Age-Based Genomic Screening during Childhood: Ethical and Practical Considerations in Public Health Genomics Implementation
Abstract
1. Introduction
2. Identifying Appropriate Conditions for Screening during Childhood
3. Sequencing Healthy Newborns and Children: Start Small, Grow with Time
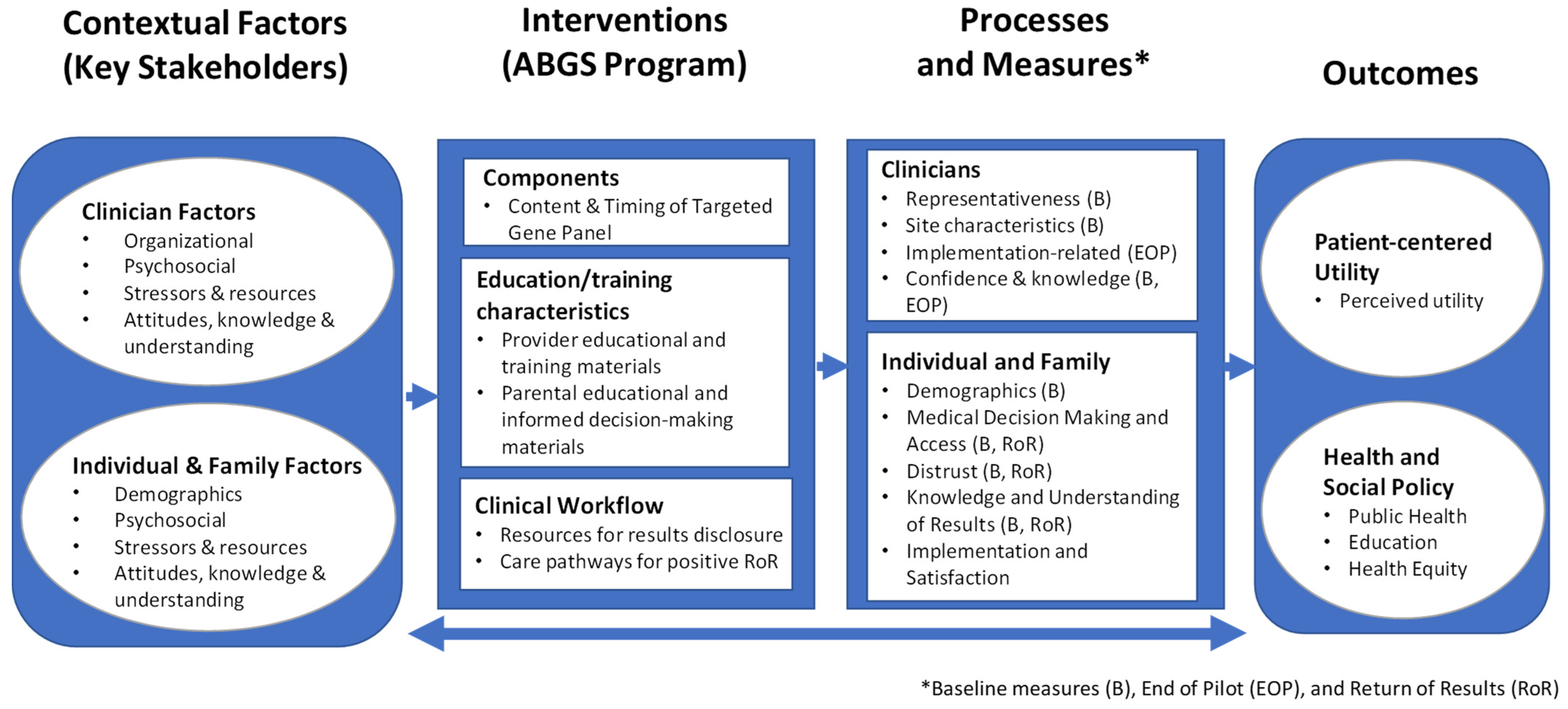
4. Using Implementation Science to Advance Innovations in Genomic Screening
5. Multilevel Partnerships with Parents and Providers
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Biesecker, L.G.; Green, R.C. Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 2014, 370, 2418–2425. [Google Scholar] [CrossRef] [PubMed]
- Strande, N.T.; Berg, J.S. Defining the Clinical Value of a Genomic Diagnosis in the Era of Next-Generation Sequencing. Annu. Rev. Genomics Hum. Genet. 2016, 17, 303–332. [Google Scholar] [CrossRef] [PubMed]
- Willig, L.K.; Petrikin, J.E.; Smith, L.D.; Saunders, C.J.; Thiffault, I.; Miller, N.A.; Soden, S.E.; Cakici, J.A.; Herd, S.M.; Twist, G.; et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: A retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med. 2015, 3, 377–387. [Google Scholar] [CrossRef]
- Berg, J.S.; Powell, C.M. Potential Uses and Inherent Challenges of Using Genome-Scale Sequencing to Augment Current Newborn Screening. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.B. A window of opportunity for newborn screening. Mol. Diagn. Ther. 2022, 26, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Kingsmore, S.F.; Smith, L.D.; Kunard, C.M.; Bainbridge, M.; Batalov, S.; Benson, W.; Blincow, E.; Caylor, S.; Chambers, C.; Del Angel, G.; et al. A genome sequencing system for universal newborn screening, diagnosis, and precision medicine for severe genetic diseases. Am. J. Hum. Genet. 2022, 109, 1605–1619. [Google Scholar] [CrossRef]
- Brothers, K.B.; Vassy, J.L.; Green, R.C. Reconciling opportunistic and population screening in clinical genomics. Mayo Clin. Proc. 2019, 94, 103–109. [Google Scholar] [CrossRef]
- Joshi, E.; Mighton, C.; Clausen, M.; Casalino, S.; Kim, T.H.M.; Kowal, C.; Birken, C.; Maguire, J.L.; Bombard, Y. Primary care provider perspectives on using genomic sequencing in the care of healthy children. Eur. J. Hum. Genet. 2020, 28, 551–557. [Google Scholar] [CrossRef]
- Kerruish, N. Parents’ experiences 12 years after newborn screening for genetic susceptibility to type 1 diabetes and their attitudes to whole-genome sequencing in newborns. Genet. Med. 2016, 18, 249–258. [Google Scholar] [CrossRef]
- Moultrie, R.R.; Paquin, R.; Rini, C.; Roche, M.I.; Berg, J.S.; Powell, C.M.; Lewis, M.A. Parental views on newborn next generation sequencing: Implications for decision support. Matern. Child Health J. 2020, 24, 856–864. [Google Scholar] [CrossRef]
