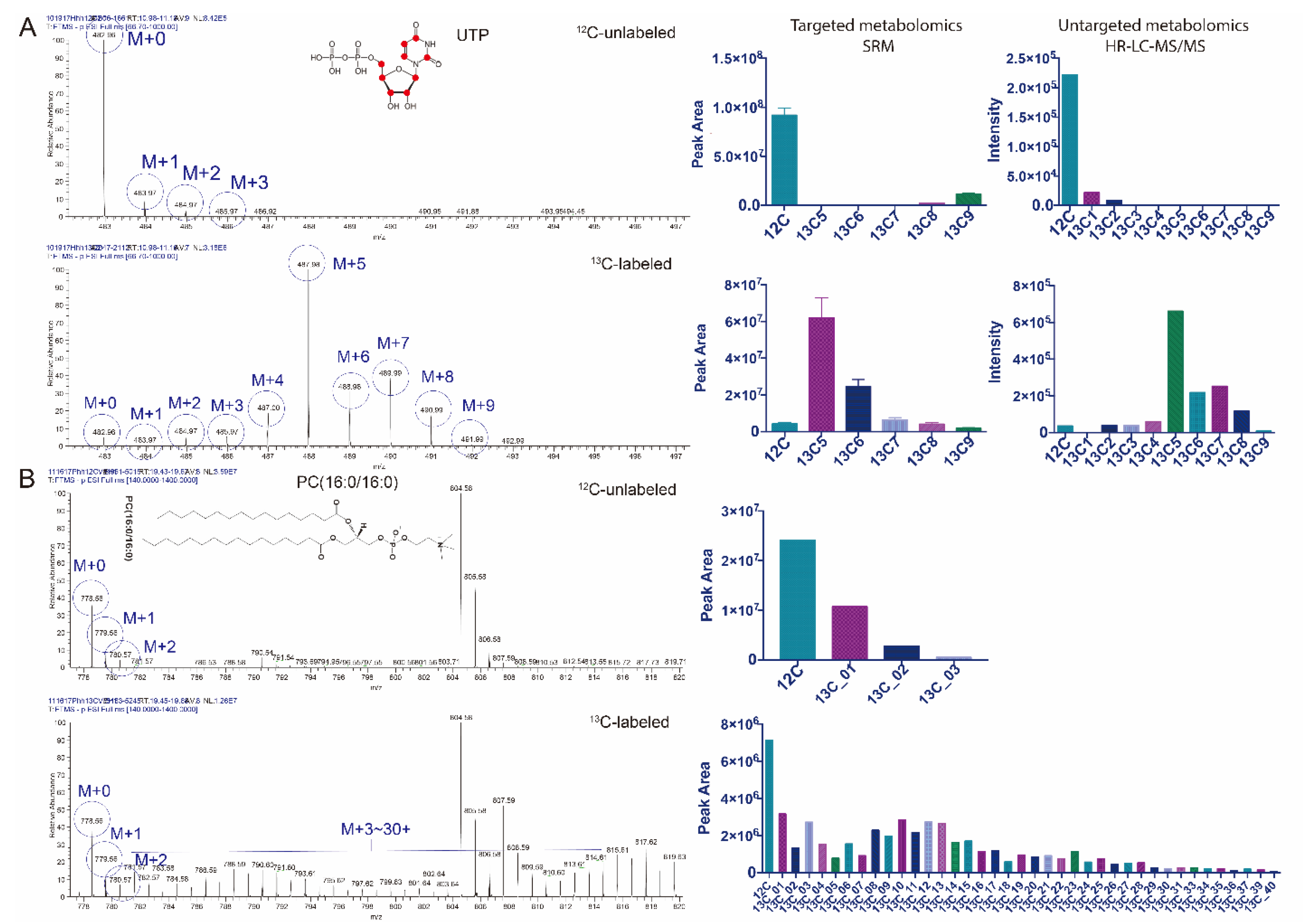
3.1. Principle of IsoSearch Based Metabolic/Lipid Isotopomer Tracing
3.2. Untargeted Isotopic Patterns Are Consistent with the Targeted Isotopomer Detection
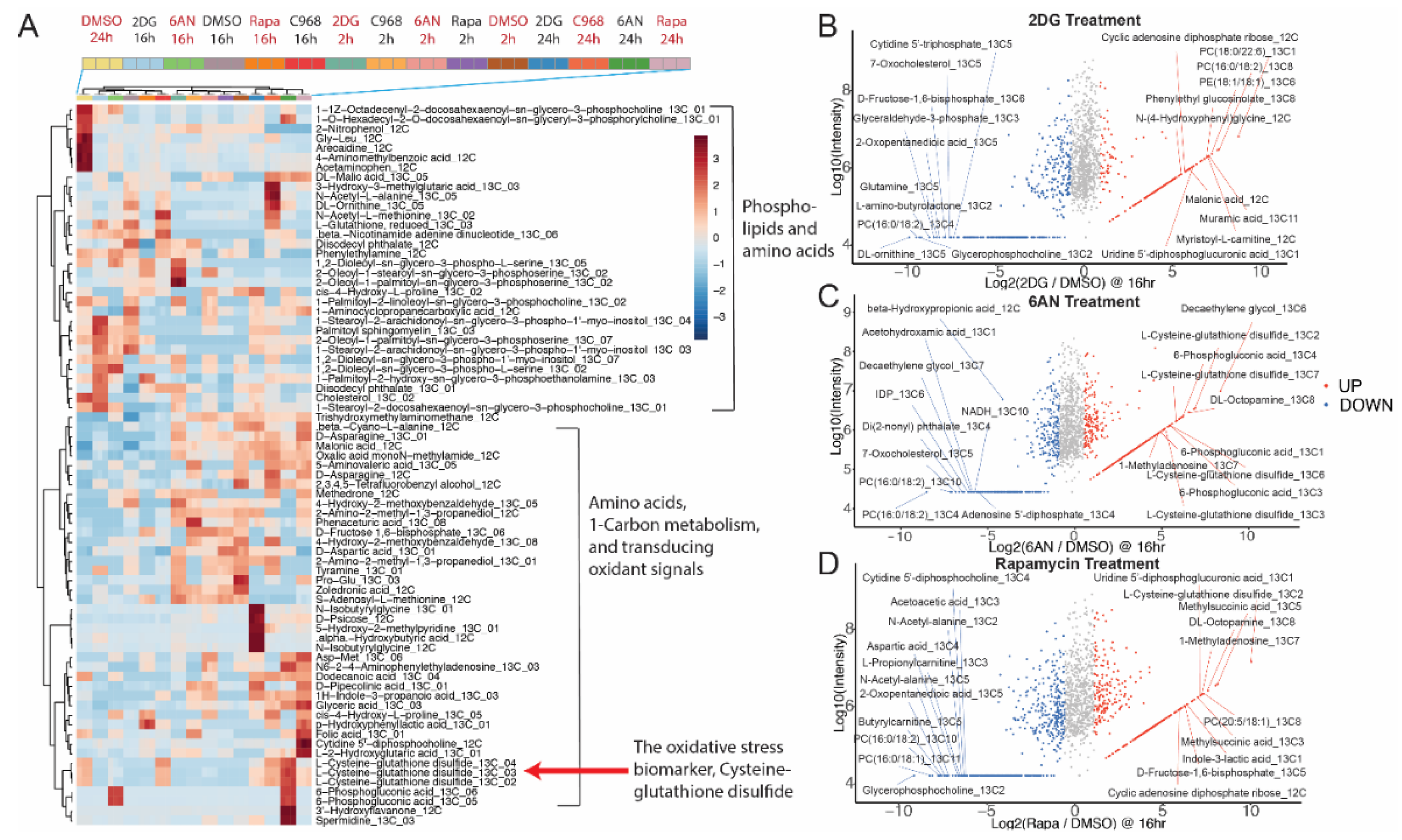
3.3. Untargeted Metabolic Time-Course Analysis Reveals Drug Stimulated 13C[6]-Glucose Flux Alteration
3.4. Drugs Induce Oxidative Responses in MCF-7 Breast Cancer Cells
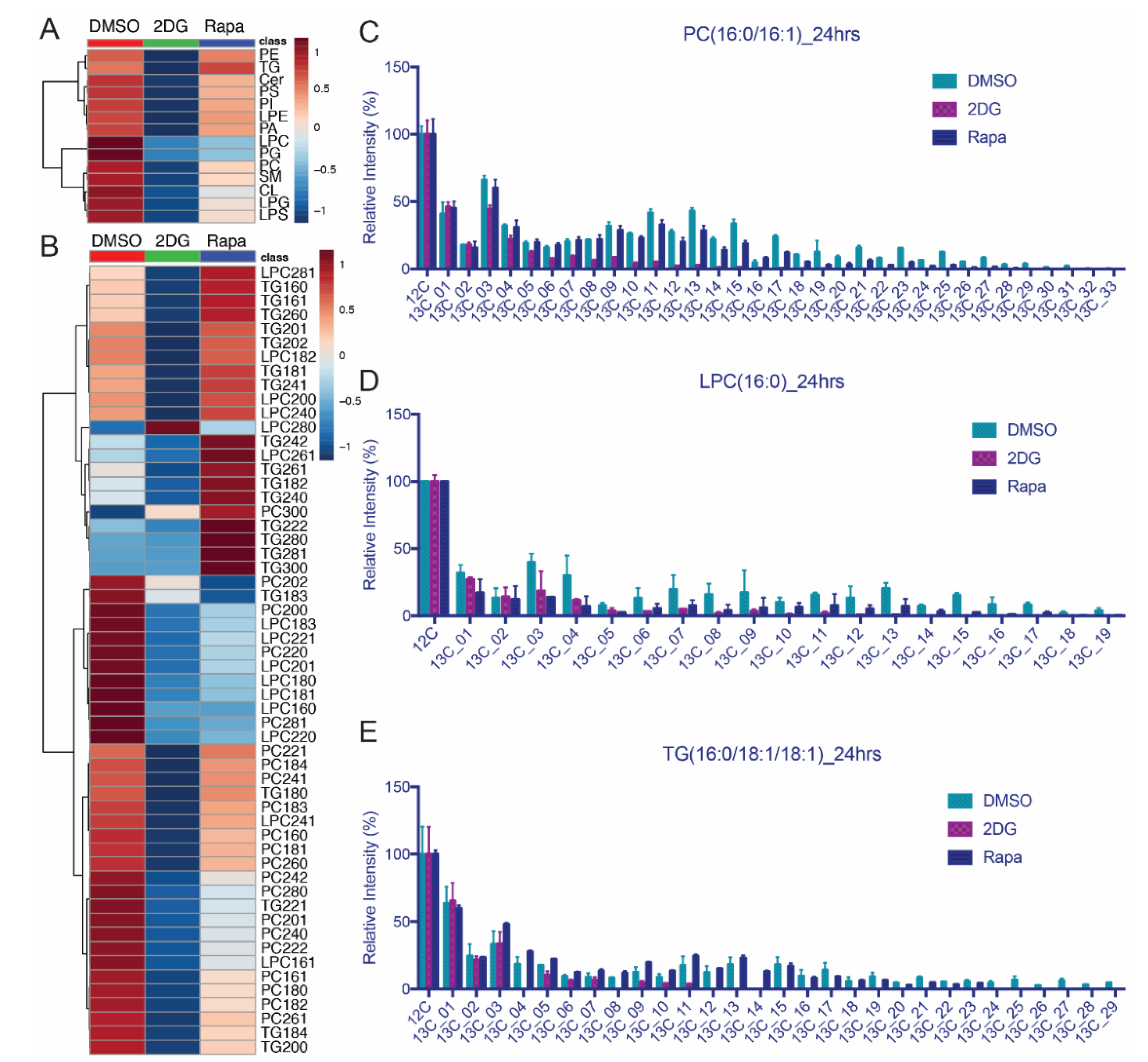
3.5. Lipid Isotopomer Regulation by Rapamycin Treatment
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Lipid and Metabolite Co-Extraction
4.4. LC-MS/MS Based Metabolomic and Lipidomic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sauer, U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2006, 2, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buescher, J.M.; Antoniewicz, M.R.; Boros, L.G.; Burgess, S.C.; Brunengraber, H.; Clish, C.B.; DeBerardinis, R.J.; Feron, O.; Frezza, C.; Ghesquiere, B.; et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015, 34, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Schwender, J.; Goffman, F.; Ohlrogge, J.B.; Shachar-Hill, Y. Rubisco without the calvin cycle improves the carbon efficiency of developing green seeds. Nature 2004, 432, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Sriram, G.; Fulton, D.B.; Iyer, V.V.; Peterson, J.M.; Zhou, R.; Westgate, M.E.; Spalding, M.H.; Shanks, J.V. Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractional (13)C labeling, two-dimensional [(13)C, (1)H] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant. Physiol. 2004, 136, 3043–3057. [Google Scholar] [CrossRef] [Green Version]
- Hellerstein, M.K.; Murphy, E. Stable isotope-mass spectrometric measurements of molecular fluxes in vivo: Emerging applications in drug development. Curr. Opin. Mol. Ther. 2004, 6, 249–264. [Google Scholar]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef] [Green Version]
- Reisz, J.A.; D’Alessandro, A. Measurement of metabolic fluxes using stable isotope tracers in whole animals and human patients. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 366–374. [Google Scholar] [CrossRef]
