An Effective Protocol for Proteome Analysis of Medaka (Oryzias latipes) after Acute Exposure to Ionizing Radiation
Abstract
1. Introduction
2. Experimental Design
2.1. Materials
- 12 wild-type adult Medaka (~6–8 months old) with a body weight of 0.45 + 0.01 g and a 14 h:10 h light-dark cycle at 25 °C;
- 4 liter of filtered water;
- Anesthetic solution: 250 mg/L of tricaine methane sulfonate (MS-222) (Fisher, Fair Lawn, NJ, USA; cat: NC0342409), buffered with sodium bicarbonate at or near to neutral pH;
- Liquid nitrogen;
- Methanol HPLC grade (Sigma-Aldrich, St. Louis, MO, USA; Cat. no.: 34860-4L-R);
- Homogenizing solution kept at 4 °C: Mix chloroform (Fisher, Fair Lawn, NJ, USA; Cat. no.: C606SK-4), methanol, and Milli-Q water at ratio of 2:4:1.5, respectively;
- Acetone (Fisher, Fair Lawn, NJ, USA; Cat. no.: C606SK-4);
- Tris-HCl buffer: 1 mM Tris base (Fisher Scientific, Fair Lawn, NJ, USA; Cat. no.: BP152-500), pH 7.2 and 2% SDS (Sigma-Aldrich, St. Louis, MO, USA; Cat. no.: L4390);
- Pierce BCA protein assay kit (Thermo Scientific Pierce, Rockford, IL, USA; Cat. no.: 23227);
- Bolt 4–12% Bis-Tris Plus gels (Thermo Scientific Pierce, Rockford, IL, USA; Cat. no.: NW04120BOX);
- Protein ladder (Thermo Scientific Pierce, Rockford, IL, USA; Cat. no.: LC5925);
- 2× Laemmli Sample Buffer (Bio-Rad, Hercules, CA, USA; Cat. no.: 1610737);
- Running buffer 1×: 50 mL of Bolt MOPS SDS running buffer (Thermo Scientific Pierce, Rockford, IL, USA; Cat. no.: B0001) mixed with 950 mL of deionized H2O (dH2O);
- Plastic container 12 × 8 × 3 cm, rinsed previously with 50% aqueous methanol twice and one time with 100% methanol;
- Instant Blue Coomassie (Expedeon, Cambridge, UK; Cat. no: ISB1L);
- Destaining solution: 50 mL methanol, 5 mL acetic acid (J.T Baker, Center Valley, PA, USA; Cat. no.: 9508-33) and 50 mL dH2O;
- Isopropanol (Fisher, Fair Lawn, NJ, USA; Cat. no.: BP2635-4);
- Ambic solution: 0.158 g of ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO, USA; Cat. no.: AG141-500G) dissolved in 20 mL of dH2O to obtain a 100 mM solution;
- Acetonitrile HPLC grade (ACN) (Fisher, Fair Lawn, NJ, USA; Cat. no.: A988-4);
- 10 mM dithiothreitol (DTT): 0.015 g of DTT (Research Products International, Mount Prospect, IL, USA; Cat. no.: D11000-25) dissolved in 10 mL of Ambic solution;
- 55 mM iodoacetamide (IDA): 0.10 g of iodoacetamide (Sigma-Aldrich, St. Louis, MO, USA; Cat. no.: I1149-5G) dissolved in 10 mL of Ambic solution;
- Trypsin solution: 20 µg of trypsin (Promega, Madison, WI, USA; Cat. no.: V5111) in 1 mL of cold Ambic solution. Final concentration 20 ng/µL;
- Extraction solution: 50% ACN / 50% dH2O and 0.1% formic acid (Sigma-Aldrich, St. Louis, MO, USA; Cat. no.: F0507-500mL);
- Buffer A (0.1% aqueous formic acid);
- Buffer B (80% ACN, 0.1% aqueous formic acid).
2.2. Equipment
- 1.5 L rectangular plastic containers (Pentair, Apopka, FL, USA; Cat. no.: PCt10);
- 1300 Ci Cs-137 source irradiator from the calibration facility at the Savannah River Nuclear Solutions (SRNS) (Aiken, SC, USA);
- Pyrex petri dish 100 × 10 mm (Sigma-Aldrich, St. Louis, MO, USA; Cat. no.: CLS3160100);
- Stainless steel disposable scalpels (Integra Miltex, York, PA, USA; Cat. no.: 4-411);
- Dressing forceps (Integra Miltex, York, PA, USA; Cat. no.: 18-184);
- Dumont #3c forceps (Fine Science Tools, Foster city, CA, USA; Cat. no.: 11231-20;
- Clear zip bag, 3″W × 4″H, 2 mL (Action health, Bensenville, IL, USA; Cat. no.: 85251-85002;
- Stereo Microscope (Olympus, Shinjuku, Tokyo, Japan; Cat. no.: (model): SZ51);
- Mortar and pestle (CoorsTek, Golden, CO, USA; Cat. No.: 60316 and 60317);
- 5 ¾″ disposable Pasteur borosilicate pipets (Fisher Scientific, Fair Lawn, NJ, USA; Cat. no.: 13-678-20B);
- Glass tube 16 × 125 mm (Corning Incorporated, Corning, NY, USA; Cat. no.: 99447-16) rinsed twice with 50% aqueous MeOH and once with 100% MeOH;
- Vertical Rocker Roto-Shake Genie (Scientific Industries, Bohemia, NY, USA; Cat. no.: SI-1100),
- Allegra 6 refrigerated centrifuge (Beckman Coulter life sciences, Brea, CA, USA; Cat. no.: 366816);
- Heto vacuum centrifuge or speed vac (Heto-Holten A/S, Allerod, Denmark; Cat. no.: 23905B VR-maxi St.a.-1);
- Vortex Genie 2 (Fisher, Fair Lawn, NJ, USA; Cat. no: 12-812);
- Lyophilizer (Labconco corporation, Kansas City, MO, USA; Cat. no.: 7960040);
- Microcentrifuge tube pestle (USA Scientific Inc., Ocala, FL, USA; Cat. no.: 1415-5390), rinsed twice with 50% MeOH and once with 100% MeOH;
- Mini Gel Tank (ThermoFisher Scientific, Rockford, IL, USA; Cat. no.: A25977);
- Horizontal rocker platform (Bellco Biotechnology, Vineland, NJ, USA; Cat. no.: 7740-10010);
- Ziploc bags quart freezer, 7″W × 711/16″H (S. C. Johnson & Son, Inc. Racine, WI, USA);
- Glass and razor precleaned with Isopropanol 100% (see Note 2);
- New clean 1.7 mL Eppendorf tubes (MIDSCI, St. Louis, MO, USA; Cat. no.: AVSS1700);
- Heating module (Thermo Fisher Scientific, Rockford, IL, USA; Cat. no.: Pierce Reacti-Therm heating stirring module 18900);
- Incubator (Fisher, Pittsburgh, PA, USA; Cat. no.: 151030513);
- Nanosep 0.2 uM centrifugal filter units (PALL Life Sciences, NY, USA; Cat. no.: ODM02C34);
- Centrifuge (Beckman Coulter life sciences, Brea, CA, USA; Cat. no.: Microfuge 18);
- Glass crimp top vials (Thermo Fisher Scientific, Rockford, IL, USA; Cat. no: C4012-15), rinsed with MeOH;
- Snap caps for glass vials (VWR, Rador, PA, USA; Cat. no.: 14235-494);
- HPLC Ultimate 3000 RSLCnano (ThermoFisher Scientific, San Jose, CA, USA; Cat. no.: ULTIM3000RSLCNANO) with a 15 cm C18 analytical PepMap column (Thermo Fisher Scientific, San Jose, CA, USA; Cat. no.: 160321);
- Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA; Cat. no.: IQLAAEGAAPFADBMBCX).