- Botkin, J.R.; Belmont, J.W.; Berg, J.S.; Berkman, B.E.; Bombard, Y.; Holm, I.A.; Levy, H.P.; Ormond, K.E.; Saal, H.M.; Spinner, N.B.; et al. Points to consider: Ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am. J. Hum. Genet. 2015, 97, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Ross, L.F.; Saal, H.M.; David, K.L.; Anderson, R.R. American Academy of Pediatrics; American College of Medical Genetics and Genomics Technical report: Ethical and policy issues in genetic testing and screening of children. Genet. Med. 2013, 15, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.; Lantos, J.D.; Goldenberg, A.; Chen, F.; Parens, E.; Koenig, B.A. Members of the NSIGHT Ethics and Policy Advisory Board Sequencing newborns: A call for nuanced use of genomic technologies. Hastings Cent. Rep. 2018, 48 Suppl 2, S2–S6. [Google Scholar] [CrossRef]
- The American Academy of Pediatrics and Bright Futures 2017 recommendations for preventive pediatric health care. Pediatrics 2017, 139, e20170254. [CrossRef]
- Mollison, L.; Berg, J.S. Genetic screening: Birthright or earned with age? Expert Rev. Mol. Diagn. 2017, 17, 735–738. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Berg, J.S.; Olshan, A.F.; Magnuson, T.; Rimer, B.K. We screen newborns, don’t we?: Realizing the promise of public health genomics. Genet. Med. 2013, 15, 332–334. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.F.; Evans, J.P.; Khoury, M.J. DNA-Based Population Screening: Potential Suitability and Important Knowledge Gaps. JAMA 2020, 323, 307–308. [Google Scholar] [CrossRef]
- Orlando, L.A.; Sperber, N.R.; Voils, C.; Nichols, M.; Myers, R.A.; Wu, R.R.; Rakhra-Burris, T.; Levy, K.D.; Levy, M.; Pollin, T.I.; et al. Developing a common framework for evaluating the implementation of genomic medicine interventions in clinical care: The IGNITE Network’s Common Measures Working Group. Genet. Med. 2018, 20, 655–663. [Google Scholar] [CrossRef]
- Weitzel, K.W.; Alexander, M.; Bernhardt, B.A.; Calman, N.; Carey, D.J.; Cavallari, L.H.; Field, J.R.; Hauser, D.; Junkins, H.A.; Levin, P.A.; et al. IGNITE Network The IGNITE network: A model for genomic medicine implementation and research. BMC Med. Genomics 2016, 9, 1. [Google Scholar] [CrossRef]
- Zebrowski, A.M.; Ellis, D.E.; Barg, F.K.; Sperber, N.R.; Bernhardt, B.A.; Denny, J.C.; Dexter, P.R.; Ginsburg, G.S.; Horowitz, C.R.; Johnson, J.A.; et al. Qualitative study of system-level factors related to genomic implementation. Genet. Med. 2019, 21, 1534–1540. [Google Scholar] [CrossRef]
- Amendola, L.M.; Berg, J.S.; Horowitz, C.R.; Angelo, F.; Bensen, J.T.; Biesecker, B.B.; Biesecker, L.G.; Cooper, G.M.; East, K.; Filipski, K.; et al. The Clinical Sequencing Evidence-Generating Research Consortium: Integrating Genomic Sequencing in Diverse and Medically Underserved Populations. Am. J. Hum. Genet. 2018, 103, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.J.; Feero, W.G.; Chambers, D.A.; Brody, L.C.; Aziz, N.; Green, R.C.; Janssens, A.C.J.W.; Murray, M.F.; Rodriguez, L.L.; Rutter, J.L.; et al. A collaborative translational research framework for evaluating and implementing the appropriate use of human genome sequencing to improve health. PLoS Med. 2018, 15, e1002631. [Google Scholar] [CrossRef]