- Yuan, M.; Kremer, D.M.; Huang, H.; Breitkopf, S.B.; Ben-Sahra, I.; Manning, B.D.; Lyssiotis, C.A.; Asara, J.M. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat. Protoc. 2019, 14, 313–330. [Google Scholar] [CrossRef]
- Ben-Sahra, I.; Howell, J.J.; Asara, J.M.; Manning, B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through MTOR and S6K1. Science 2013, 339, 1323–1328. [Google Scholar] [CrossRef] [Green Version]
- Son, J.; Lyssiotis, C.A.; Ying, H.; Wang, X.; Hua, S.; Ligorio, M.; Perera, R.M.; Ferrone, C.R.; Mullarky, E.; Shyh-Chang, N.; et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013, 496, 101–105. [Google Scholar] [CrossRef]
- Ying, H.; Kimmelman, A.C.; Lyssiotis, C.A.; Hua, S.; Chu, G.C.; Fletcher-Sananikone, E.; Locasale, J.W.; Son, J.; Zhang, H.; Coloff, J.L.; et al. Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012, 149, 656–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolay, B.N.; Gameiro, P.A.; Tschop, K.; Korenjak, M.; Heilmann, A.M.; Asara, J.M.; Stephanopoulos, G.; Iliopoulos, O.; Dyson, N.J. Loss of RBF1 changes glutamine catabolism. Genes Dev. 2013, 27, 182–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiechert, W. 13C metabolic flux analysis. Metab. Eng. 2001, 3, 195–206. [Google Scholar] [CrossRef]
- Zamboni, N.; Fendt, S.-M.; Ruhl, M.; Sauer, U. (13)C-based metabolic flux analysis. Nat. Protoc. 2009, 4, 878–892. [Google Scholar] [CrossRef]
- Nanchen, A.; Fuhrer, T.; Sauer, U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: Protocol and principles. Methods Mol. Biol. 2007, 358, 177–197. [Google Scholar]
- Crown, S.B.; Long, C.P.; Antoniewicz, M.R. Optimal tracers for parallel labeling experiments and (13)C metabolic flux analysis: A new precision and synergy scoring system. Metab. Eng. 2016, 38, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Kogadeeva, M.; Zamboni, N. SUMOFLUX: A generalized method for targeted 13C metabolic flux ratio analysis. PLoS Comput. Biol. 2016, 12, e1005109. [Google Scholar] [CrossRef] [Green Version]
- Weitzel, M.; Nöh, K.; Dalman, T.; Niedenführ, S.; Stute, B.; Wiechert, W. 13CFLUX2—High-performance software suite for (13)C-metabolic flux analysis. Bioinformatics 2013, 29, 143–145. [Google Scholar] [CrossRef]