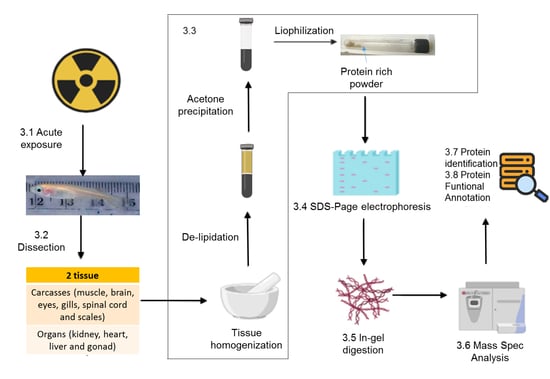
3. Procedures
3.1. Exposure to Radiation (Time for Completion: 48 h)
- Divide the fish into 2 groups: 6 adult fish for the control group and 6 for the treatment group.
- Place each group in small plastic containers with 0.5 L of filtered water.
- Expose the treatment group to ionizing radiation (0.5 Gy) at the Savannah River site calibration facility using a 1300 Ci CS-137 source calibrated to a dose rate of 0.028 Gy/minute, for a total exposure of 17.9 min obtaining a total dose of 0.5 Gy. An additional sham control group (no exposure) is subjected to the same protocol to account for handling stress.
- After exposure fish are returned to the laboratory and kept in tanks for 24 h.
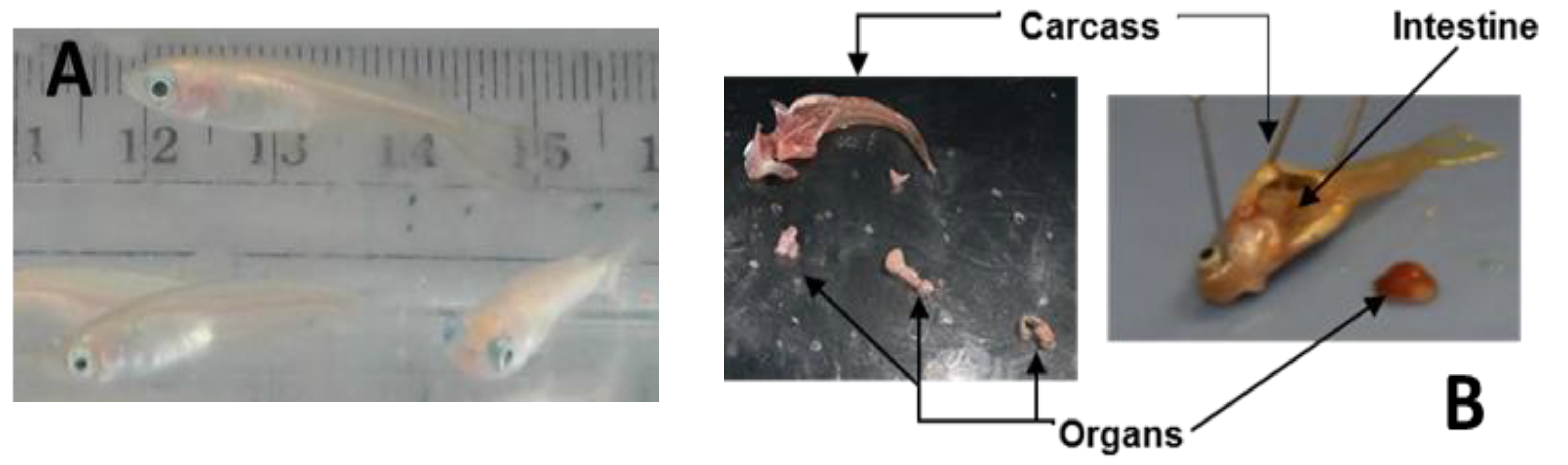
3.2. Dissection (for 10 Fish 2–3 h)
- Euthanize the fish at 24 h post exposure according to the requirements of Animal Care and Use at the University of Georgia, AUP #A201305-018-Y1-A0 part C: Experimental procedures: “Euthanasia of animals: Animals sacrificed for proteomic tissue research (or sick diseased fish) will be euthanatized by an overdose via immersion in anesthetic solution. A concentration of 250–500 mg/L (5–10 times the anesthetic dosage) is effective for Medaka according to AVMA 2013 guidelines [30]. Medaka will be left in the anesthetic solution for a minimum of 10 min after cessation of opercular movement. Tissues used for the radiation/proteomics study will be frozen in liquid nitrogen and stored at −80 °C until extracted for proteomics analysis. Euthanasia of animals will occur only at the Savannah River Ecology Laboratory”.Note: The full AUP document can be found in the Supplementary Material File S1.
- Place the fish into a glass petri dish (bottom or cover) and using a dissecting microscope open the fish with a scalpel, starting from the anus and continuing to the beginning of the head. Note: All the instruments and glassware must be clean, pre-washed with 50% methanol twice and 100% methanol once in order to avoid contamination of the samples. Use of plastic should be avoided, as it may result in contamination of the tissues with phthalates, complicating the mass spectrometry analysis.
- Using dressing forceps, open the ventral area of the fish and take out the kidney, heart, liver, and gonads and put together in a plastic zip bag previously labeled. This will be the organs group. CRITICAL STEP The tissues have to be keep on ice until they are frozen to avoid degradation and/or expression of proteins associated with death.
- Separate the intestines and stomach and place in another zip bag, and finally place the carcass (muscle, brain, eyes, gills, spinal cord, fins, and scales) in a third plastic bag. Figure 1 shows a dissected Medaka highlighting the different tissue groups.
- Using liquid nitrogen freeze all the tissues for 30–60 s. PAUSE STEP The samples are stored at −80 °C until the next step.
3.3. Preparing Protein-Rich Powder (7 h)
- In a mortar and pestle, add the sample and 3 mL of the homogenizing solution using glass Pasteur pipets and homogenize the tissue.
- Transfer the homogenized sample into a 15 mL glass tube. Rinse the mortar and pestle with 3 mL of the homogenizing solution and add the rinse to the homogenized sample.
- Allow the sample to incubate at room temperature on a vertical rocker for 3 h.
- Centrifuge the sample for 15 min at 4 °C at 3500 rpm. CRITICAL STEP The centrifugation generates heat, and thus refrigeration is necessary to avoid degradation of proteins.
- Decant the supernatant (glycosphingolipids) and, then, dry down the protein pellet using vacuum centrifugation for approximately 15–20 min. CRITICAL STEP Do not over dry. Over drying will result in an incomplete/difficult homogenization and can cause degradation of the samples. Note: If there is any interest in analyzing the glycosphingolipids, the supernatant from step 5 and 8 should be preserved in a pre-cleaned glass tube, dried under nitrogen, and kept at −20 °C for further analyses.
