Real-Time Detection of Meningiomas by Image Segmentation: A Very Deep Transfer Learning Convolutional Neural Network Approach
Abstract
:1. Introduction
- Conventional CT and MRI are reliable for diagnosing meningiomas, with CT demonstrating an accuracy of around 83% [26]. MRI is highly accurate for diagnosing meningiomas, offering sensitivities and positive predictive values generally of 82.6% and above [27]. However, MRI alone may not always differentiate between benign and malignant meningiomas, requiring further investigation like biopsy for a firm diagnosis [28]. Also, while generally effective, MRI accuracy can be lower for smaller lesions or in specific locations like the skull base [27]. Although some researchers have demonstrated that MRI could provide valuable information for the evaluation of meningiomas, the radiological performance of different grades largely overlaps, which could lead to misdiagnosis and inappropriate treatment strategies. Therefore, improving the preoperative classification of meningiomas is a prerogative. ML, an intersection of statistics and computer science, is a branch of artificial intelligence as it enables the extraction of meaningful patterns from examples, which is a component of human intelligence. Over the last decade, it has been successfully applied in the field of radiology, particularly in automatically detecting disease and discriminating tumors. Recently, some studies demonstrated that ML based on MRI was a promising tool in grading meningiomas. However, a few radiomics studies combined with deep learning (DL) features were conducted using a pretrained convolutional neural network (CNN) [29,30].
- This research proposes a novel approach that significantly advances the application of DL for medical diagnostics by employing a very deep transfer learning CNN model (VGG-16) enhanced by CUDA optimization for the accurate and timely real-time identification of meningiomas.
- We have employed FLAIR (Fluid-Attenuated Inversion Recovery) structural magnetic resonance imaging (sMRI) in this case. Using quantitative and qualitative rendering of different brain subregions, sMRI measures variations in the brain’s water constitution, which are represented as different shades of gray. These data are then utilized to depict and characterize the location and size of tumors. For effective skull stripping (three-dimensional views of brain slices, axial, coronal, and sagittal), FLAIR images guarantee that surrounding fluids are not magnetized and that CSF (cerebrospinal fluid) is suppressed.
- In the practice of radiology, error is inevitable. In everyday practice, the amount of evidence collected during the plain film era is thought to be between 3–5% [31]. Interpretative error rates in cross-sectional imaging are reported to be much greater, ranging from 20–30% [32,33]. The clinical implications of accurate brain tumor grading classification are significant, as they can inform treatment decisions and improve patient outcomes. Discussing the method’s integration into clinical workflows has offered insights into its practical applications and impact on patient care, enhancing the paper’s relevance to healthcare professionals, thus providing a practical tool for streamlined medical analysis and decision making.
- A comparison has been made with recent state-of-the-art technique research propositions in the literature review.
2. Literature Review
3. Methods