- Huang, X.; Chen, Y.-J.; Cho, K.; Nikolskiy, I.; Crawford, P.A.; Patti, G.J. X13CMS: Global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 2014, 86, 1632–1639. [Google Scholar] [CrossRef]
- Noh, K.; Droste, P.; Wiechert, W. Visual workflows for 13C-metabolic flux analysis. Bioinformatics 2015, 31, 346–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puchalska, P.; Huang, X.; Martin, S.E.; Han, X.; Patti, G.J.; Crawford, P.A. Isotope tracing untargeted metabolomics reveals macrophage polarization-state-specific metabolic coordination across intracellular compartments. iScience 2018, 9, 298–313. [Google Scholar] [CrossRef] [Green Version]
- Puchalska, P.; Martin, S.E.; Huang, X.; Lengfeld, J.E.; Daniel, B.; Graham, M.J.; Han, X.; Nagy, L.; Patti, G.J.; Crawford, P.A. Hepatocyte-macrophage acetoacetate shuttle protects against tissue fibrosis. Cell Metab. 2019, 29, 383–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-J.; Mahieu, N.G.; Huang, X.; Singh, M.; Crawford, P.A.; Johnson, S.L.; Gross, R.W.; Schaefer, J.; Patti, G.J. Lactate metabolism is associated with mammalian mitochondria. Nat. Chem. Biol. 2016, 12, 937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitzer, P.M.; Searle, B.C. Incorporating in-source fragment information improves metabolite identification accuracy in untargeted LC–MS data sets. J. Proteome Res. 2019, 18, 791–796. [Google Scholar] [CrossRef]
- Breitkopf, S.B.; Ricoult, S.J.H.; Yuan, M.; Xu, Y.; Peake, D.A.; Manning, B.D.; Asara, J.M. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 2017, 13, 30. [Google Scholar] [CrossRef] [Green Version]
- Gibbons, H.; Brennan, L. Metabolomics as a tool in the identification of dietary biomarkers. Proc. Nutr. Soc. 2017, 76, 42–53. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Nam, I.; Lee, J.K.; Nam, H.G.; Zare, R.N. Abiotic production of sugar phosphates and uridine ribonucleoside in aqueous microdroplets. Proc. Natl. Acad. Sci. USA 2017, 114, 12396–12400. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, A.K.; DeBerardinis, R.J. Applications of metabolomics to study cancer metabolism. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 2–14. [Google Scholar] [CrossRef]