- Using Pasteur pipets, cover the sample in the bottom of the tube with homogenizing solution and incubate on the rocker for an additional 2 h at room temperature.
- Centrifuge sample for 15 min at 4 °C at 3500 rpm.
- Decant supernatant (glycosphingolipids) and, then, dry down the protein pellet using vacuum centrifugation.
- Add 1 mL of cold (4 °C) Milli-Q water, and mix using the vortex. Add 4 mL of cold (4 °C) acetone, mix using the vortex, and incubate on ice for 15 min.
- Centrifuge sample for 15 min at 4 °C at 3500 rpm. Decant supernatant into waste and dry down the protein pellet.
- Repeat steps 9–10.
- Freeze protein powder (−80 °C) and lyophilize overnight.
- Once dry, store protein powder at −20 °C. PAUSE STEP the protein-rich powder can be keep at −20 °C for several months (In our case we have stored samples for up to 3 years with no significant change in the analyses). Note: the glass tubes have to be well capped to avoid humidity getting into the samples.
3.4. SDS-Electrophoresis (1.5 h)
- Weigh 3–5 mg of protein-rich powder and resuspend with Tris-HCl buffer. Insoluble material is removed by centrifugation. Note: If necessary, use a microcentrifuge tube pestle before centrifugation to get a better homogenization of the sample. Prior to use, clean the pestle with 70% ethanol.
- Determine the protein concentration using the Pierce BCA protein assay kit with bovine serum albumin as standard.
- Prepare aliquots of 100 µg of protein and dry under vacuum centrifugation.
- Add 15 µL of Milli-Q water to dissolve the dry sample and add the same volume of the 2× Laemmli Sample Buffer. Mix with the vortex and centrifuge. Note: The final volume cannot be more than 35 µL, this is due to the capacity of the loading wells being 40 µL. CRITICAL STEP Observe the color of the mix, if yellowish, add 2 µL 100 mM NaOH at a time and mix until it turns blue. Mix using the vortex and centrifuge again.
- Boil the samples for 5 min and then put the samples into a refrigerator set at 7 °C for 5 min. Note: Be sure to cap the tubes well, or the sample will evaporate.
- Add 10 µL of protein ladder in the first well. Add protein samples leaving an empty well between samples, this will simplify cutting out the individual gel sections for the in-gel digestion step.
- Run the gel at 200 volts for 30–60 min.
- Place the gel in a clean clear plastic container and add enough deionized water to cover it, swish back and forth 5 times. Pour out the water. Repeat the wash at least 3 times. Note: The plastic container must be dust and detergent free. It should be cleaned prior to use with 70% ethanol and allowed to dry.
- Pour off the last water wash and add enough Instant Blue Coomasie stain to cover the gel, leave for 30 min to 1 h with gentle shaking. Note: Be sure that the gel can move freely in the staining solution to facilitate diffusion. Usually a 100 µg of protein will stain well after 30 min.
- Discard the stain solution and wash 2–3 times with deionized water.
- PAUSE STEP Keep the gel in water inside a Ziploc bag until the next step to avoid the gel drying out.
3.5. In-Gel Digestion (48 h after Full Distain of Gels Pieces)
- Place the gel on sanitized glass. Use a razor blade to remove top and bottom of the gel. Note: prior to use, clean the glass with 50% methanol and 100% methanol, and then one time with isopropanol, then, let it dry.
- Carefully cut each lane sample run into 10 equally sized sections and then cut each section into smaller pieces (1 × 1 mm2). Place all the gel pieces for each section into an Eppendorf tube. Note: Label the tube with sample and fraction information, i.e., control, fraction 5 can be CF5.
- Add 500 µL destaining solution to the gels and put on a rocker. Replace the solution 2–3 times during the day or let it rock overnight at room temperature. Repeat this until the gels are completely destained. NOTE: The time to completely destain the pieces of gel will depend on the frequency of changing the destaining solution, but 24 h is the fastest that the gel pieces can be destained.
- Once that the gel pieces are completely destained, remove destaining solution from each tube, using a different tip for each tube, then add 150 µL of HPLC grade water, and wait 5–10 min. Pull off water. Note: Starting at this point the tips and tubes used should be new and not been autoclaved, due to concerns of contamination that are detectable in the mass spectrometer.
- Add 150 µL of 30% aqueous ACN and wait 5–10 min. Pull off the solution. Repeat.
- Add 150 µL of Ambic solution, wait 5 min. Add 150 µL ACN 100% and wait 5–10 min. Pull off the solution.
- Add 150 µL ACN, wait 5–10 min. Pull off the solution. Samples are then dried under vacuum centrifugation (50–60 min).
- PAUSE STEP Properly capped tubes containing dried gel pieces can be stored at room temperature until the next step.
- Add 150 µL of 100 mM DTT, incubate at 65 °C for 1 h. Remove the samples from bath, let cool to room temperature and pull off the solution.
- Add 150 µL of 55 mM iodoacetamide for 1 h at room temperature in the dark. Pull off the solution.
- Wash gel pieces with 150 µL of Ambic solution for 5–10 min. Add 150 µL ACN, wait 5–10 min. Pull off the solution.
- Dry the gels under vacuum centrifugation for 45–60 min.
3.6. Tryptic Digestion (20 h)
- Add trypsin solution 50:1 ww (protein/trypsin ratio) and, then, add enough Ambic to a final volume of 125 µL to ensure that the dry gel pieces are completely submerged. Note: For 100 µg of protein use 2 µg of trypsin (100 µL of trypsin solution). To ensure a better distribution and absorption of trypsin into the gels, we mix 100 µL of trypsin solution with 2.4 mL of Ambic to obtain a total of 2.5 mL (125 µL per sample × 20 samples = 2.5 mL). Vortex and add 125 µL of the mix.
- Incubate over night at 37 °C (maximum 18 h).
- After incubation, spin tubes, collect the supernatants and transfer each into a new prewashed tube (tube A). Change pipette tips between each tube.
- Add 150 µL of extraction solution to the tubes containing the gel pieces and wait 5 min. Transfer the liquid to a fresh set of tubes (tube A). Repeat these extractions two more times.
- Transfer all the liquid from the set of tube A’s to a set of Nanosep centrifugal filter units. Centrifuge at 12,000 rpm for 15–30 min.
- The filtrates, containing tryptic peptides, are then dried on a speed vacuum, usually overnight. The samples can be stored at −20 °C until MS analyses.
3.7. LC-MS/MS Analysis of Tryptic Peptides (Mass Spectral Analysis) (8 h)
- Suspend the dried peptide in 19 µL of buffer A and 1 µL of buffer B and, then, transfer the dilute peptides into glass crimp top vials pre-cleaned with methanol 50% and 100%.
- Load the sample vials into the autosampler of an Ultimate 3000 LC System (Thermo Scientific Dionex).