3.1. Justification for Choosing VGGNet CNN Model
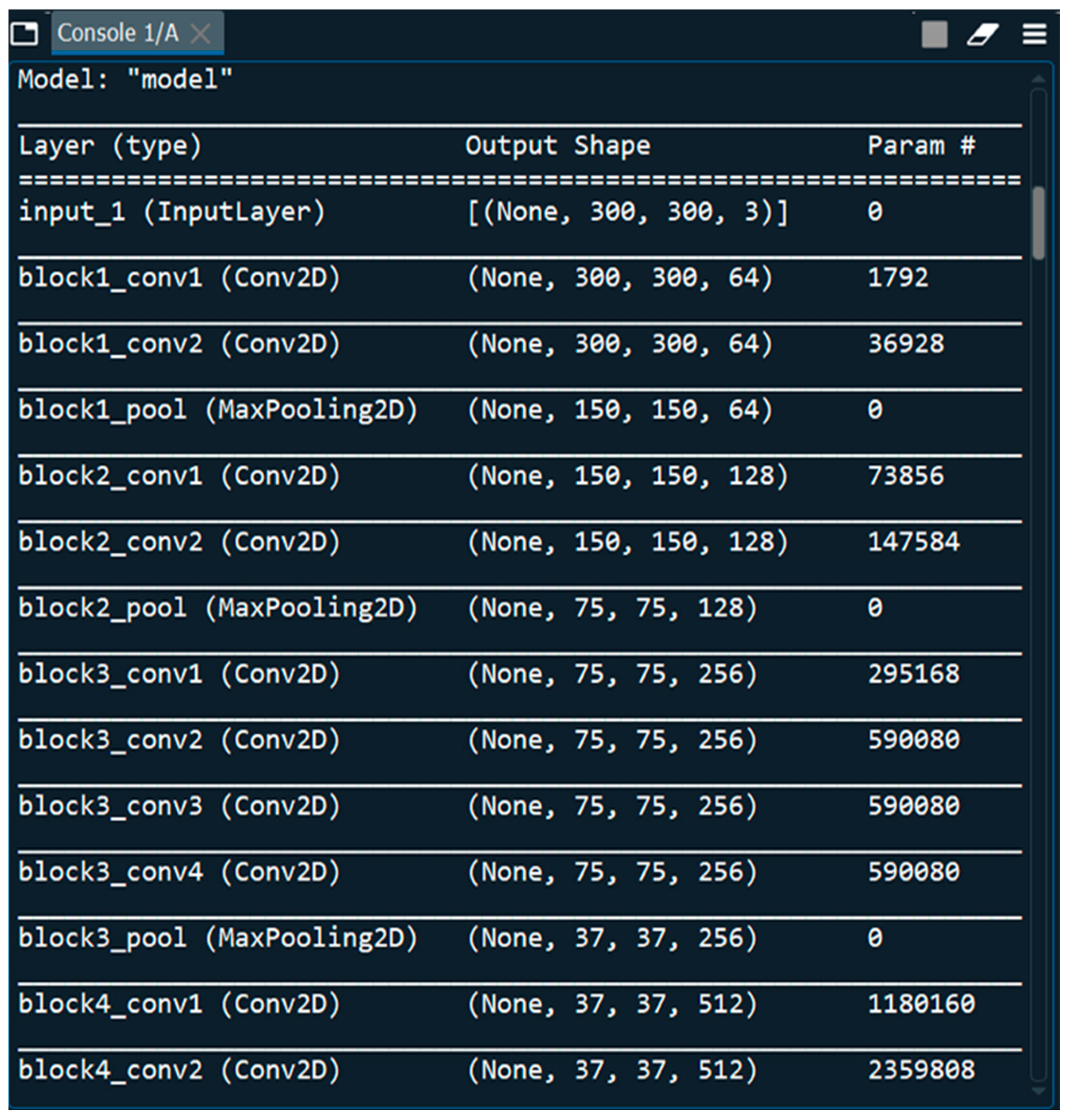
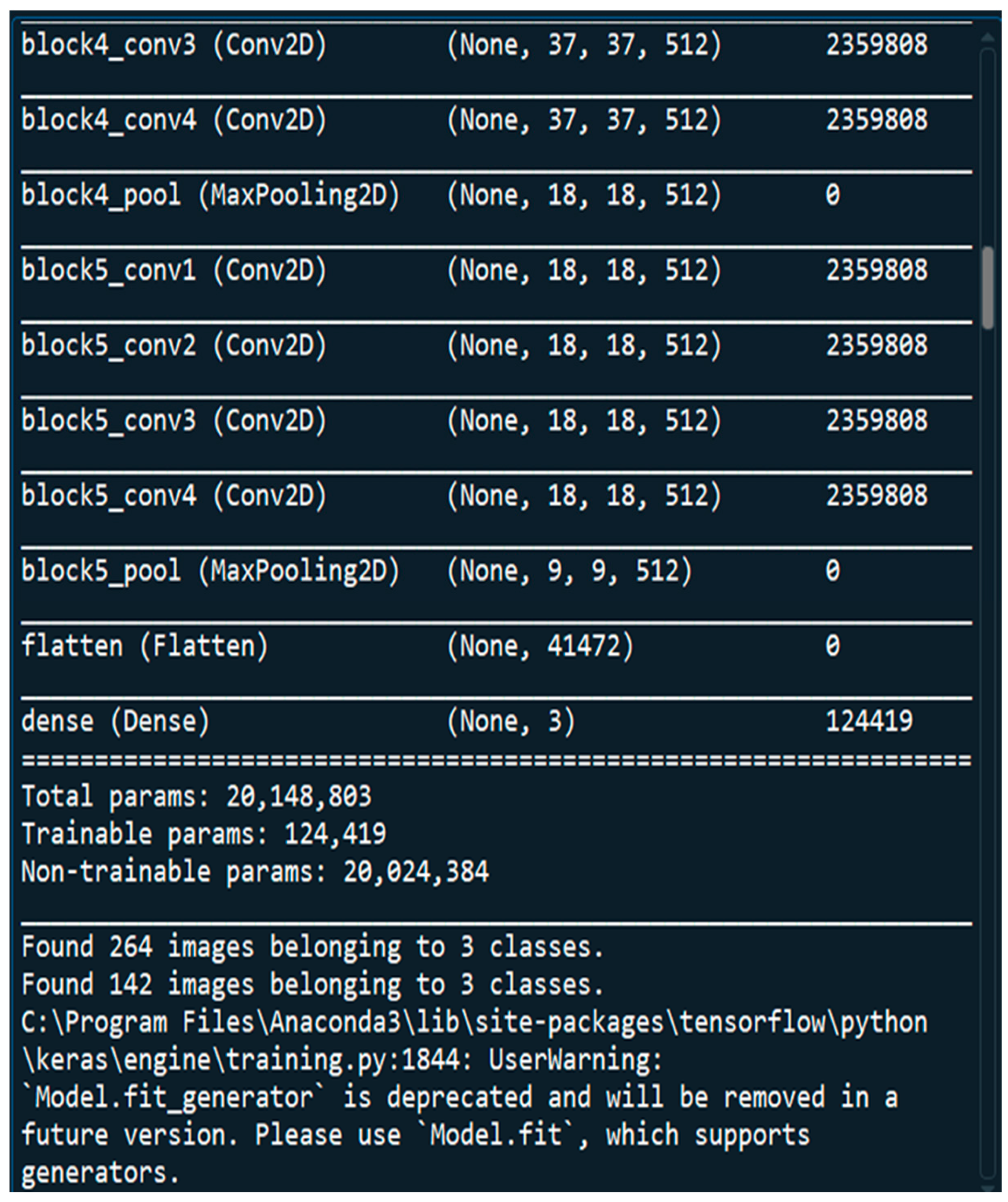
3.2. Architecture of VGGNet CNN Model
3.3. Dataset
3.4. Activation Functions
3.5. Optimization Algorithm
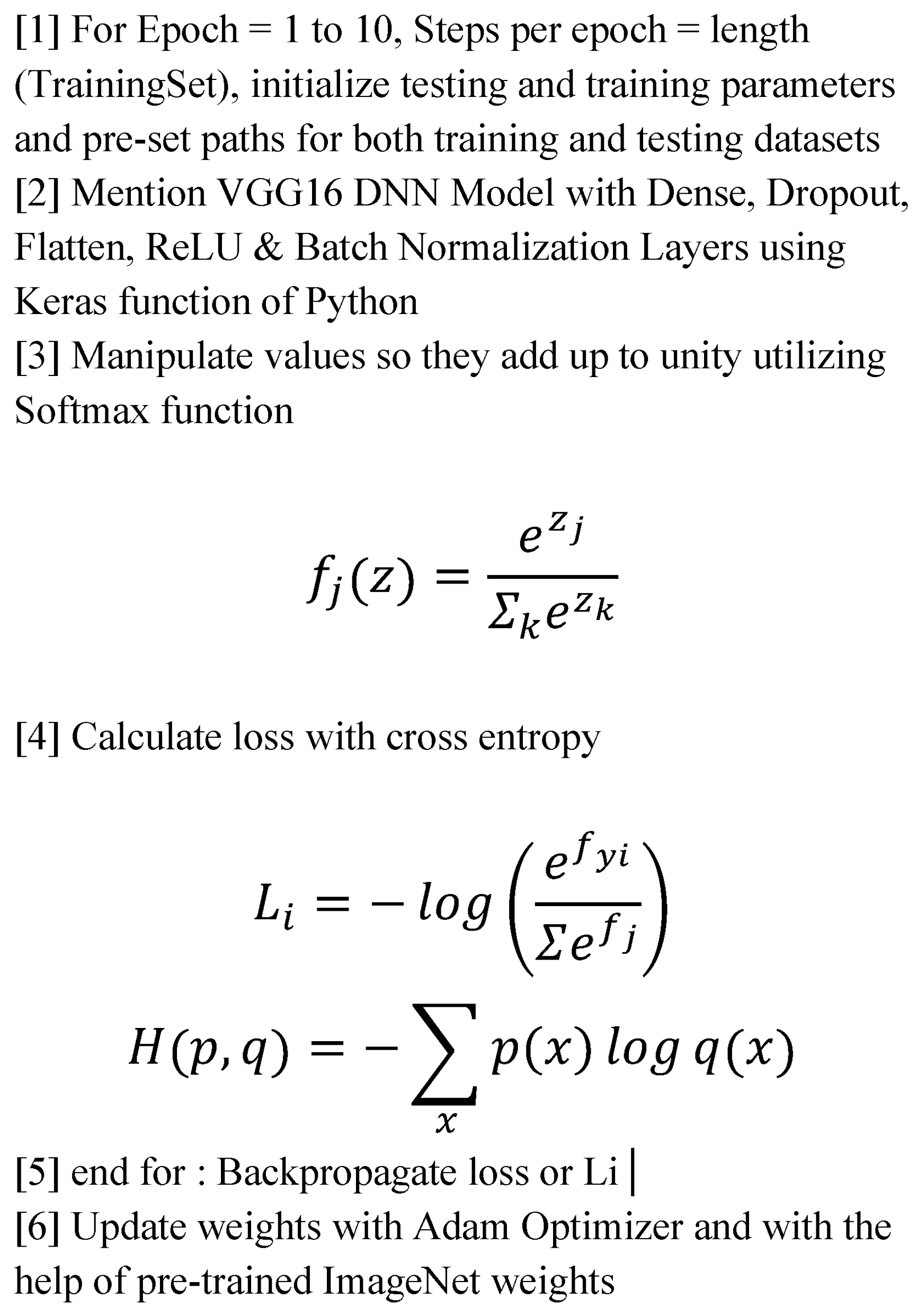
3.6. DNN Implementation Algorithm
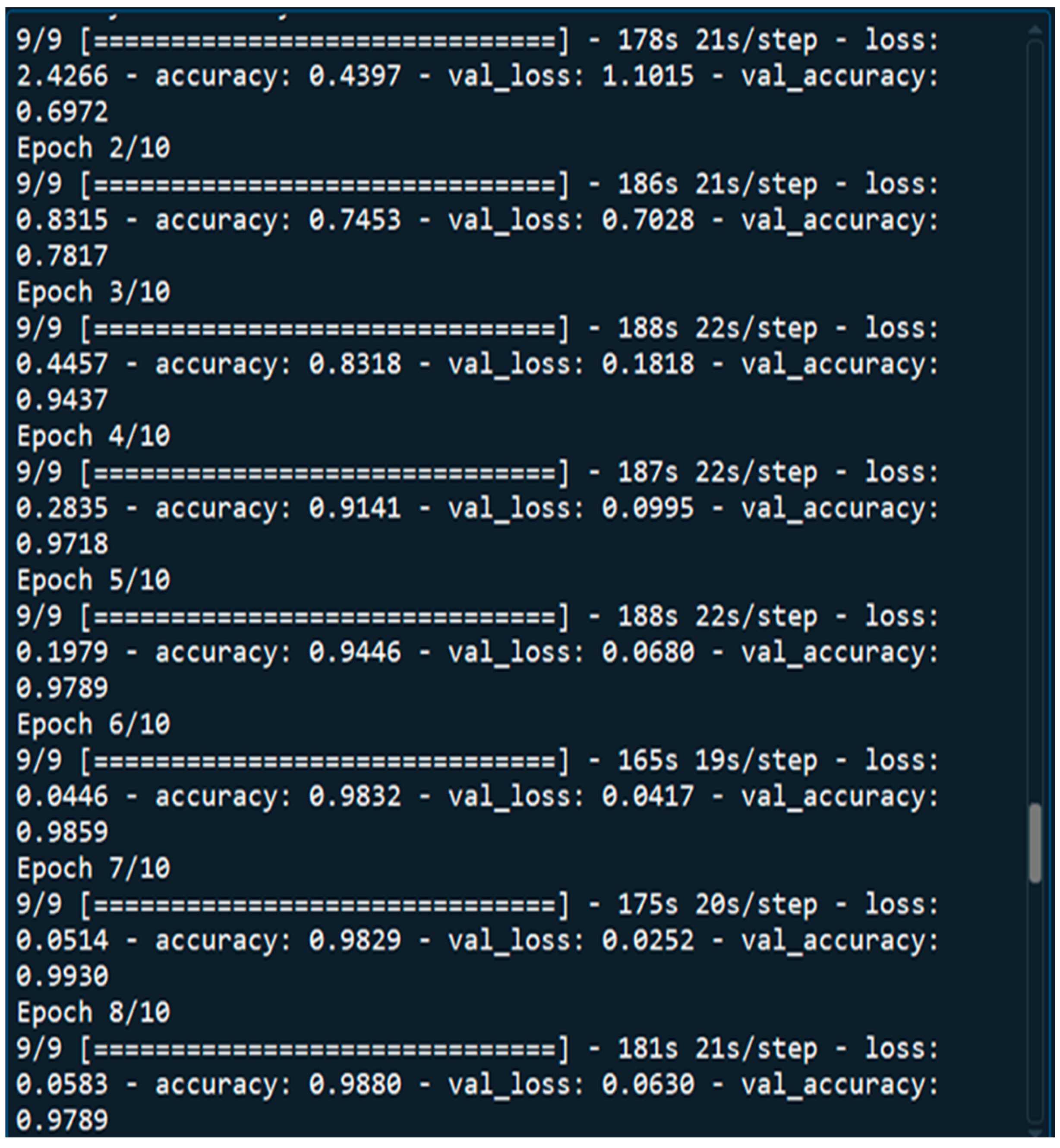
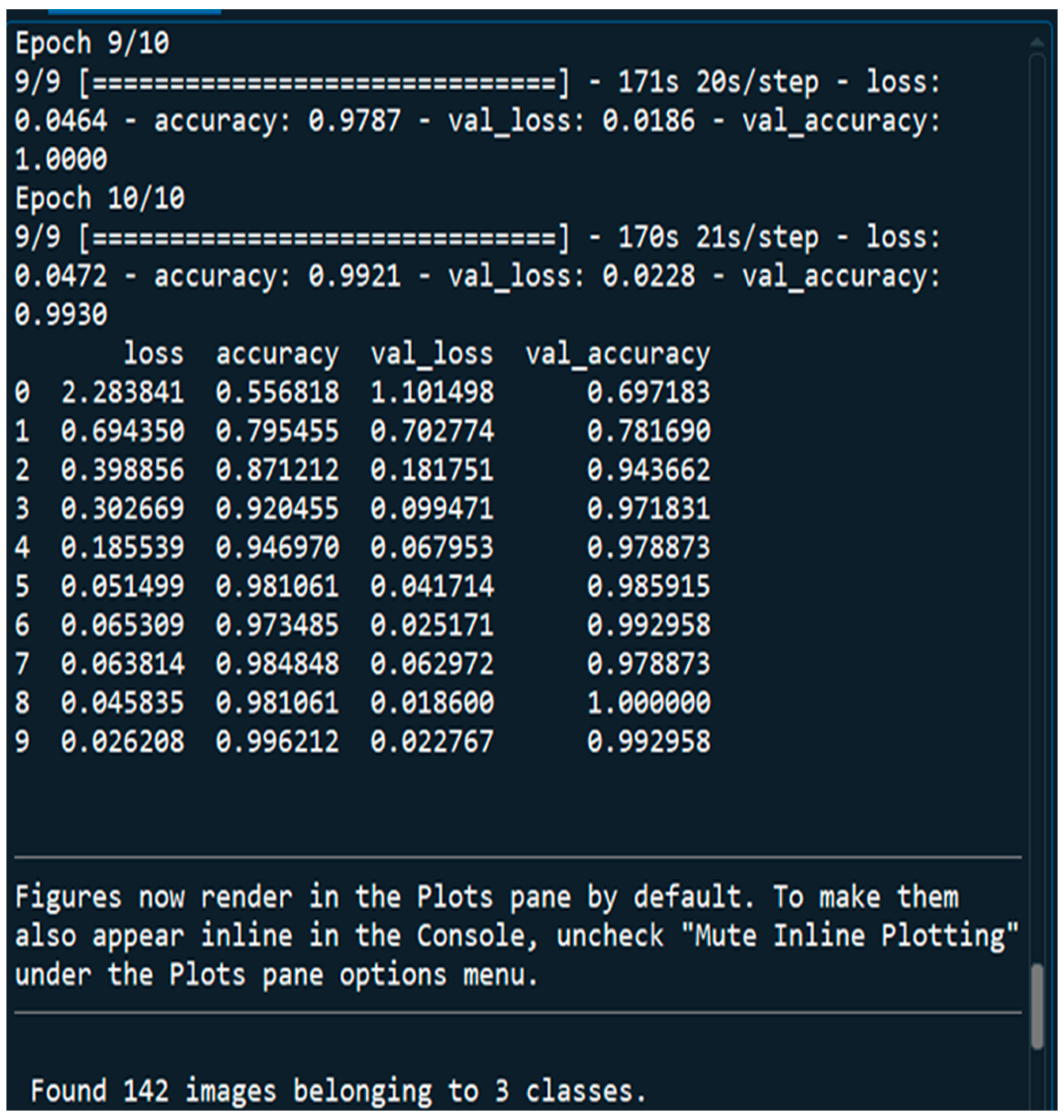
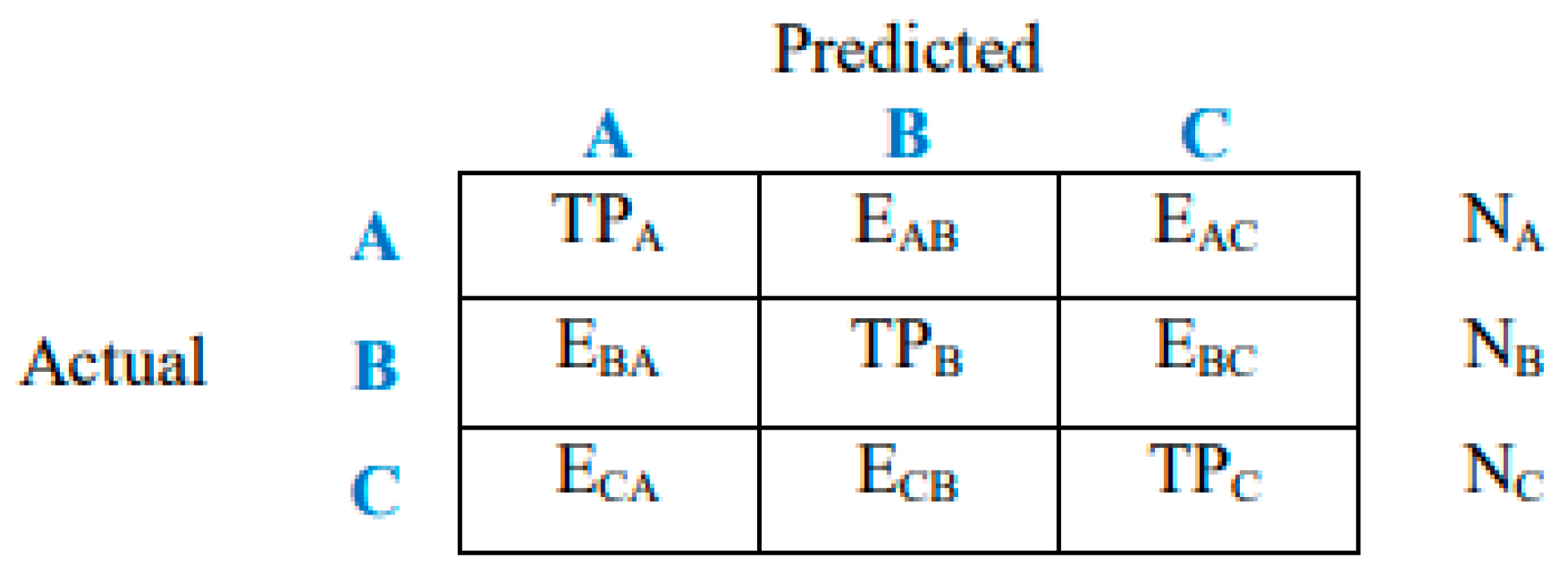
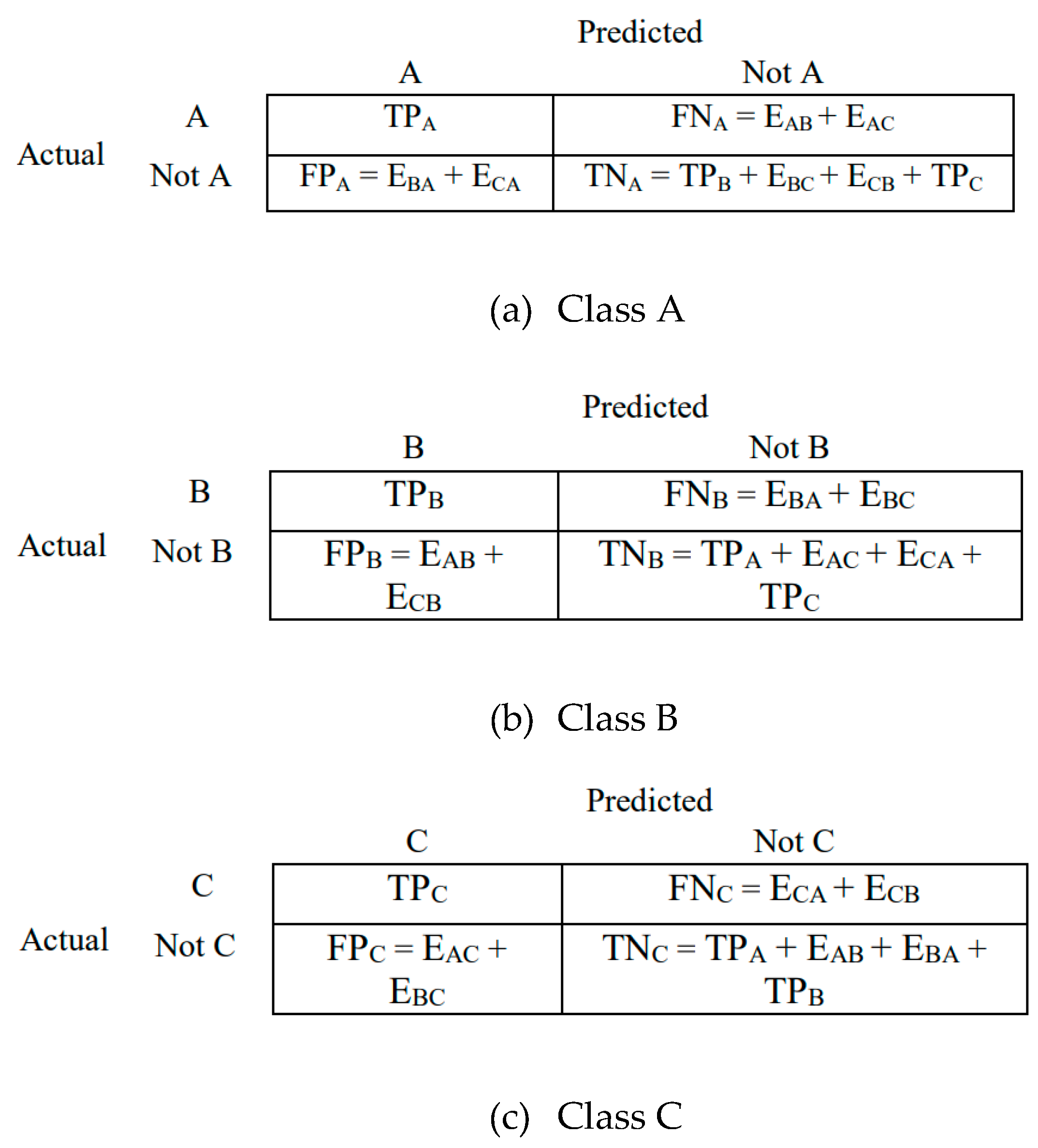
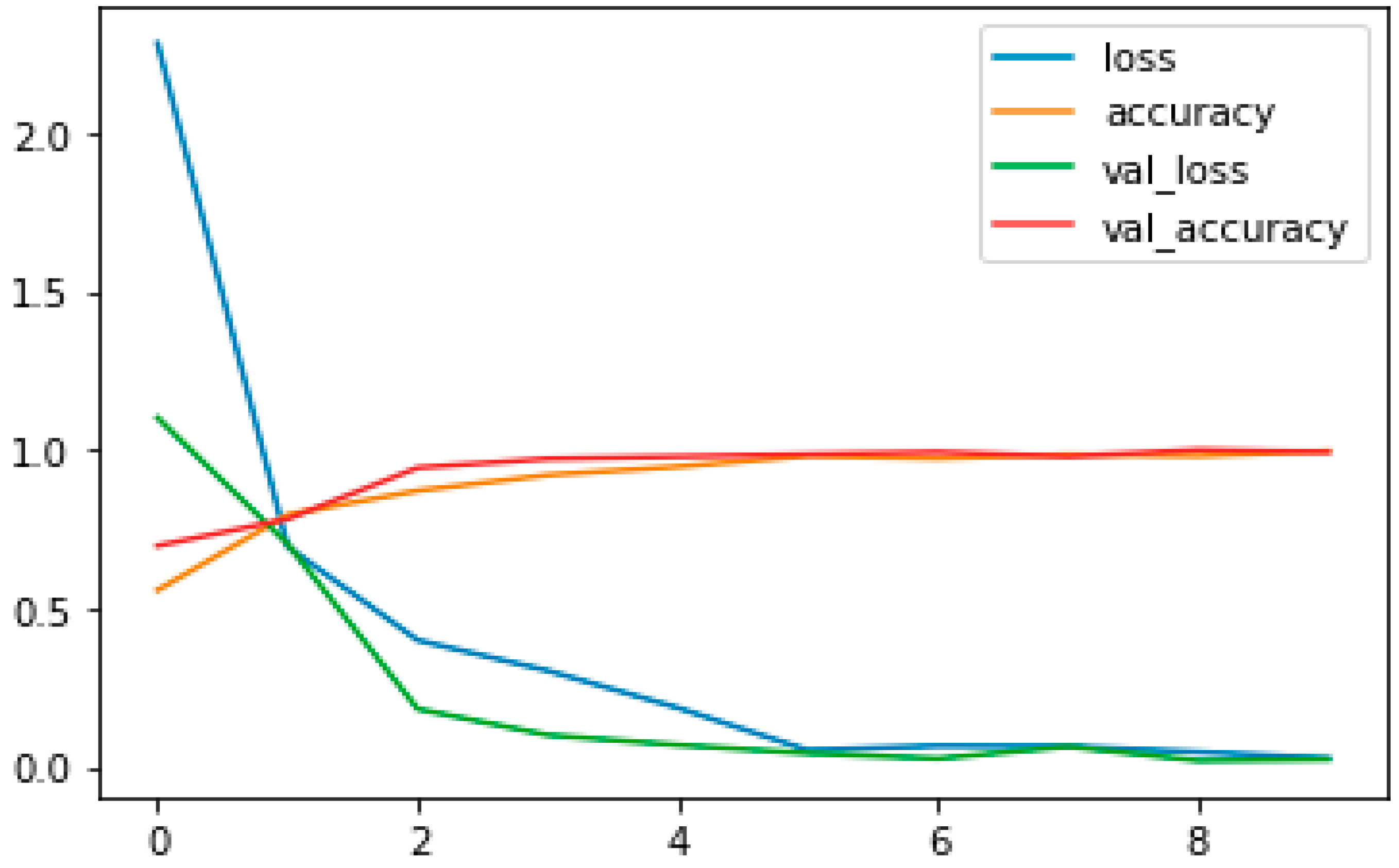
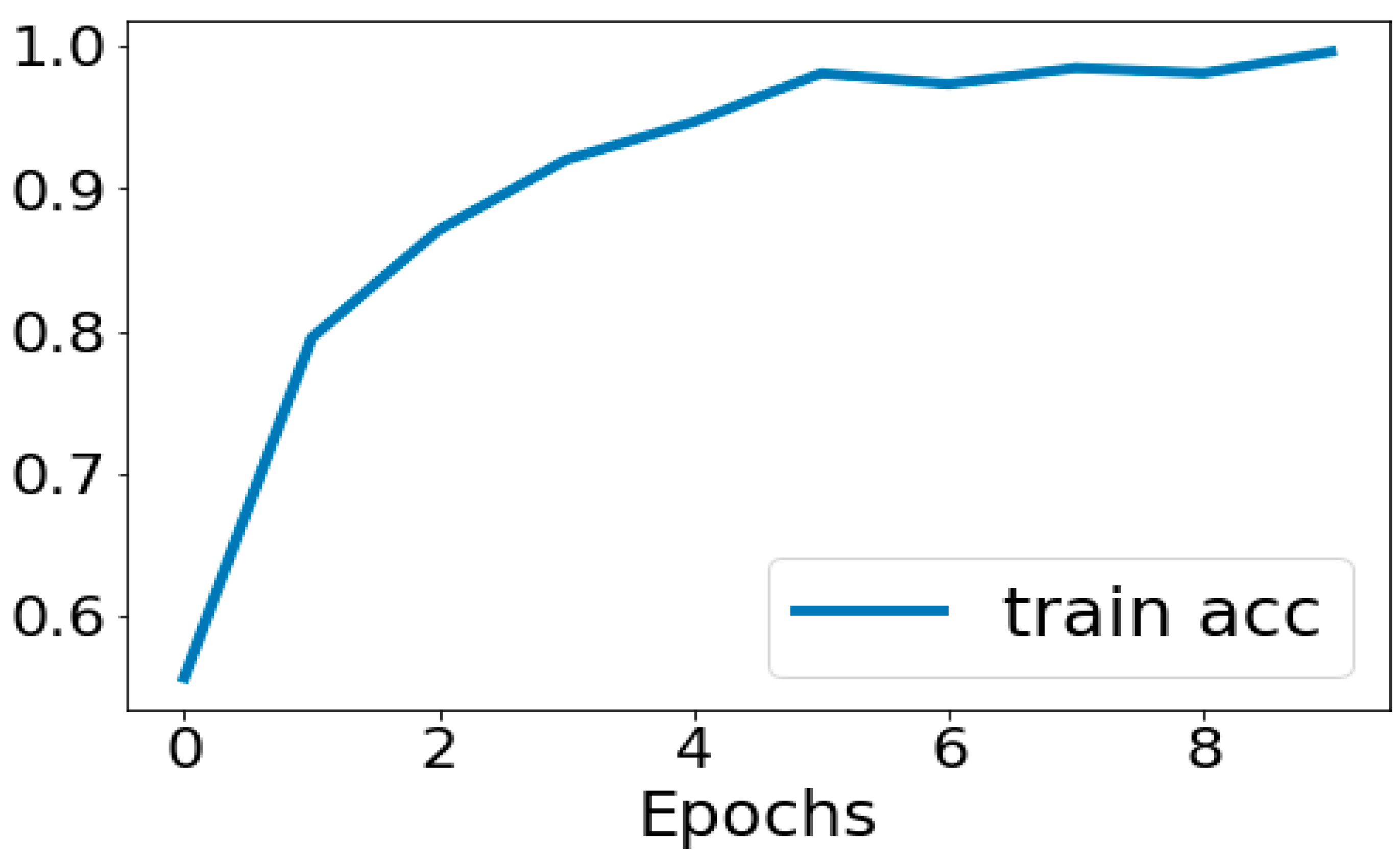
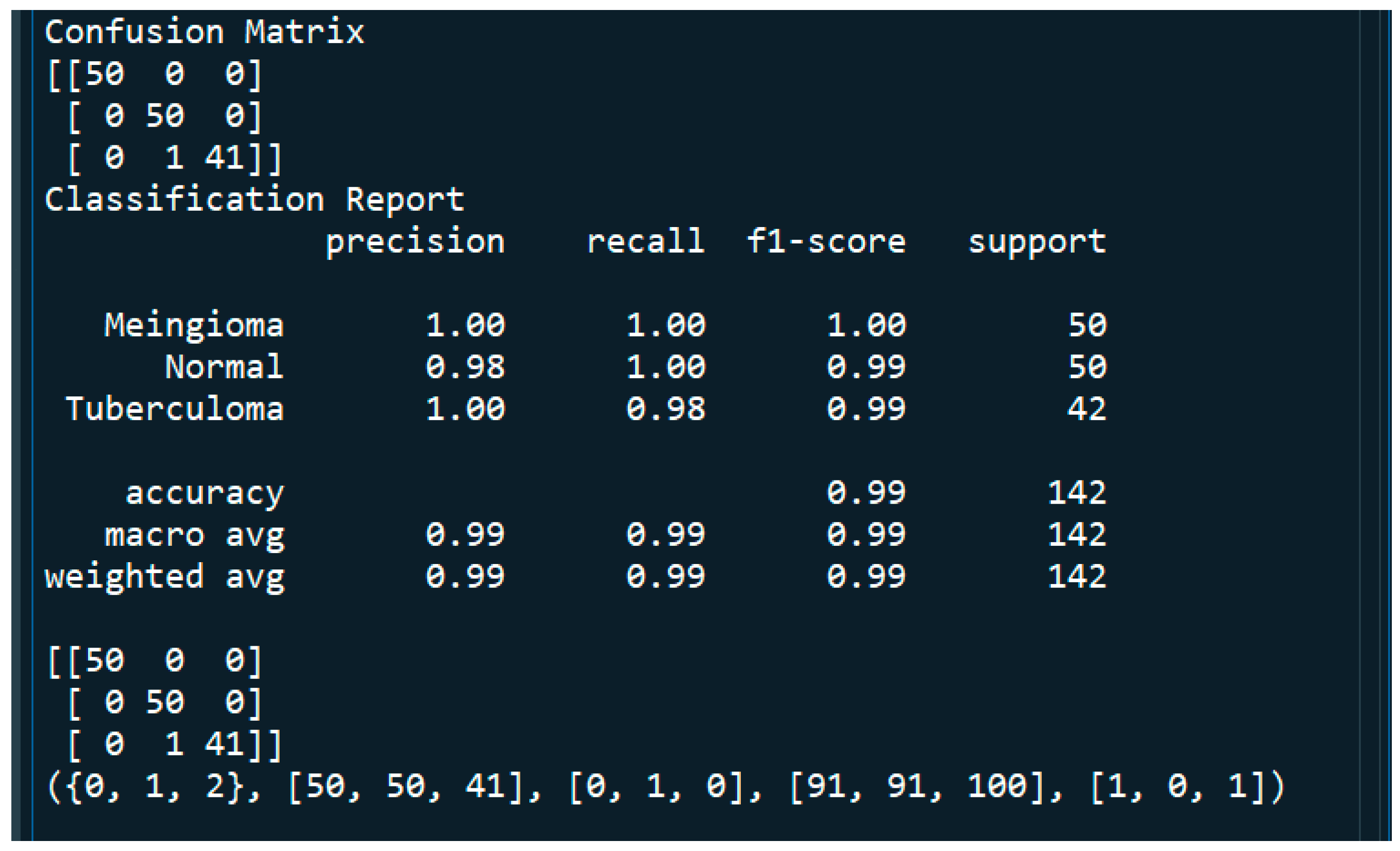
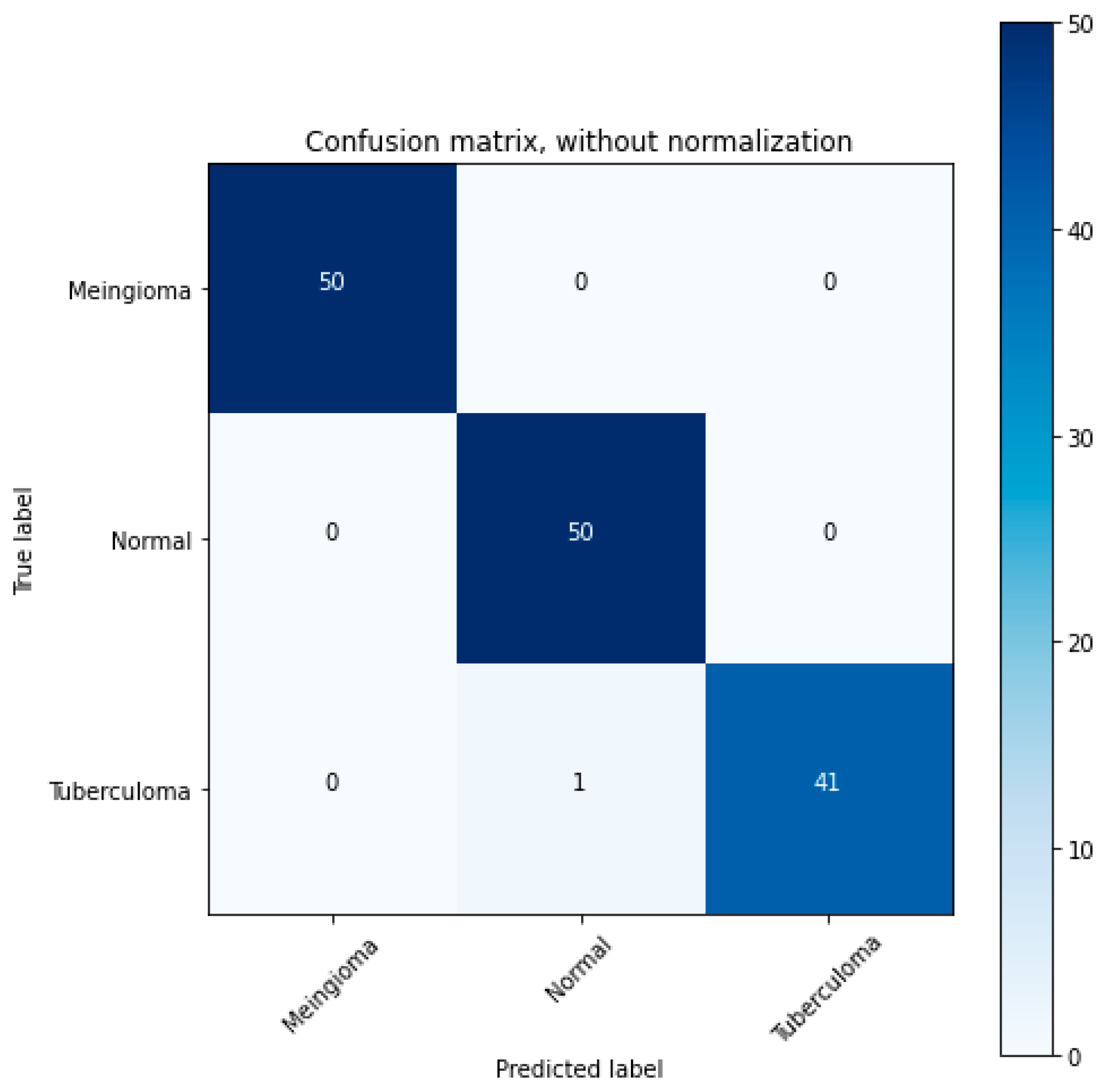
3.7. Evaluation of Model Performance
- True positive (TP): Correctly classified as belonging to a specific class.
- True negative (TN): Correctly classified as not belonging to a specific class.
- False positive (FP): Incorrectly classified as belonging to a specific class when it actually belongs to another.
- False negative (FN): Incorrectly classified as not belonging to a specific class when it actually belongs to that class.
3.8. Comparison of the Model with Human Experts
4. Output
4.1. Performance Metrics Analysis and Discussion
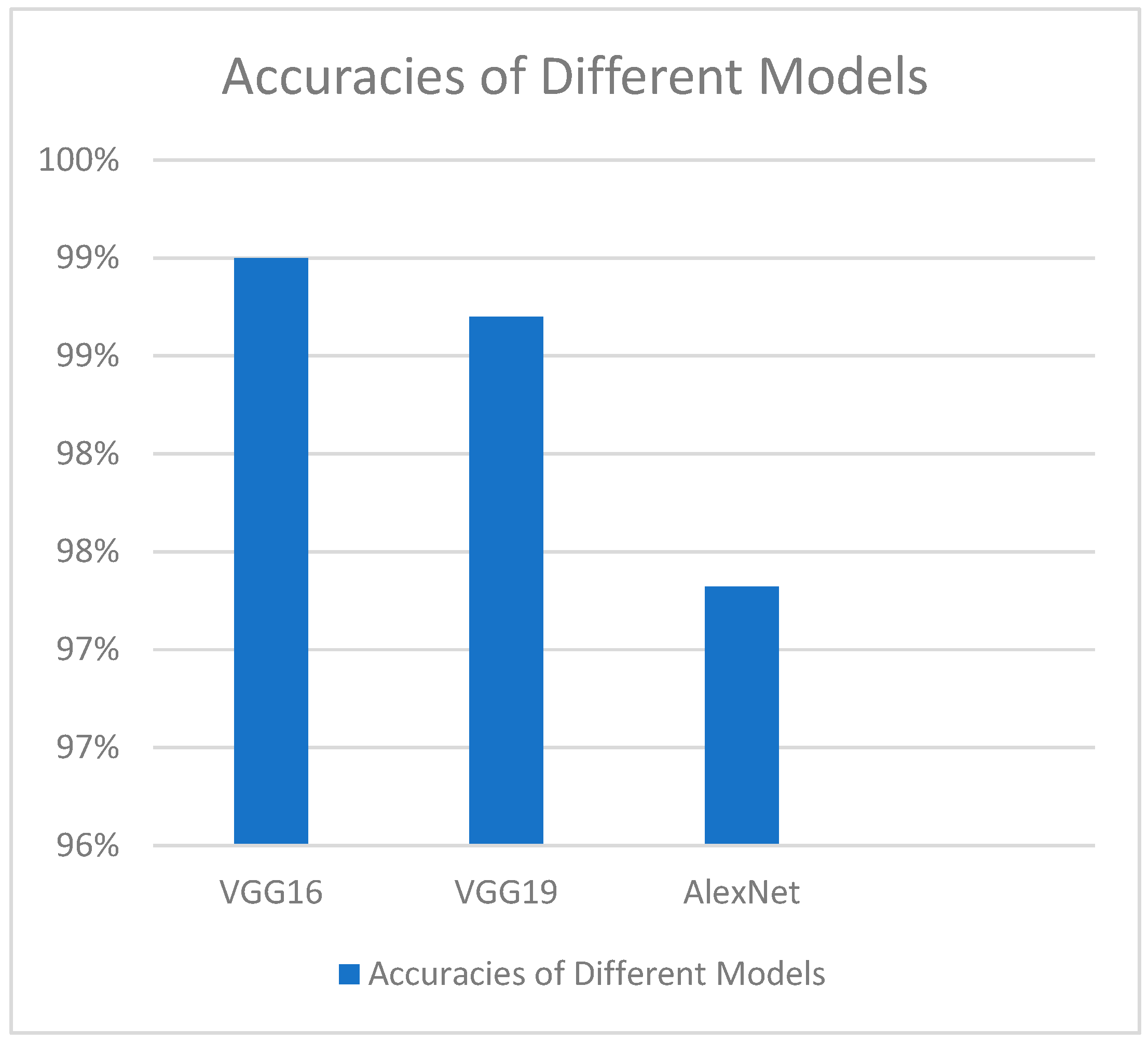
4.2. Comparison of VGG-16 Model Performance with Other Models
5. Conclusions and Future Work
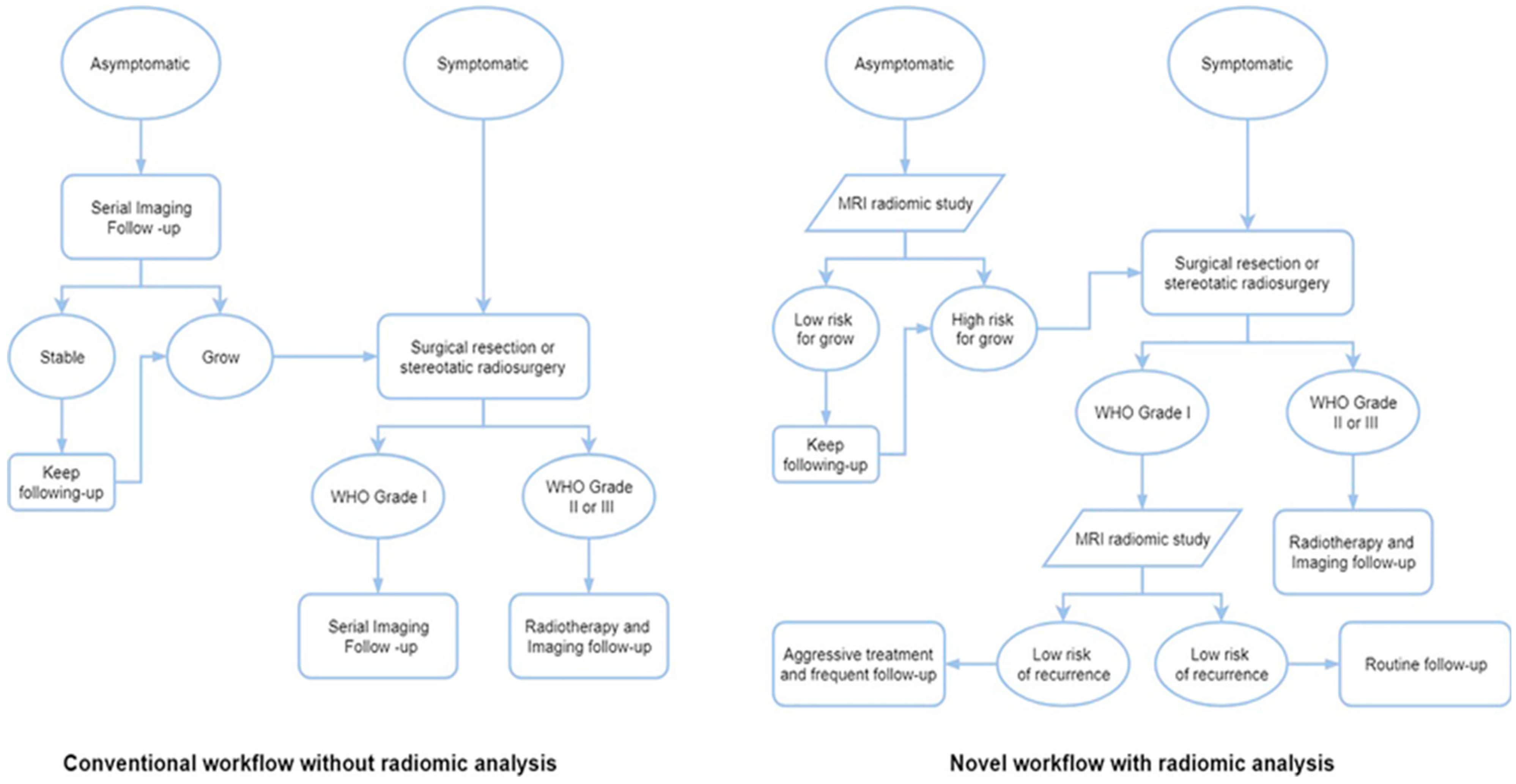
5.1. Integration of Output of This Very Deep Transfer CNN Based Real-Time Meningioma Detection Methodology Within the Clinico-Radiomics Workflow
5.2. Limitations of Our Study
Computational Complexity of the Model: Trade-Offs Between Accuracy and Computational Demands
5.3. Potential Future Research Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WHO CNS5 | The 2021 World Health Organization Classification of Tumors of the Central Nervous System, is an update to the previous 2016 classification. It incorporates findings from the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy (cIMPACT-NOW) and emphasizes the role of genetic and molecular changes in tumor characteristics. This fifth edition includes new tumor types, revised nomenclature, and refined grading systems |
| sMRI | structural Magnetic Resonance Imaging is a non-invasive imaging technique for examining the anatomy and morphological pathology of the brain, |
| CUDA | Compute Unified Device Architecture is a parallel computing platform and application programming interface (API) developed by NVIDIA for general computing on Graphical Processing Units (GPUs) with dramatic escalation of computing application speeds, |
| CNN | Convolutional Neural Network is an advanced version of artificial neural networks (ANNs), primarily designed to extract features from grid-like matrix datasets. This is particularly useful for visual datasets such as images or videos, where data patterns play a crucial role, |
| FLAIR | Fluid-Attenuated Inversion Recovery MRI highlights regions of tissue with T2 prolongation while suppressing (darkening) the signal from cerebrospinal fluid (CSF). This makes it easier to see brain lesions, particularly in locations near CSF. The goal of FLAIR MRI is to reduce the strong signal from CSF, which can mask mild aberrations in the brain, particularly in regions close to the brain surface and the periventricular region (around the ventricles). It effectively nullifies the CSF signal by using a unique inversion recovery pulse sequence with a long inversion time (TI). In order to generate strong T2 weighting, which identifies regions of tissue T2 prolongation (bright signal), it also uses a long echo duration (TE), |