- Kang, Y.P.; Ward, N.P.; DeNicola, G.M. Recent advances in cancer metabolism: A technological perspective. Exp. Mol. Med. 2018, 50, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa, C.M.; Kimmelman, A.C. The complex landscape of pancreatic cancer metabolism. Carcinogenesis 2014, 35, 1441–1450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, A.N.; Drury, D.R.; Nakada, H.I.; Wolfe, J.B. Localization of the primary metabolic block produced by 2-deoxyglucose. J. Biol. Chem. 1957, 224, 963–969. [Google Scholar] [PubMed]
- Salimi, A.; Paeezi, M.; Yousefsani, B.S.; Shadnia, S.; Hassanian-Moghaddam, H.; Zamani, N.; Pourahmad, J. Inhibition of glucose-6-phosphate dehydrogenase protects hepatocytes from aluminum phosphide-induced toxicity. Pestic. Biochem. Physiol. 2017, 143, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Sheng, X.; Clark, L.H.; Zhang, L.; Guo, H.; Jones, H.M.; Willson, A.K.; Gehrig, P.A.; Zhou, C.; Bae-Jump, V.L. Glutaminase inhibitor compound 968 inhibits cell proliferation and sensitizes paclitaxel in ovarian cancer. Am. J. Transl. Res. 2016, 8, 4265–4277. [Google Scholar]
- Wang, D.; Meng, G.; Zheng, M.; Zhang, Y.; Chen, A.; Wu, J.; Wei, J. The glutaminase-1 inhibitor 968 enhances dihydroartemisinin-mediated antitumor efficacy in hepatocellular carcinoma cells. PLoS ONE 2016, 11, e0166423. [Google Scholar] [CrossRef]
- Caron, A.; Richard, D.; Laplante, M. The roles of MTOR complexes in lipid metabolism. Annu. Rev. Nutr. 2015, 35, 321–348. [Google Scholar] [CrossRef]
- Ben-Sahra, I.; Hoxhaj, G.; Ricoult, S.J.H.; Asara, J.M.; Manning, B.D. MTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 2016, 351, 728–733. [Google Scholar] [CrossRef] [Green Version]
- Wright, P.; Felskie, A.; Anderson, P. Induction of ornithine-urea cycle enzymes and nitrogen metabolism and excretion in rainbow trout (oncorhynchus mykiss) during early life stages. J. Exp. Biol. 1995, 198, 127–135. [Google Scholar]
- Camardella, L.; Damonte, G.; Carratore, V.; Benatti, U.; Tonetti, M.; Moneti, G. Glucose 6-phosphate dehydrogenase from human erythrocytes: Identification of N-acetyl-alanine at the N-terminus of the mature protein. Biochem. Biophys. Res. Commun. 1995, 207, 331–338. [Google Scholar] [CrossRef]