- Mass spectrometry parameters: Peptides are separated on a 15 cm C18 analytical PepMap Column (Thermo Fisher Scientific) and eluted into an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) utilizing a nanoelectrospray ionization source via a 90 min gradient of increasing buffer B at a flow rate of approximately 200 nL/min. The gradient goes from 1% to 99% of buffer B between 3–60 min and holds at 99% for 5 min, then, there is a ramp back down to 1% over 5 min and holding 1% for the last 20 min for equilibration. Full MS scans are acquired at 60K resolution and MS2 scans following collision-induced dissociation are collected in the ion trap for the most intense ions in top-speed mode within a three second cycle using Fusion instrumentation software (version 4.1, Thermo Fisher Scientific). Dynamic exclusion is utilized to exclude precursor ions from the selection process for 60 s following a second selection within a 10 s window. We perform ”blank-runs” (only buffer B) in between samples injections to ensure no carryover from sample to sample.
- Results of the mass spectral analysis are in Raw format and are ready for the bioinformatics analysis that the user chooses. Below are summarized the bioinformatics and search options that we performed. Note: As an example, the raw data corresponding to the carcasses samples can be found in the public JPOST repository [32] under the Announced ID JPST000608.
3.8. Database Searching and Protein Identification (6 h)
- Raw files obtained from the mass spectra analysis following each preparation/separation protocol were converted to mzXML files and then to pkl (peak list format) using Trans-Proteomic Pipeline Software (Seattle Proteome Center, Seattle, WA, USA). Each pkl file was searched for protein identification against concatenated database (normal and reverse database) containing proteins from the following species: Oryzias latipes and Dario rerio, from the Broad Institute and National Center for Biotechnology Information (NCBI) using MASCOT (Matrix Scientific, Boston, MA, USA). The reverse database is created by reversing all protein sequences from the target database using an in-house utility. Note: The concatenated fasta file can be found in the Supplementary Material File S2.
- Mascot settings were as follows: tryptic enzymatic cleavages allowing for up to 2 missed cleavages, peptide tolerance of 20 parts-per-million, fragment ion tolerance of 0.5 Da, fixed modification due to carboxyamidomethylation of cysteine (+57 Da), and variable modifications of oxidation of methionine (+16 Da) and deamidation of asparagine or glutamine (+0.98 Da). Note: the pkl and mascot files corresponding to all the tissue groups can be found in the public JPOST repository under the Announced ID JPST000608.
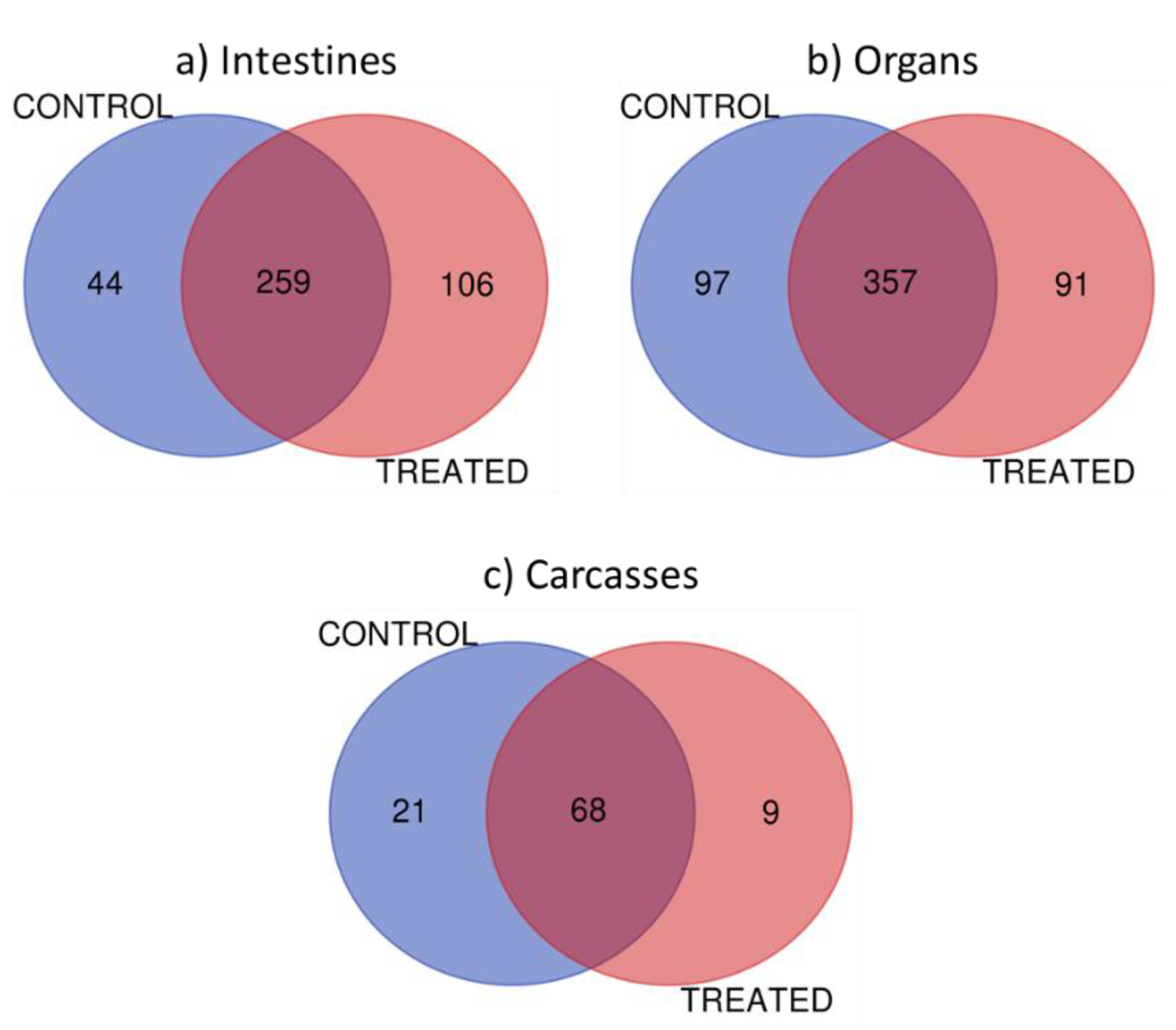
- Proteins were organized and filtered using a 1% protein false discovery rate applied, minimum 2 peptides, and 40 score in proteins via ProteoIQ software (Provalt_3.1.12_03-21-18, NuSep, Bogart, GA, USA) to obtain a nonredundant list of homologous protein groups [33], by loading Mascot target and decoy search files into the software program. (See Table 1 in results sections for an example of the list of some identified protein in Carcasses).
3.9. Protein Functional Annotation
- Gene Ontology terms are extracted from the Interpro and ProteoFun web sites (https://www.ebi.ac.uk/interpro/ and http://www.cbs.dtu.dk/services/ProtFun/).
- Signal peptides in the deduced amino acid sequences are examined using the SignalP Web site (http://www.cbs.dtu.dk/services/SignalP/) and the SecretomeP 2.0 Web site (http://www.cbs.dtu.dk/services/SecretomeP/).