| CSF | Cerebrospinal Fluid which is a clear, colorless fluid that surrounds the brain and spinal cord, acting as a cushion and providing nutrients and waste removal, |
| GBM | Glioblastoma multiforme, also known as glioblastoma, is the most common and aggressive type of primary brain tumor in adults, |
| ReLU | Rectified Linear Unit is one of the most popular activation functions for artificial neural networks, and finds application in biomedical image processing, computer vision and speech recognition using deep neural nets and computational neuroscience, |
| DNN | Deep neural networks are a type of artificial neural network with multiple hidden layers, which makes them more complex and resource-intensive compared to conventional neural networks. They are used for various applications and work best with GPU-based architectures for faster training times, |
| ADAM | Adaptive Moment Estimation optimizer is an adaptive learning rate algorithm designed to improve training speeds in deep neural networks and reach convergence quickly. It customizes each parameter’s learning rate based on its gradient history, and this adjustment helps the neural network learn efficiently as a whole, |
| RNN | Recurrent Neural Network is a type of deep learning model specifically designed to process sequential data like text, speech, or time series, |
| SVM | A Support Vector Machine is a supervised machine learning algorithm used for both classification and regression tasks. It works by finding the optimal hyperplane that separates data points into different classes, maximizing the margin between them. SVMs are particularly effective for binary classification and can handle both linear and non-linear data using kernel functions, |
| VGG-16 | refering to Visual Geometry Group of the University of Oxford, is a convolutional neural network (CNN) model primarily used for image classification and object recognition. It’s known for its simplicity and effectiveness, making it a foundational model in the field of computer vision. The “16” in VGG16 refers to the number of layers in the network that have learnable parameters, including convolutional and fully connected layers, |
| DLR | Deep Learning Radiomics is a fusion of deep learning and radiomics which are powerful techniques for extracting and analyzing quantitative features from medical images, enabling precision imaging in various applications. These techniques can help in diagnosis, prognosis, and treatment planning by identifying patterns and biomarkers that might not be readily apparent to the human eye. |
References
- Soni, N.; Ora, M.; Bathla, G.; Szekeres, D.; Desai, A.; Pillai, J.J.; Agarwal, A. Meningioma: Molecular Updates from the 2021 World Health Organization Classification of CNS Tumors and Imaging Correlates. Am. J. Neuroradiol. 2025, 46, 240–250. [Google Scholar] [CrossRef]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classification of Central Nervous System tumours: A practical update on what neurosurgeons need to know-a minireview. Acta Neurochir. 2022, 164, 2453–2464. [Google Scholar] [CrossRef]
- Baumgarten, P.; Gessler, F.; Schittenhelm, J.; Skardelly, M.; Tews, D.S.; Senft, C.; Dunst, M.; Imoehl, L.; Plate, K.H.; Wagner, M.; et al. Brain invasion in otherwise benign meningiomas does not predict tumor recurrence. Acta Neuropathol. 2016, 132, 479–481. [Google Scholar] [CrossRef]