- Dena, R.; Fabbro, M.; Rigoni, F. Formation and utilization of 3-hydroxy-3-methylglutarate in liver mitochondria of starved and streptozotocin-diabetic rats. Biochem. J. 1978, 172, 371–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moorman, A.F.; de Boer, P.A.; Watford, M.; Dingemanse, M.A.; Lamers, W.H. Hepatic glutaminase MRNA is confined to part of the urea cycle domain in the adult rodent liver lobule. FEBS Lett. 1994, 356, 76–80. [Google Scholar] [CrossRef] [Green Version]
- Häussinger, D.; Gerok, W. Regulation of hepatic glutamate metabolism. Eur. J. Biochem. 1984, 143, 491–497. [Google Scholar] [PubMed]
- Chen, Y.; Xu, Q.; Ji, D.; Wei, Y.; Chen, H.; Li, T.; Wan, B.; Yuan, L.; Huang, R.; Chen, G. Inhibition of pentose phosphate pathway suppresses acute myelogenous leukemia. Tumour Biol. 2016, 37, 6027–6034. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.H.; Kauffman, F.C. Metabolism via the pentose phosphate pathway in rat pheochromocytoma PC12 cells: Effects of nerve growth factor and 6-aminonicotinamide. Neurochem. Res. 1987, 12, 521–527. [Google Scholar] [CrossRef]
- Panieri, E.; Santoro, M.M. ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016, 7, e2253. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Burns, J.S.; Manda, G. Metabolic pathways of the warburg effect in health and disease: Perspectives of choice, chain or chance. Int. J. Mol. Sci. 2017, 18, 2755. [Google Scholar] [CrossRef] [Green Version]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Dabritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 2016, 167, 457–470. [Google Scholar] [CrossRef] [Green Version]
- Bezawork-Geleta, A.; Rohlena, J.; Dong, L.; Pacak, K.; Neuzil, J. Mitochondrial complex II: At the crossroads. Trends Biochem. Sci. 2017, 42, 312–325. [Google Scholar] [CrossRef]
- Mailloux, R.J.; Beriault, R.; Lemire, J.; Singh, R.; Chenier, D.R.; Hamel, R.D.; Appanna, V.D. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE 2007, 2, e690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijevic, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Ord, E.N.J.