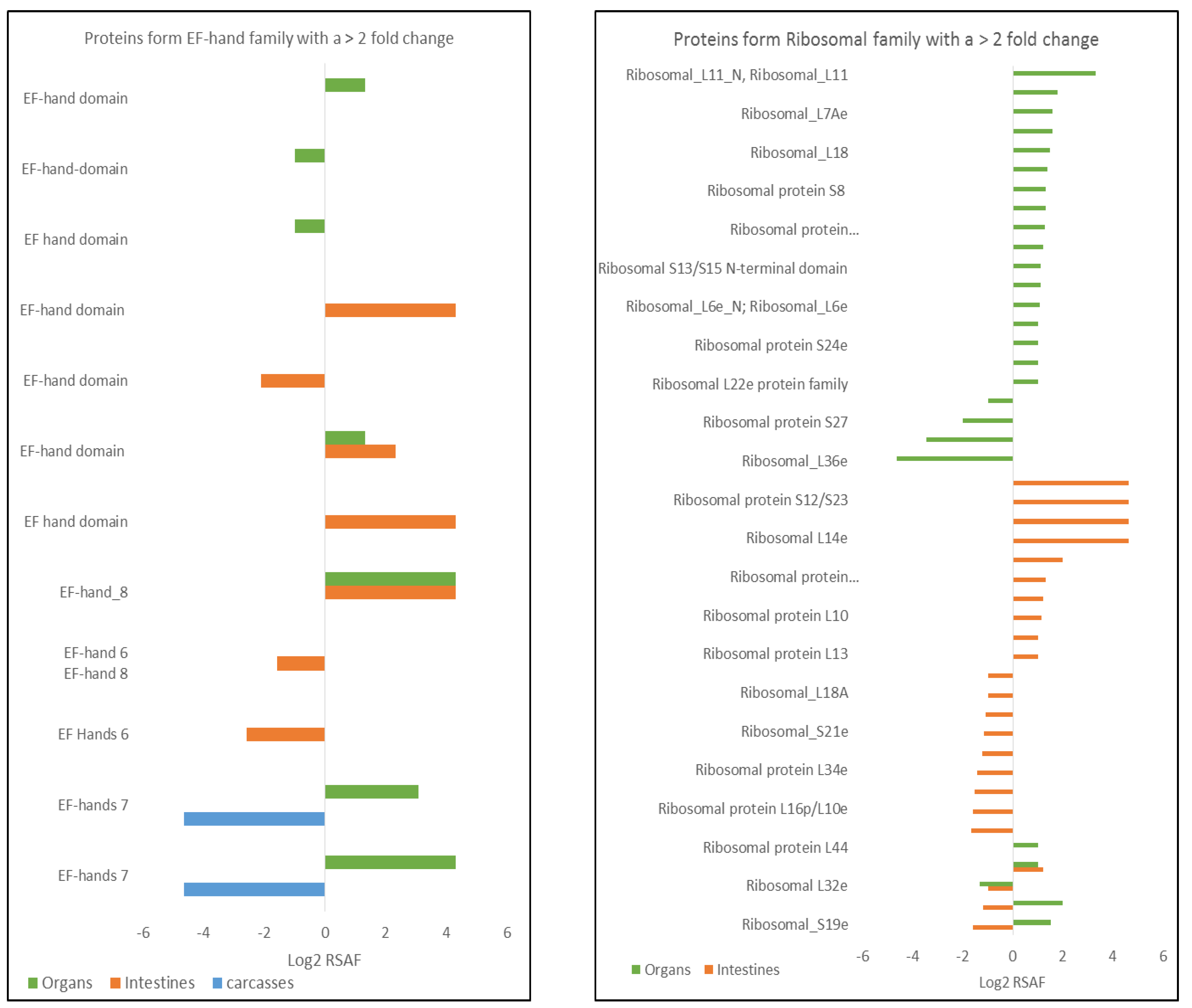
- The family classification and functional category was obtained by using the pFam database (https://pfam.xfam.org/). (see Table 2 in results sections for an example of biological information of some proteins identified in carcasses).
4. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- UNSCEAR. Sources and Effects of Ionizing Radiation. Report to the General Assembly with Scientific Annexes; Nations, U., Ed.; United Nations Scientific Committee on the Effects of Atomic Radiation Vienna: New York, NY, USA, 2008; Volume 1, p. 221. [Google Scholar]
- Reisz, J.A.; Bansal, N.; Qian, J.; Zhao, W.; Furdui, C.M. Effects of ionizing radiation on biological molecules-mechanisms of damage and emerging methods of detection. Antioxid. Redox Signal. 2014, 21, 260–292. [Google Scholar] [CrossRef] [PubMed]
- Dubrova, Y.E. Radiation-induced transgenerational instability. Oncogene 2003, 22, 7087–7093. [Google Scholar] [CrossRef] [PubMed]
- Goodhead, D.T. Initial events in the cellular effects of ionizing radiations: Clustered damage in DNA. Int. J. Radiat. Biol. 1994, 65, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Mavragani, I.V.; Nikitaki, Z.; Souli, M.P.; Aziz, A.; Nowsheen, S.; Aziz, K.; Rogakou, E.; Georgakilas, A.G. Complex DNA Damage: A Route to Radiation-Induced Genomic Instability and Carcinogenesis. Cancers 2017, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Magnander, K.; Elmroth, K. Biological consequences of formation and repair of complex DNA damage. Cancer Lett. 2012, 327, 90–96. [Google Scholar] [CrossRef]
- Desouky, O.; Ding, N.; Zhou, G.M. Targeted and non-targeted effects of ionizing radiation. J. Radiat. Res. Appl. Sci. 2015, 8, 247–254. [Google Scholar] [CrossRef]
- Mothersill, C.; Seymour, C. Radiation-induced bystander and other non-targeted effects: Novel intervention points in cancer therapy? Curr. Cancer Drug Targets 2006, 6, 447–454. [Google Scholar] [CrossRef]
- Mothersill, C.; Bucking, C.; Smith, R.W.; Agnihotri, N.; Oneill, A.; Kilemade, M.; Seymour, C.B. Communication of radiation-induced stress or bystander signals between fish in vivo. Environ. Sci. Technol. 2006, 40, 6859–6864. [Google Scholar] [CrossRef]
- Smith, R.W.; Mothersill, C.; Hinton, T.; Seymour, C.B. Exposure to low level chronic radiation leads to adaptation to a subsequent acute X-ray dose and communication of modified acute X-ray induced bystander signals in medaka (Japanese rice fish, Oryzias latipes). Int. J. Radiat. Biol. 2011, 87, 1011–1022. [Google Scholar] [CrossRef]
- Vo, N.T.K.; Seymour, C.B.; Mothersill, C.E. Radiobiological characteristics of descendant progeny of fish and amphibian cells that survive the initial ionizing radiation dose. Environ. Res. 2019, 169, 494–500. [Google Scholar] [CrossRef]
- Xie, A.; Odate, S.; Chandramouly, G.; Scully, R. H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell Cycle 2010, 9, 3602–3610. [Google Scholar] [CrossRef] [PubMed]
- Chaze, T.; Slomianny, M.-C.; Milliat, F.; Tarlet, G.; Lefebvre-Darroman, T.; Gourmelon, P.; Bey, E.; Benderitter, M.; Michalski, J.-C.; Guipaud, O. Alteration of the Serum N-glycome of Mice Locally Exposed to High Doses of Ionizing Radiation. Mol. Cell. Proteom. 2013, 12, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.V.; Azimzadeh, O.; Barjaktarovic, Z.; Kempf, S.J.; Merl-Pham, J.; Hauck, S.M.; Buratovic, S.; Eriksson, P.; Atkinson, M.J.; Tapio, S. Total body exposure to low-dose ionizing radiation induces long-term alterations to the liver proteome of neonatally exposed mice. J. Proteome Res. 2015, 14, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.V.; Barjaktarovic, Z.; Azimzadeh, O.; Kempf, S.J.; Merl, J.; Hauck, S.M.; Eriksson, P.; Buratovic, S.; Atkinson, M.J.; Tapio, S. Long-term effects of acute low-dose ionizing radiation on the neonatal mouse heart: A proteomic study. Radiat. Environ. Biophys. 2013, 52, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Little, M.P.; Tawn, E.J.; Tzoulaki, I.; Wakeford, R.; Hildebrandt, G.; Paris, F.; Tapio, S.; Elliott, P. Review and meta-analysis of epidemiological associations between low/moderate doses of ionizing radiation and circulatory disease risks, and their possible mechanisms. Radiat. Environ. Biophys. 2010, 49, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, O.; Scherthan, H.; Sarioglu, H.; Barjaktarovic, Z.; Conrad, M.; Vogt, A.; Calzada-Wack, J.; Neff, F.; Aubele, M.; Buske, C.; et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 2011, 11, 3299–3311. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.W.; Wang, J.; Mothersill, C.E.; Hinton, T.G.; Aizawa, K.; Seymour, C.B. Proteomic changes in the gills of wild-type and transgenic radiosensitive medaka following exposure to direct irradiation and to X-ray induced bystander signals. Biochim. Biophys. Acta 2011, 1814, 290–298. [Google Scholar] [CrossRef]
- Wittbrodt, J.; Shima, A.; Schartl, M. Medaka—A model organism from the far East. Nat. Rev. Genet. 2002, 3, 53–64. [Google Scholar] [CrossRef]