- Murray, M.F.; Evans, J.P.; Angrist, M.; Uhlmann, W.R.; Lochner Doyle, D.; Fullerton, S.M.; Ganiats, T.G.; Hagenkord, J.; Imhof, S.; Rim, S.H.; et al. A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults. NAM Perspectives 2018. [Google Scholar] [CrossRef]
- Shen, E.C.; Srinivasan, S.; Passero, L.E.; Allen, C.G.; Dixon, M.; Foss, K.; Halliburton, B.; Milko, L.V.; Smit, A.K.; Carlson, R.; et al. Barriers and facilitators for population genetic screening in healthy populations: A systematic review. Front. Genet. 2022, 13, 865384. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Cox, L.; Kruger, L.; Channamsetty, V.; Haga, S.B. Evaluating primary care providers’ readiness for delivering genetic and genomic services to underserved populations. Public Health Genomics 2021, 25, 1–10. [Google Scholar] [CrossRef]
- Kaphingst, K.A.; Bather, J.R.; Daly, B.M.; Chavez-Yenter, D.; Vega, A.; Kohlmann, W.K. Interest in cancer predisposition testing and carrier screening offered as part of routine healthcare among an ethnically diverse sample of young women. Front. Genet. 2022, 13, 866062. [Google Scholar] [CrossRef] [PubMed]
- Griesemer, I.; Staley, B.S.; Lightfoot, A.F.; Bain, L.; Byrd, D.; Conway, C.; Grant, T.L.; Leach, B.; Milko, L.; Mollison, L.; et al. Engaging community stakeholders in research on best practices for clinical genomic sequencing. Per. Med. 2020, 17, 435–444. [Google Scholar] [CrossRef]
- Berg, J.S.; Khoury, M.J.; Evans, J.P. Deploying whole genome sequencing in clinical practice and public health: Meeting the challenge one bin at a time. Genet. Med. 2011, 13, 499–504. [Google Scholar] [CrossRef]
- Berg, J.S.; Foreman, A.K.M.; O’Daniel, J.M.; Booker, J.K.; Boshe, L.; Carey, T.; Crooks, K.R.; Jensen, B.C.; Juengst, E.T.; Lee, K.; et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet. Med. 2016, 18, 467–475. [Google Scholar] [CrossRef]
- Hunter, J.E.; Irving, S.A.; Biesecker, L.G.; Buchanan, A.; Jensen, B.; Lee, K.; Martin, C.L.; Milko, L.; Muessig, K.; Niehaus, A.D.; et al. A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet. Med. 2016, 18, 1258–1268. [Google Scholar] [CrossRef]
- Hunter, J.E.; Jenkins, C.L.; Bulkley, J.E.; Gilmore, M.J.; Lee, K.; Pak, C.M.; Wallace, K.E.; Buchanan, A.H.; Foreman, A.K.M.; Freed, A.S.; et al. ClinGen ClinGen’s Pediatric Actionability Working Group: Clinical actionability of secondary findings from genome-scale sequencing in children and adolescents. Genet. Med. 2022, 24, 1328–1335. [Google Scholar] [CrossRef] [PubMed]
- Milko, L.V.; O’Daniel, J.M.; DeCristo, D.M.; Crowley, S.B.; Foreman, A.K.M.; Wallace, K.E.; Mollison, L.F.; Strande, N.T.; Girnary, Z.S.; Boshe, L.J.; et al. An Age-Based Framework for Evaluating Genome-Scale Sequencing Results in Newborn Screening. J. Pediatr. 2019, 209, 68–76. [Google Scholar] [CrossRef] [PubMed]