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heman-Ackah, S.E.; Juhn, S.K.; Huang, T.C.; Wiedmann, T.S. A combination antioxidant therapy prevents age-related hearing loss in C57BL/6 mice. Otolaryngol. Head. Neck Surg. 2010, 143, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-H.; Qanungo, S.; Pai, H.V.; Starke, D.W.; Steller, K.M.; Fujioka, H.; Lesnefsky, E.J.; Kerner, J.; Rosca, M.G.; Hoppel, C.L.; et al. Aging-dependent changes in rat heart mitochondrial glutaredoxins—Implications for redox regulation. Redox Biol. 2013, 1, 586–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkeley, L.I.; Cohen, J.F.; Crankshaw, D.L.; Shirota, F.N.; Nagasawa, H.T. Hepatoprotection by L-cysteine-glutathione mixed disulfide, a sulfhydryl-modified prodrug of glutathione. J. Biochem. Mol. Toxicol. 2003, 17, 95–97. [Google Scholar] [CrossRef]
- Hong, S.Y.; Yu, F.-X.; Luo, Y.; Hagen, T. Oncogenic activation of the PI3K/Akt pathway promotes cellular glucose uptake by downregulating the expression of thioredoxin-interacting protein. Cell. Signal. 2016, 28, 377–383. [Google Scholar] [CrossRef]
- Cao, B.-Y.; Li, R.; Tian, H.-H.; Ma, Y.-J.; Hu, X.-G.; Jia, N.; Wang, Y.-Y. PI3K-GLUT4 signal pathway associated with effects of EX-B3 electroacupuncture on hyperglycemia and insulin resistance of T2DM rats. Evid. Based Complement. Alternat. Med. 2016, 2016, 7914387. [Google Scholar] [CrossRef] [Green Version]
- Ricoult, S.J.H.; Yecies, J.L.; Ben-Sahra, I.; Manning, B.D. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through MTORC1 and SREBP. Oncogene 2016, 35, 1250–1260. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xiong, X.; Sa, N.; Roje, S.; Chen, S. Metabolic engineering of enhanced Glycerol-3-phosphate synthesis to increase lipid production in synechocystis sp. PCC 6803. Appl. Microbiol. Biotechnol. 2016, 100, 6091–6101. [Google Scholar] [CrossRef]
- Xue, L.-L.; Chen, H.-H.; Jiang, J.-G. Implications of glycerol metabolism for lipid production. Prog. Lipid Res. 2017, 68, 12–25. [Google Scholar] [CrossRef]