- Gritsch, S.; Batchelor, T.T.; Gonzalez Castro, L.N. Diagnostic, therapeutic, and prognostic implications of the 2021 World Health Organization classification of tumors of the central nervous system. Cancer 2022, 128, 47–58. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Rogers, C.L.; Perry, A.; Pugh, S.; Vogelbaum, M.A.; Brachman, D.; McMillan, W.; Jenrette, J.; Barani, I.; Shrieve, D.; Sloan, A.; et al. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOG Trial 0539. Neuro Oncol. 2016, 18, 565–574. [Google Scholar] [CrossRef]
- Goldbrunner, R.; Stavrinou, P.; Jenkinson, M.D.; Sahm, F.; Mawrin, C.; Weber, D.C.; Preusser, M.; Minniti, G.; Lund-Johansen, M.; Lefranc, F.; et al. EANO guideline on the diagnosis and management of meningiomas. Neuro Oncol. 2021, 23, 1821–1834. [Google Scholar] [CrossRef]
- Harter, P.N.; Braun, Y.; Plate, K.H. Classification of meningiomas-advances and controversies. Chin. Clin. Oncol. 2017, 6 (Suppl. S1), S2. [Google Scholar] [CrossRef]
- Maas, S.L.; Stichel, D.; Hielscher, T.; Sievers, P.; Berghoff, A.S.; Schrimpf, D.; Sill, M.; Euskirchen, P.; Blume, C.; Patel, A.; et al. Integrated molecular-morphologic meningioma classification: A multicenter retrospective analysis, retrospectively and prospectively validated. J. Clin. Oncol. 2021, 39, 3839–3852. [Google Scholar] [CrossRef]
- Trybula, S.J.; Youngblood, M.W.; Karras, C.L.; Murthy, N.K.; Heimberger, A.B.; Lukas, R.V.; Sachdev, S.; Kalapurakal, J.A.; Chandler, J.P.; Brat, D.J.; et al. The evolving classification of meningiomas: Integration of Molecular discoveries to inform patient care. Cancers 2024, 16, 1753. [Google Scholar] [CrossRef]
- Wang, E.J.; Haddad, A.F.; Young, J.S.; Morshed, R.A.; Wu, J.P.; Salha, D.M.; Butowski, N.; Aghi, M.K. Recent advances in the molecular prognostication of meningiomas. Front. Oncol. 2023, 12, 910199. [Google Scholar] [CrossRef] [PubMed]
- Birzu, C.; Peyre, M.; Sahm, F. Molecular alterations in meningioma: Prognostic and therapeutic perspectives. Curr. Opin. Oncol. 2020, 32, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Gauchotte, G.; Peyre, M.; Pouget, C.; Cazals-Hatem, D.; Polivka, M.; Rech, F.; Varlet, P.; Loiseau, H.; Lacomme, S.; Mokhtari, K.; et al. Prognostic value of histopathological features and loss of H3K27me3 immunolabeling in anaplastic meningioma: A multicenter retrospective study. J. Neuropathol. Exp. Neurol. 2020, 79, 754–762. [Google Scholar] [CrossRef]
- Sahm, F.; Schrimpf, D.; Stichel, D.; Jones, D.T.W.; Hielscher, T.; Schefzyk, S.; Okonechnikov, K.; Koelsche, C.; Reuss, D.E.; Capper, D.; et al. DNA methylation-based classification and grading system for meningioma: A multicentre, retrospective analysis. Lancet Oncol. 2017, 18, 682–694. [Google Scholar] [CrossRef]
- Nassiri, F.; Liu, J.; Patil, V.; Mamatjan, Y.; Wang, J.Z.; Hugh-White, R.; Macklin, A.M.; Khan, S.; Singh, O.; Karimi, S.; et al. A clinically applicable integrative molecular classification of meningiomas. Nature 2021, 597, 119–125. [Google Scholar] [CrossRef]
- Buerki, R.A.; Horbinski, C.M.; Kruser, T.; Horowitz, P.M.; James, C.D.; Lukas, R.V. An overview of meningiomas. Future Oncol. 2018, 14, 2161–2177. [Google Scholar] [CrossRef]
- Nowosielski, M.; Galldiks, N.; Iglseder, S.; Kickingereder, P.; Von Deimling, A.; Bendszus, M.; Wick, W.; Sahm, F. Diagnostic challenges in meningioma. Neuro-Oncology 2017, 19, 1588–1598. [Google Scholar] [CrossRef]