- Winn, R.N.; Norris, M.B.; Brayer, K.J.; Torres, C.; Muller, S.L. Detection of mutations in transgenic fish carrying a bacteriophage lambda cII transgene target. Proc. Natl. Acad. Sci. USA 2000, 97, 12655–12660. [Google Scholar] [CrossRef]
- Metcalfe, C. Test for predicting carcinogenicity in fish. CRC Rev. Aquat. Sci. 1989, 1, 111–129. [Google Scholar]
- Law, J.M. Mechanistic considerations in small fish carcinogenicity testing. Ilar J. 2001, 42, 274–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ding, L.; Kuhne, W.W.; Hinton, D.E.; Song, J.; Dynan, W.S. Quantifiable biomarkers of normal aging in the Japanese medaka fish (Oryzias latipes). PLoS ONE 2010, 5, e13287. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Shimada, A. The art of medaka genetics and genomics: What makes them so unique? Annu. Rev. Genet. 2010, 44, 217–241. [Google Scholar] [CrossRef] [PubMed]
- Chaze, T.; Hornez, L.; Chambon, C.; Haddad, I.; Vinh, J.; Peyrat, J.P.; Benderitter, M.; Guipaud, O. Serum Proteome Analysis for Profiling Predictive Protein Markers Associated with the Severity of Skin Lesions Induced by Ionizing Radiation. Proteomes 2013, 1, 40–69. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Maxwell, C.A.; Barcellos-Hoff, M.H.; Parvin, B. Geometric approach to segmentation and protein localization in cell culture assays. J. Microsc. 2007, 225, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Ostling, O.; Johanson, K.J. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 1984, 123, 291–298. [Google Scholar] [CrossRef]
- Tsyusko, O.; Glenn, T.; Yi, Y.; Joice, G.; Jones, K.; Aizawa, K.; Coughlin, D.; Zimbrick, J.; Hinton, T. Differential genetic responses to ionizing irradiation in individual families of Japanese medaka, Oryzias latipes. Mutat. Res. 2011, 718, 18–23. [Google Scholar] [CrossRef]
- Members of the Panel on Euthanasia. AVMA Guidelines for the Euthanasia of Animals, 2013th ed.; American Veterinary Medical Association: Schaumburg, IL, USA, 2013; ISBN 978-1-882691-21-0. [Google Scholar]
- Aoki, K.; Perlman, M.; Lim, J.M.; Cantu, R.; Wells, L.; Tiemeyer, M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 2007, 282, 9127–9142. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef]
- Weatherly, D.B.; Atwood, J.A., 3rd; Minning, T.A.; Cavola, C.; Tarleton, R.L.; Orlando, R. A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol. Cell. Proteom. 2005, 4, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, J.S. Protein identification using MS/MS data. J. Proteom. 2011, 74, 1842–1851. [Google Scholar] [CrossRef] [PubMed]
- Lubec, G.; Afjehi-Sadat, L.; Yang, J.W.; John, J.P. Searching for hypothetical proteins: Theory and practice based upon original data and literature. Prog. Neurobiol. 2005, 77, 90–127. [Google Scholar] [CrossRef] [PubMed]
- Alcantara-Martinez, N.; Figueroa-Martinez, F.; Rivera-Cabrera, F.; Gutierrez-Sanchez, G.; Volke-Sepulveda, T. An endophytic strain of Methylobacterium sp. increases arsenate tolerance in Acacia farnesiana (L.) Willd: A proteomic approach. Sci. Total Environ. 2018, 625, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Volke-Sepulveda, T.; Salgado-Bautista, D.; Bergmann, C.; Wells, L.; Gutierrez-Sanchez, G.; Favela-Torres, E. Secretomic Insight into Glucose Metabolism of Aspergillus brasiliensis in Solid-State Fermentation. J. Proteome Res. 2016, 15, 3856–3871. [Google Scholar] [CrossRef] [PubMed]
- Donato, R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001, 33, 637–668. [Google Scholar] [CrossRef]
- Orre, L.M.; Pernemalm, M.; Lengqvist, J.; Lewensohn, R.; Lehtio, J. Up-regulation, modification, and translocation of S100A6 induced by exposure to ionizing radiation revealed by proteomics profiling. Mol. Cell. Proteom. 2007, 6, 2122–2131. [Google Scholar] [CrossRef]
- Braunstein, S.; Badura, M.L.; Xi, Q.; Formenti, S.C.; Schneider, R.J. Regulation of protein synthesis by ionizing radiation. Mol. Cell. Biol. 2009, 29, 5645–5656. [Google Scholar] [CrossRef]
- Sekihara, K.; Saitoh, K.; Yang, H.; Kawashima, H.; Kazuno, S.; Kikkawa, M.; Arai, H.; Miida, T.; Hayashi, N.; Sasai, K.; et al. Low-dose ionizing radiation exposure represses the cell cycle and protein synthesis pathways in in vitro human primary keratinocytes and U937 cell lines. PLoS ONE 2018, 13, e0199117. [Google Scholar] [CrossRef]
- Zhao, Y.; Tan, M.; Liu, X.; Xiong, X.; Sun, Y. Inactivation of ribosomal protein S27-like confers radiosensitivity via the Mdm2-p53 and Mdm2-MRN-ATM axes. Cell Death Dis. 2018, 9, 145. [Google Scholar] [CrossRef]
- Tian, W.N.; Braunstein, L.D.; Pang, J.; Stuhlmeier, K.M.; Xi, Q.C.; Tian, X.; Stanton, R.C. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem. 1998, 273, 10609–10617. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.B. Physics and Radiobiology of Nuclear Medicine, 4th ed.; Springer: New York, NY, USA; London, UK, 2012. [Google Scholar]