- Moessinger, C.; Klizaite, K.; Steinhagen, A.; Philippou-Massier, J.; Shevchenko, A.; Hoch, M.; Ejsing, C.S.; Thiele, C. Two different pathways of phosphatidylcholine synthesis, the kennedy pathway and the lands cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 2014, 15, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, C.W.; Kim, B.H.; Kim, I.-H.; Lee, M.-W. Modeling and optimization of phospholipase A(1)-catalyzed hydrolysis of phosphatidylcholine using response surface methodology for lysophosphatidylcholine production. Biotechnol. Prog. 2015, 31, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Tsao, F.H.; Zachman, R.D. Phosphatidylcholine-lysophosphatidylcholine cycle pathway enzymes in rabbit lung. II. marked differences in the effect of gestational age on activity compared to the CDP-choline pathway. Pediatr. Res. 1977, 11, 858–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasiske, B.L.; de Mattos, A.; Flechner, S.M.; Gallon, L.; Meier-Kriesche, H.-U.; Weir, M.R.; Wilkinson, A. Mammalian target of rapamycin inhibitor dyslipidemia in kidney transplant recipients. Am. J. Transplant. 2008, 8, 1384–1392. [Google Scholar] [CrossRef]
- Morrisett, J.D.; Abdel-Fattah, G.; Hoogeveen, R.; Mitchell, E.; Ballantyne, C.M.; Pownall, H.J.; Opekun, A.R.; Jaffe, J.S.; Oppermann, S.; Kahan, B.D. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J. Lipid Res. 2002, 43, 1170–1180. [Google Scholar] [CrossRef] [Green Version]
- Breitkopf, S.B.; Taveira, M.D.O.; Yuan, M.; Wulf, G.M.; Asara, J.M. Serial-omics of P53-/-, Brca1-/- mouse breast tumor and normal mammary gland. Sci. Rep. 2017, 7, 14503. [Google Scholar] [CrossRef] [Green Version]

| Function | Parameters | Description |
| Flux_result() | input_negative, input_positive, referInput, score = 0.61 (default) | main function to generate the final result of the flux analysis |
| ref_13C_neg() | refInput, refOutput | create the reference list using negative mode unlabeled samples (12C) |
| ref_13C_pos() | refInput, refOutput | create the reference list using positive mode unlabeled samples (12C) |
| msMatch() | mzFile, refile | searching process |
| resultWrap_neg() | inputFile_neg, refInput | wrapping the flux 13C-labeling results of negative ion mode samples |
| resultWrap_pos() | inputFile_pos, refInput | wrapping the flux 13C-labeling results of positive ion mode samples |
| Sgrade() | inputResult | scoring function using Equation (1) to calculate the filtering scores |