- Maggio, I.; Franceschi, E.; Tosoni, A.; Nunno, V.D.; Gatto, L.; Lodi, R.; Brandes, A.A. Meningioma: Not always a benign tumor. A review of advances in the treatment of meningiomas. CNS Oncol. 2021, 10, CNS72. [Google Scholar] [CrossRef]
- Qureshi, S.A.; Raza, S.E.; Hussain, L.; Malibari, A.A.; Nour, M.K.; Rehman, A.U.; Al-Wesabi, F.N.; Hilal, A.M. Intelligent ultra-light deep learning model for multi-class brain tumor detection. Appl. Sci. 2022, 12, 3715. [Google Scholar] [CrossRef]
- Qureshi, S.A.; Hussain, L.; Ibrar, U.; Alabdulkreem, E.; Nour, M.K.; Alqahtani, M.S.; Nafie, F.M.; Mohamed, A.; Mohammed, G.P.; Duong, T.Q. Radiogenomic classification for MGMT promoter methylation status using multi-omics fused feature space for least invasive diagnosis through mpMRI scans. Sci. Rep. 2023, 13, 3291. [Google Scholar]
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in brain tumor: Image assessment, quantitative feature descriptors, and machine-learning approaches. Am. J. Neuroradiol. 2018, 39, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Mitra, S.; Shankar, B.U. Synergetic neuro-fuzzy feature selection and classification of brain tumors. In Proceedings of the 2017 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), Naples, Italy, 9–12 July 2017; IEEE: Piscataway, NJ, USA; pp. 1–6. [Google Scholar]
- De Coene, B.; Hajnal, J.V.; Gatehouse, P.; Longmore, D.B.; White, S.J.; Oatridge, A.; Pennock, J.M.; Young, I.R.; Bydder, G.M. MR of the brain using fluid-attenuated inversion recovery (FLAIR) pulse sequences. AJNR Am. J. Neuroradiol. 1992, 13, 1555–1564. [Google Scholar]
- Rydberg, J.N.; Hammond, C.A.; Grimm, R.C.; Erickson, B.J.; Jack, C.R., Jr.; Huston, J., 3rd; Riederer, S.J. Initial clinical experience in MR imaging of the brain with a fast fluid-attenuated inversion recovery pulse sequence. Radiology 1994, 193, 173–180. [Google Scholar] [CrossRef]
- Hajnal, J.V.; Bryant, D.J.; Kasuboski, L.; Pattany, P.M.; De Coene, B.; Lewis, P.D.; Pennock, J.M.; Oatridge, A.; Young, I.R.; Bydder, G.M. Use of fluid attenuated inversion recovery (FLAIR) pulse sequences in MRI of the brain. J. Comput. Assist. Tomogr. 1992, 16, 841–844. [Google Scholar] [CrossRef]
- Getachew, A.; Senait, A.; Tadios, M. Meningiomas: Clinical correlates, skull x-ray, CT and pathological evaluations. Ethiop. Med. J. 2006, 44, 263–267. [Google Scholar]
- Yan, P.F.; Yan, L.; Zhang, Z.; Salim, A.; Wang, L.; Hu, T.T.; Zhao, H.Y. Accuracy of conventional MRI for preoperative diagnosis of intracranial tumors: A retrospective cohort study of 762 cases. Int. J. Surg. 2016, 36, 109–117. [Google Scholar] [CrossRef]
- Ugga, L.; Spadarella, G.; Pinto, L.; Cuocolo, R.; Brunetti, A. Meningioma radiomics: At the nexus of imaging, pathology and biomolecular characterization. Cancers 2022, 14, 2605. [Google Scholar] [CrossRef]
- Yang, L.; Xu, P.; Zhang, Y.; Cui, N.; Wang, M.; Peng, M.; Gao, C.; Wang, T. A deep learning radiomics model may help to improve the prediction performance of preoperative grading in meningioma. Neuroradiology 2022, 64, 1373–1382. [Google Scholar] [CrossRef]
- Shedbalkar, J.; Prabhushetty, D.K. Convolutional Neural Network based Classification of Brain Tumors images: An Approach. Int. J. Eng. Res. Technol. 2021, 10, 639–644. [Google Scholar]
- Itri, J.N.; Tappouni, R.R.; McEachern, R.O.; Pesch, A.J.; Patel, S.H. Fundamentals of diagnostic error in imaging. Radiographics 2018, 38, 1845–1865. [Google Scholar] [CrossRef]