- Jovine, L.; Qi, H.; Williams, Z.; Litscher, E.; Wassarman, P.M. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat. Cell Biol. 2002, 4, 457–461. [Google Scholar] [CrossRef] [PubMed]
| Sequence Id | Sequence Name | Gene | Total Score | Total Peptides | Total Spectra | Control Score | Control Peptides | Control Spectral Count | Treated Score | Treated Peptides | Treated Spectral Count |
|---|---|---|---|---|---|---|---|---|---|---|---|
| gi|116062147|dbj|BAF34704.1| | fast skeletal myosin heavy chain isoform mMYH-7 [Oryzias latipes] | LOC110015468 | 7561.29 | 102 | 1935 | 7104.5 | 97 | 1061 | 5896.33 | 85 | 874 |
| gi|116062139|dbj|BAF34700.1| | fast skeletal myosin heavy chain isoform mMYH-5 [Oryzias latipes] | LOC101163631 | 7442.38 | 100 | 1852 | 7069.86 | 96 | 1022 | 5726.27 | 82 | 830 |
| gi|116062137|dbj|BAF34699.1| | fast skeletal myosin heavy chain isoform mMYH-6 [Oryzias latipes] | LOC111947749 | 7317.12 | 99 | 1755 | 6798.53 | 92 | 960 | 5818.96 | 84 | 795 |
| gi|116062145|dbj|BAF34703.1| | fast skeletal myosin heavy chain isoform mMYH-3 [Oryzias latipes] | LOC101163661 | 6961.55 | 96 | 1640 | 6463.32 | 89 | 896 | 5464.79 | 80 | 744 |
| gi|116062143|dbj|BAF34702.1| | fast skeletal myosin heavy chain isoform mMYH-2 [Oryzias latipes] | LOC101163414 | 6885.75 | 94 | 1697 | 6514.88 | 90 | 953 | 5332.89 | 78 | 744 |
| gi|116062149|dbj|BAF34705.1| | fast skeletal myosin heavy chain mMYH-9 [Oryzias latipes] | LOC101163903 | 6816.79 | 92 | 1675 | 6449.61 | 88 | 933 | 5242.95 | 76 | 742 |
| gi|1174695518|ref|XP_020560645.1| | myosin heavy chain, fast skeletal muscle-like [Oryzias latipes] | LOC101163903 | 5923.87 | 82 | 1426 | 5592.76 | 78 | 811 | 4426.38 | 66 | 615 |
| gi|116062151|dbj|BAF34706.1| | fast skeletal myosin heavy chain isoform mMYH-11 [Oryzias latipes] | LOC101164155 | 5713.97 | 77 | 1363 | 5411.98 | 74 | 779 | 4382.73 | 64 | 584 |
| gi|432868092|ref|XP_004071407.1| | myosin heavy chain, fast skeletal muscle [Oryzias latipes] | LOC101158198 | 5675.58 | 79 | 1400 | 5396.89 | 76 | 771 | 4490.4 | 65 | 629 |
| gi|239735374|dbj|BAH70477.1| | myosin heavy chain embryonic type 1 [Oryzias latipes] | mmyhemb1 | 4594.2 | 64 | 1254 | 4369.27 | 62 | 703 | 3572.81 | 55 | 551 |
| gi|1040677427|ref|XP_017208743.1| | myosin heavy chain, fast skeletal muscle [Danio rerio] | LOC113076616 | 4293.4 | 60 | 1003 | 4122.75 | 59 | 574 | 3208.54 | 49 | 429 |
| gi|528483089|ref|XP_001339206.5| | myosin heavy chain, fast skeletal muscle [Danio rerio] | myhb | 3651.99 | 51 | 839 | 3460.57 | 48 | 477 | 2656.29 | 42 | 362 |
| gi|239735378|dbj|BAH70479.1| | myosin heavy chain larval type 2 [Oryzias latipes] | mmyhl2 | 3505.06 | 45 | 752 | 3270.63 | 44 | 422 | 2650.25 | 37 | 330 |
| gi|239735376|dbj|BAH70478.1| | myosin heavy chain larval type 1 [Oryzias latipes] | mmyhl1 | 3434.68 | 44 | 767 | 3216.83 | 43 | 425 | 2614.22 | 37 | 342 |
| gi|28422303|gb|AAH46881.1| | Zgc:66156 protein, partial [Danio rerio] | zgc:66156 | 3092.15 | 43 | 586 | 2917.85 | 42 | 340 | 2349.64 | 36 | 246 |
| gi|432864495|ref|XP_004070322.1| | intermediate filament protein ON3-like [Oryzias latipes] | LOC101167707 | 1287.9 | 21 | 120 | 1007.83 | 16 | 59 | 1107.19 | 18 | 61 |
| gi|432920251|ref|XP_004079911.1| | alpha-actinin-3 [Oryzias latipes] | actn3 | 1221.75 | 22 | 61 | 1108.52 | 20 | 44 | 286.58 | 6 | 17 |
| gi|1174681026|ref|XP_020566641.1| | actin, alpha skeletal muscle isoform X1 [Oryzias latipes] | acta1 | 1102.66 | 17 | 268 | 833.62 | 12 | 148 | 917.13 | 14 | 120 |
| gi|1174662893|ref|XP_020566626.1| | myosin-7 [Oryzias latipes] | LOC100125526 | 1055.2 | 16 | 265 | 979.22 | 15 | 148 | 793.85 | 13 | 117 |
| gi|432895621|ref|XP_004076079.1| | creatine kinase M-type [Oryzias latipes] | LOC101166239 | 932.48 | 13 | 135 | 900.94 | 13 | 88 | 528.82 | 8 | 47 |
| gi|1174683558|ref|XP_020557031.1| | vitellogenin 1 isoform X1 [Oryzias latipes] | ol-vit1 | 903.99 | 14 | 71 | 712.57 | 11 | 29 | 647.26 | 10 | 42 |
| gi|1207193593|ref|XP_021329509.1| | actin, alpha cardiac muscle 1 [Danio rerio] | LOC108941121 | 862.86 | 13 | 213 | 760.34 | 11 | 118 | 668.01 | 10 | 95 |
| gi|432852666|ref|XP_004067324.1| | tropomyosin alpha-1 chain isoform X1 [Oryzias latipes] | LOC101164789 | 855.51 | 14 | 127 | 825.11 | 14 | 75 | 658.6 | 11 | 52 |
| gi|432922695|ref|XP_004080348.1| | sarcoplasmic/endoplasmic reticulum calcium ATPase 1 isoform X1 [Oryzias latipes] | LOC101171864 | 821.21 | 13 | 96 | 575.85 | 8 | 46 | 630.84 | 11 | 50 |
| gi|190338754|gb|AAI63562.1| | Myosin, heavy polypeptide 6, cardiac muscle, alpha [Danio rerio] | myh6 | 806.87 | 13 | 183 | 742.39 | 12 | 99 | 633.76 | 10 | 84 |
| gi|432864501|ref|XP_004070325.1| | keratin, type II cytoskeletal 8-like isoform X1 [Oryzias latipes] | LOC101168366 | 799.55 | 14 | 43 | 516.97 | 9 | 24 | 605.5 | 11 | 19 |
| gi|432847946|ref|XP_004066228.1| | keratin, type I cytoskeletal 13-like [Oryzias latipes] | LOC101159648 | 795.99 | 12 | 91 | 729.77 | 12 | 38 | 644.12 | 9 | 53 |