- DeCristo, D.M.; Milko, L.V.; O’Daniel, J.M.; Foreman, A.K.M.; Mollison, L.F.; Powell, B.C.; Powell, C.M.; Berg, J.S. Actionability of commercial laboratory sequencing panels for newborn screening and the importance of transparency for parental decision-making. Genome Med. 2021, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- van der Pal, S.M.; Wins, S.; Klapwijk, J.E.; van Dijk, T.; Kater-Kuipers, A.; van der Ploeg, C.P.B.; Jans, S.M.P.J.; Kemp, S.; Verschoof-Puite, R.K.; van den Bosch, L.J.M.; et al. Parents’ views on accepting, declining, and expanding newborn bloodspot screening. PLoS ONE 2022, 17, e0272585. [Google Scholar] [CrossRef]
- Brownson, R.C.; Shelton, R.C.; Geng, E.H.; Glasgow, R.E. Revisiting concepts of evidence in implementation science. Implement. Sci. 2022, 17, 26. [Google Scholar] [CrossRef]
- Milko, L.V.; Khoury, M.J. Editorial: DNA-based population screening for precision public health. Front. Genet. 2022, 13, 1061329. [Google Scholar] [CrossRef]
- Kwan, B.M.; Brownson, R.C.; Glasgow, R.E.; Morrato, E.H.; Luke, D.A. Designing for dissemination and sustainability to promote equitable impacts on health. Annu. Rev. Public Health 2022, 43, 331–353. [Google Scholar] [CrossRef]
- Shelton, R.C.; Adsul, P.; Oh, A.; Moise, N.; Griffith, D.M. Application of an antiracism lens in the field of implementation science (IS): Recommendations for reframing implementation research with a focus on justice and racial equity. Implement. Res. Pract. 2021, 2, 263348952110494. [Google Scholar] [CrossRef]
- Horowitz, C.R.; Orlando, L.A.; Slavotinek, A.M.; Peterson, J.; Angelo, F.; Biesecker, B.; Bonham, V.L.; Cameron, L.D.; Fullerton, S.M.; Gelb, B.D.; et al. The genomic medicine integrative research framework: A conceptual framework for conducting genomic medicine research. Am. J. Hum. Genet. 2019, 104, 1088–1096. [Google Scholar] [CrossRef]
- Ginsburg, G.S.; Horowitz, C.R.; Orlando, L.A. What will it take to implement genomics in practice? Lessons from the IGNITE Network. Per. Med. 2019, 16, 259–261. [Google Scholar] [CrossRef]
- Damschroder, L.J.; Aron, D.C.; Keith, R.E.; Kirsh, S.R.; Alexander, J.A.; Lowery, J.C. Fostering implementation of health services research findings into practice: A consolidated framework for advancing implementation science. Implement. Sci. 2009, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Kirk, M.A.; Kelley, C.; Yankey, N.; Birken, S.A.; Abadie, B.; Damschroder, L. A systematic review of the use of the Consolidated Framework for Implementation Research. Implement. Sci. 2016, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- IGNITE Toolbox. Available online: https://gmkb.org/ignite-toolbox-overview/ (accessed on 3 March 2020).
- Clinical Sequencing Evidence-Generating Research CSER Research Materials. Available online: https://cser-consortium.org/cser-research-materials (accessed on 12 August 2020).