- Maskell, G. Error in radiology—Where are we now? Br. J. Radiol. 2019, 92, 20180845. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Gnocchi, G.; Quarrella, C.; Sorce, A.; Nicosia, L.; Mariano, L.; Bozzini, A.C.; Marinucci, I.; Priolo, F.; Abbate, F.; et al. Errors in Radiology: A Standard Review. J. Clin. Med. 2024, 13, 4306. [Google Scholar] [CrossRef] [PubMed]
- Histed, S.N.; Lindenberg, M.L.; Mena, E.; Turkbey, B.; Choyke, P.L.; Kurdziel, K.A. Review of functional/anatomical imaging in oncology. Nucl. Med. Commun. 2012, 33, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Hricak, H.; Abdel-Wahab, M.; Atun, R.; Lette, M.M.; Paez, D.; Brink, J.A.; Donoso-Bach, L.; Frija, G.; Hierath, M.; Holmberg, O.; et al. Medical imaging and nuclear medicine: A Lancet Oncology Commission. Lancet Oncol. 2021, 22, e136–e172. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-“how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Castellano, G.; Bonilha, L.; Li, L.M.; Cendes, F. Texture analysis of medical images. Clin. Radiol. 2004, 59, 1061–1069. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H.R. Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Zanfardino, M.; Franzese, M.; Pane, K.; Cavaliere, C.; Monti, S.; Esposito, G.; Salvatore, M.; Aiello, M. Bringing radiomics into a multiomics framework for a comprehensive genotype–phenotype characterization of oncological diseases. J. Transl. Med. 2019, 17, 337. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The applications of radiomics in precision diagnosis and treatment of oncology: Opportunities and challenges. Theranostics 2019, 9, 1303–1322. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21 (Suppl. S5), v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.; Barani, I.; Chamberlain, M.; Kaley, T.J.; McDermott, M.; Raizer, J.; Schiff, D.; Weber, D.C.; Wen, P.Y.; Vogelbaum, M.A. Meningiomas: Knowledge base, treatment outcomes, and uncertainties. A RANO review. J. Neurosurg. 2015, 122, 4–23. [Google Scholar] [CrossRef] [PubMed]
- van Alkemade, H.; de Leau, M.; Dieleman, E.M.; Kardaun, J.W.; van Os, R.; Vandertop, W.P.; van Furth, W.R.; Stalpers, L.J. Impaired survival and long-term neurological problems in benign meningioma. Neuro Oncol. 2012, 14, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Pettersson-Segerlind, J.; Orrego, A.; Lönn, S.; Mathiesen, T. Long-term 25-year follow-up of surgically treated parasagittal meningiomas. World Neurosurg. 2011, 76, 564–571. [Google Scholar] [CrossRef]
- Lagman, C.; Bhatt, N.S.; Lee, S.J.; Bui, T.T.; Chung, L.K.; Voth, B.L.; Barnette, N.E.; Pouratian, N.; Lee, P.; Selch, M.; et al. Adjuvant radiosurgery versus serial surveillance following subtotal resection of atypical meningioma: A systematic analysis. World Neurosurg. 2017, 98, 339–346. [Google Scholar] [CrossRef]
- Stessin, A.M.; Schwartz, A.; Judanin, G.; Pannullo, S.C.; Boockvar, J.A.; Schwartz, T.H.; Stieg, P.E.; Wernicke, A.G. Does adjuvant external beam radiotherapy improve outcomes for nonbenign meningiomas? A surveillance, epidemiology, and end results (SEER)–based analysis. J. Neurosurg. 2012, 117, 669–675. [Google Scholar] [CrossRef]
- Riemenschneider, M.J.; Perry, A.; Reifenberger, G. Histological classification and molecular genetics of meningiomas. Lancet Neurol. 2006, 5, 1045–1054. [Google Scholar] [CrossRef]