| gi|765137894|ref|XP_011480537.1| | creatine kinase M-type [Oryzias latipes] | LOC101163677 | 794.05 | 11 | 122 | 789.15 | 11 | 85 | 410.13 | 6 | 37 |
| gi|1174691476|ref|XP_020559720.1| | tropomyosin alpha-1 chain [Oryzias latipes] | LOC112151854 | 697.97 | 12 | 102 | 687.97 | 12 | 62 | 466.42 | 8 | 40 |
| gi|628601863|ref|NP_001278765.1| | fructose-bisphosphate aldolase A [Oryzias latipes] | aldoa | 697.38 | 9 | 133 | 622.22 | 8 | 68 | 593.27 | 9 | 65 |
| Sequence Id | Sequence Name | Gene | RATIO RSAF | pFAM | Secretome | Signal IP Score | Biological Process Involve in | Molecular Function Enables | Cellular Component Part of | |
|---|---|---|---|---|---|---|---|---|---|---|
| gi|1174671686|ref|XP_004080935.3| | LOW QUALITY PROTEIN: Ras-related protein Rab-18 [Oryzias latipes] | rab18 | NC | Ras family | 0.377 | 0.118 | NO | NP | 3924 GTPase activity 5525 GTP binding | NP |
| gi|765158221|ref|XP_011488823.1| | uncharacterized protein LOC101162088 [Oryzias latipes] | LOC101162088 | NC | Zona pellucida-like domain | 0.535 | 0.735 | YES | GO:2000344 positive regulation of acrosome reaction GO:0005803 egg coat formation GO:0007339 binding of sperm to zona pellucida | GO:0032190 acrosin binding | NP |
| gi|1174681297|ref|XP_020567346.1| | 60S ribosomal protein L12 [Oryzias latipes] | rpl12 | 10 | Ribosomal_L11_N, Ribosomal_L11 | 0.853 | 0.331 | NO | GO:0006412 translation | GO:0003735 structural constituent of ribosome | GO:0005840 ribosome |
| gi|765151708|ref|XP_011486186.1| | parvalbumin beta-like [Oryzias latipes] | LOC101173896 | 8.5 | EF-hands 7 | 0.328 | 0.176 | NO | NP | GO:0005509 calcium ion binding GO:0046872 metal ion binding | GO:0005737 cytoplasm GO:0005634 nucleus |
| gi|157278241|ref|NP_001098220.1| | ZPC domain containing protein 4 precursor [Oryzias latipes] | LOC100049336 | 5.33333 | zona pellucida | 0.721 | 0.659 | YES | GO:0007339 binding of sperm to zona pellucida GO:0035803 egg coat formation GO:2000344 positive regulation of acrosome reaction | GO:0035804 structural constituent of egg coat GO:0032190 acrosin binding | NP |
| gi|1174689816|ref|XP_011474004.2| | 60S ribosomal protein L22-like [Oryzias latipes] | LOC101166956 | 3.5 | Ribosomal_L22e | 0.75 | 0.128 | NO | GO:0033077 T cell differentiation in thymus GO:0006412 translation | GO:0003723 RNA binding GO:0003735 structural constituent of ribosome | GO:0005840 ribosome |
| gi|432867558|ref|XP_004071242.1| | cytochrome c oxidase subunit NDUFA4 [Oryzias latipes] | LOC101155111 | 3 | B12D | 0.884 | 0.239 | NO | GO:0002290 electron transport chain GO:1902600 proton transmembrane transport | GO:0004129 cytochrome-c oxidase activity | GO:0016020 membrane GO:0016021 integral component of membrane GO:0005751 mitochondrial respiratory chain complex IV |
| gi|765145559|ref|XP_004077973.2| | 60S ribosomal protein L18 [Oryzias latipes] | rpl18 | 2.8 | Ribosomal_L18 | 0.614 | 0.202 | NO | GO:0006412 translation | GO:0003735 structural constituent of ribosome | GO:0005840 ribosome |
| gi|1174693985|ref|XP_020560297.1| | myosin light polypeptide 6 isoform X1 [Oryzias latipes] | LOC101167617 | 2.5 | EF-hand domain | 0.4 | 0.106 | NO | NP | GO:0005509 calcium ion binding | NP |
| gi|182889290|gb|AAI64893.1| | Calm1b protein [Danio rerio] | calm1 | 2.5 | EF-hand domain | 0.676 | 0.101 | NO | GO:0019722 calcium-mediated signaling | GO:0005509 calcium ion binding GO:0046872 metal ion binding | NP |
| gi|1174655990|ref|XP_020564957.1| | uncharacterized protein LOC101172014 isoform X2 [Oryzias latipes] | LOC110014571 | 2.27273 | NP | 0.896 | 0.814 | YES | NP | NP | NP |
| gi|765142574|ref|XP_011482440.1| | uncharacterized protein LOC101162625 isoform X2 [Oryzias latipes] | LOC101162625 | 2.26667 | NP | 0.687 | 0.738 | YES | NP | NP | NP |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Gélvez, Y.; Unger, S.; Gutiérrez-Sánchez, G.; Bridger, R.; Rhodes, O.E., Jr.; Bergmann, C. An Effective Protocol for Proteome Analysis of Medaka (Oryzias latipes) after Acute Exposure to Ionizing Radiation. Methods Protoc. 2019, 2, 66. https://doi.org/10.3390/mps2030066
Pérez-Gélvez Y, Unger S, Gutiérrez-Sánchez G, Bridger R, Rhodes OE Jr., Bergmann C. An Effective Protocol for Proteome Analysis of Medaka (Oryzias latipes) after Acute Exposure to Ionizing Radiation. Methods and Protocols. 2019; 2(3):66. https://doi.org/10.3390/mps2030066
Chicago/Turabian StylePérez-Gélvez, Yeni, Shem Unger, Gerardo Gutiérrez-Sánchez, Robert Bridger, Olin E. Rhodes, Jr., and Carl Bergmann. 2019. "An Effective Protocol for Proteome Analysis of Medaka (Oryzias latipes) after Acute Exposure to Ionizing Radiation" Methods and Protocols 2, no. 3: 66. https://doi.org/10.3390/mps2030066
APA StylePérez-Gélvez, Y., Unger, S., Gutiérrez-Sánchez, G., Bridger, R., Rhodes, O. E., Jr., & Bergmann, C. (2019). An Effective Protocol for Proteome Analysis of Medaka (Oryzias latipes) after Acute Exposure to Ionizing Radiation. Methods and Protocols, 2(3), 66. https://doi.org/10.3390/mps2030066