- Halley, M.C.; Young, J.L.; Fernandez, L.; Kohler, J.N.; Undiagnosed Diseases Network; Bernstein, J.A.; Wheeler, M.T.; Tabor, H.K. Perceived utility and disutility of genomic sequencing for pediatric patients: Perspectives from parents with diverse sociodemographic characteristics. Am. J. Med. Genet. A 2022, 188, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Downie, L.; Halliday, J.; Lewis, S.; Amor, D.J. Principles of genomic newborn screening programs: A systematic review. JAMA Netw. Open 2021, 4, e2114336. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, A.J. Considering equity in assessing familial benefit from the return of genomic research results. Pediatrics 2019, 144, e20193111. [Google Scholar] [CrossRef]
- Henderson, G.E.; Ewing, M.; Kuczynski, K.J.; Cadigan, R.J.; Waltz, M.; Butterfield, R.M.; Rini, C.; Weck, K.; Berg, J.S.; Edwards, T.P. Development and validation of a measure of comprehension of genomic screening-negative results (CoG-NR). Eur. J. Hum. Genet. 2020, 28, 1394–1402. [Google Scholar] [CrossRef]
- Hendricks-Sturrup, R.M.; Linsky, A.; Lu, C.Y.; Vassy, J.L. Genomic testing is best integrated into clinical practice when it is actionable. Per. Med. 2020, 17, 5–8. [Google Scholar] [CrossRef]
- Waltz, M.; Meagher, K.M.; Henderson, G.E.; Goddard, K.A.; Muessig, K.; Berg, J.S.; Weck, K.E.; Cadigan, R.J. Assessing the implications of positive genomic screening results. Per. Med. 2020, 17, 101–109. [Google Scholar] [CrossRef]
- Borry, P.; Goffin, T.; Nys, H.; Dierickx, K. Attitudes regarding predictive genetic testing in minors: A survey of European clinical geneticists. Am. J. Med. Genet. C Semin. Med. Genet. 2008, 148C, 78–83. [Google Scholar] [CrossRef]
- Borry, P.; Evers-Kiebooms, G.; Cornel, M.C.; Clarke, A.; Dierickx, K. Public and Professional Policy Committee (PPPC) of the European Society of Human Genetics (ESHG) Genetic testing in asymptomatic minors: Background considerations towards ESHG Recommendations. Eur. J. Hum. Genet. 2009, 17, 711–719. [Google Scholar] [CrossRef]
- Knoppers, B.M.; Sénécal, K.; Borry, P.; Avard, D. Whole-genome sequencing in newborn screening programs. Sci. Transl. Med. 2014, 6, 229cm2. [Google Scholar] [CrossRef] [PubMed]
- Shkedi-Rafid, S.; Fenwick, A.; Dheensa, S.; Lucassen, A.M. Genetic testing of children for adult-onset conditions: Opinions of the British adult population and implications for clinical practice. Eur. J. Hum. Genet. 2015, 23, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.N.; Byfield, G.; Bennetone, A.; Frantz, A.M.; Harrison, L.K.; James-Crook, E.R.; Osborne, H.; Owens, T.H.; Shaw, J.L.; O’Daniel, J.; et al. Parental guidance suggested: Engaging parents as partners in research studies of genomic screening for a pediatric population. Front. Genet. 2022, 13, 867030. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milko, L.V.; Berg, J.S. Age-Based Genomic Screening during Childhood: Ethical and Practical Considerations in Public Health Genomics Implementation. Int. J. Neonatal Screen. 2023, 9, 36. https://doi.org/10.3390/ijns9030036
Milko LV, Berg JS. Age-Based Genomic Screening during Childhood: Ethical and Practical Considerations in Public Health Genomics Implementation. International Journal of Neonatal Screening. 2023; 9(3):36. https://doi.org/10.3390/ijns9030036
Chicago/Turabian StyleMilko, Laura V., and Jonathan S. Berg. 2023. "Age-Based Genomic Screening during Childhood: Ethical and Practical Considerations in Public Health Genomics Implementation" International Journal of Neonatal Screening 9, no. 3: 36. https://doi.org/10.3390/ijns9030036
APA StyleMilko, L. V., & Berg, J. S. (2023). Age-Based Genomic Screening during Childhood: Ethical and Practical Considerations in Public Health Genomics Implementation. International Journal of Neonatal Screening, 9(3), 36. https://doi.org/10.3390/ijns9030036