- Bayley, J.C.; Hadley, C.C.; Harmanci, A.O.; Harmanci, A.S.; Klisch, T.J.; Patel, A.J. Multiple approaches converge on three biological subtypes of meningioma and extract new insights from published studies. Sci. Adv. 2022, 8, eabm6247. [Google Scholar] [CrossRef]
- Driver, J.; Hoffman, S.E.; Tavakol, S.; Woodward, E.; Maury, E.A.; Bhave, V.; Greenwald, N.F.; Nassiri, F.; Aldape, K.; Zadeh, G.; et al. A molecularly integrated grade for meningioma. Neuro Oncol. 2022, 24, 796–808. [Google Scholar] [CrossRef]
- Magill, S.T.; Vasudevan, H.N.; Seo, K.; Villanueva-Meyer, J.E.; Choudhury, A.; Liu, S.J.; Pekmezci, M.; Findakly, S.; Hilz, S.; Lastella, S.; et al. Multiplatform genomic profiling and magnetic Resonance imaging identify mechanisms underlying intratumor heterogeneity in meningioma. Nat. Commun. 2020, 11, 4803. [Google Scholar] [CrossRef]
- Horbinski, C.; Ligon, K.L.; Brastianos, P.; Huse, J.T.; Venere, M.; Chang, S.; Buckner, J.; Cloughesy, T.; Jenkins, R.B.; Giannini, C.; et al. The medical necessity of advanced molecular testing in the diagnosis and treatment of brain tumor patients. Neuro Oncol. 2019, 21, 1498–1508. [Google Scholar] [CrossRef] [PubMed]
- Aldape, K.; Brindle, K.M.; Chesler, L.; Chopra, R.; Gajjar, A.; Gilbert, M.R.; Gottardo, N.; Gutmann, D.H.; Hargrave, D.; Holland, E.C.; et al. Challenges to curing primary brain tumours. Nat. Rev. Clin. Oncol. 2019, 16, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Bhandari, A.; Koppen, J.; Agzarian, M. Convolutional neural networks for brain tumour segmentation. Insights Imaging 2020, 11, 77. [Google Scholar] [CrossRef]
- Liu, Z.; Tong, L.; Chen, L.; Jiang, Z.; Zhou, F.; Zhang, Q.; Zhang, X.; Jin, Y.; Zhou, H. Deep learning based brain tumor segmentation: A survey. Complex. Intell. Syst. 2023, 9, 1001–1026. [Google Scholar] [CrossRef]
- Anwar, R.W.; Abrar, M.; Ullah, F. Transfer Learning in Brain Tumor Classification: Challenges, Opportunities, and Future Prospects. In Proceedings of the 2023 14th International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Republic of Korea, 11–13 October 2023; IEEE: Piscataway, NJ, USA; pp. 24–29. [Google Scholar] [CrossRef]
- Ullah, F.; Nadeem, M.; Abrar, M.; Al-Razgan, M.; Alfakih, T.; Amin, F.; Salam, A. Brain Tumor Segmentation from MRI Images Using Handcrafted Convolutional Neural Network. Diagnostics 2023, 13, 2650. [Google Scholar] [CrossRef]
- Ullah, F.; Nadeem, M.; Abrar, M. Revolutionizing Brain Tumor Segmentation in MRI with Dynamic Fusion of Handcrafted Features and Global Pathway-based Deep Learning. KSII Trans. Internet Inf. Syst. 2024, 18, 105–125. [Google Scholar] [CrossRef]
- Ullah, F.; Nadeem, M.; Abrar, M.; Amin, F.; Salam, A.; Khan, S. Enhancing brain tumor segmentation accuracy through scalable federated learning with advanced data privacy and security measures. Mathematics 2023, 11, 4189. [Google Scholar] [CrossRef]
- Irmak, E. Multi-classification of brain tumor MRI images using deep convolutional neural network with fully optimized framework. Iran. J. Sci. Technol. Trans. Electr. Eng. 2021, 45, 1015–1036. [Google Scholar] [CrossRef]
- Ullah, F.; Nadeem, M.; Abrar, M.; Amin, F.; Salam, A.; Alabrah, A.; AlSalman, H. Evolutionary Model for Brain Cancer-Grading and Classification. IEEE Access 2023, 11, 126182–126194. [Google Scholar] [CrossRef]