| refLibPos_ls() | refInput | preserved function for lipidSearch |
| refLibNeg_ls() | refInput | preserved function for lipidSearch |
| refLibPos_ele() | refInput | preserved function for Elements |
| refLibNeg_ele() | refInput | preserved function for Elements |
| Column Heading | Description |
| mz1 | mass to charge ratio of the searched file |
| rt1 | Retention time value of the data file |
| Intensity | feature peak intensity |
| mz2 | m/z value of the 13C-labeled experimental file |
| rt2 | retention time value of the data file |
| Metabolite/Lipid | name of the feature |
| fattyAcid, lipidClass, lipidForm | fatty acid chain, lipid class and chemical formula of the lipid (lipid fluxomics only) |
| Accession | accession number assigned by the database |
| Theoretical_mz | isotopomer theoretical m/z value |
| Adduct | adduct ion (+H, −H, etc.) |
| Charge (z) | ion charge mode |
| Annotation | isotopomer notation (M+1, M+2, M+3, etc.) |
| Score | score used for feature screening |
| Δmz_ppm | the difference between experimental and theoretical m/z in ppm |
| Grades | Quality associated with the score where A is best |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Yuan, M.; Seitzer, P.; Ludwigsen, S.; Asara, J.M. IsoSearch: An Untargeted and Unbiased Metabolite and Lipid Isotopomer Tracing Strategy from HR-LC-MS/MS Datasets. Methods Protoc. 2020, 3, 54. https://doi.org/10.3390/mps3030054
Huang H, Yuan M, Seitzer P, Ludwigsen S, Asara JM. IsoSearch: An Untargeted and Unbiased Metabolite and Lipid Isotopomer Tracing Strategy from HR-LC-MS/MS Datasets. Methods and Protocols. 2020; 3(3):54. https://doi.org/10.3390/mps3030054
Chicago/Turabian StyleHuang, He, Min Yuan, Phillip Seitzer, Susan Ludwigsen, and John M. Asara. 2020. "IsoSearch: An Untargeted and Unbiased Metabolite and Lipid Isotopomer Tracing Strategy from HR-LC-MS/MS Datasets" Methods and Protocols 3, no. 3: 54. https://doi.org/10.3390/mps3030054
APA StyleHuang, H., Yuan, M., Seitzer, P., Ludwigsen, S., & Asara, J. M. (2020). IsoSearch: An Untargeted and Unbiased Metabolite and Lipid Isotopomer Tracing Strategy from HR-LC-MS/MS Datasets. Methods and Protocols, 3(3), 54. https://doi.org/10.3390/mps3030054