- Abuqadumah, M.M.; Ali, M.A.; Abd Almisreb, A.; Durakovic, B. Deep transfer learning for human identification based on footprint: A comparative study. Period. Eng. Nat. Sci. 2019, 7, 1300–1307. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A deep learning-based framework for automatic brain tumors classification using transfer learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]
- Rao, D.D.; Ramana, K.V. Accelerating training of deep neural networks on GPU using CUDA. Int. J. Intell. Syst. Appl. 2019, 11, 18. [Google Scholar] [CrossRef]
- Patel, R.V.; Yao, S.; Huang, R.Y.; Bi, W.L. Application of radiomics to meningiomas: A systematic review. Neuro-Oncology 2023, 25, 1166. [Google Scholar] [CrossRef]
- Du, X.; Sun, Y.; Song, Y.; Sun, H.; Yang, L. A comparative study of different CNN models and transfer learning effect for underwater object classification in side-scan sonar images. Remote Sens. 2023, 15, 593. [Google Scholar] [CrossRef]
- Gikunda, P.K.; Jouandeau, N. State-of-the-Art Convolutional Neural Networks for Smart Farms: A Review. In Intelligent Computing. CompCom 2019. Advances in Intelligent Systems and Computing; Arai, K., Bhatia, R., Kapoor, S., Eds.; Springer: Cham, Switzerland, 2019; Volume 997. [Google Scholar] [CrossRef]
- Gikunda, P.K.; Jouandeau, N. Modern CNNs for IoT based farms. In Proceedings of the Information and Communication Technology for Development for Africa: Second International Conference, ICT4DA 2019, Bahir Dar, Ethiopia, 28–30 May 2019; Revised Selected Papers. 2. Springer International Publishing: Cham, Switzerland, 2019; pp. 68–79. [Google Scholar]
- Kora, P.; Ooi, C.P.; Faust, O.; Raghavendra, U.; Gudigar, A.; Chan, W.Y.; Meenakshi, K.; Swaraja, K.; Plawiak, P.; Acharya, U.R. Transfer learning techniques for medical image analysis: A review. Biocybern. Biomed. Eng. 2022, 42, 79–107. [Google Scholar] [CrossRef]
- Khanam, N.; Kumar, R. Recent applications of artificial intelligence in early cancer detection. Curr. Med. Chem. 2022, 29, 4410–4435. [Google Scholar] [CrossRef]
- Singh, A.; Arjunaditya; Tripathy, B.K. Detection of Cancer Using Deep Learning Techniques. In Deep Learning Applications in Image Analysis; Springer Nature: Singapore, 2023; pp. 187–210. [Google Scholar]
- Mall, P.K.; Singh, P.K.; Srivastav, S.; Narayan, V.; Paprzycki, M.; Jaworska, T.; Ganzha, M. A comprehensive review of deep neural networks for medical image processing: Recent developments and future opportunities. Healthc. Anal. 2023, 4, 100216. [Google Scholar] [CrossRef]
- Panyaping, T.; Punpichet, M.; Tunlayadechanont, P.; Tritanon, O. Usefulness of a rim-enhancing pattern on the contrast-enhanced 3D-FLAIR sequence and MRI characteristics for distinguishing meningioma and malignant dural-based tumor. Am. J. Neuroradiol. 2023, 44, 247–253. [Google Scholar] [CrossRef]
- Oner, A.Y.; Tokgöz, N.; Tali, E.T.; Uzun, M.; Isik, S. Imaging meningiomas: Is there a need for post-contrast FLAIR? Clin. Radiol. 2005, 60, 1300–1305. [Google Scholar] [CrossRef]
- Amoo, M.; Henry, J.; Farrell, M.; Javadpour, M. Meningioma in the elderly. Neuro-Oncol. Adv. 2023, 5 (Suppl. S1), i13–i25. [Google Scholar] [CrossRef] [PubMed]
- Baldi, I.; Engelhardt, J.; Bonnet, C.; Bauchet, L.; Berteaud, E.; Grüber, A.; Loiseau, H. Epidemiology of meningiomas. Neurochirurgie 2018, 64, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Chinga, A.; Bendezu, W.; Angulo, A. Comparative Study of CNN Architectures for Brain Tumor Classification Using MRI: Exploring GradCAM for Visualizing CNN Focus. Eng. Proc. 2025, 83, 22. [Google Scholar] [CrossRef]
- Reyes, D.; Sánchez, J. Performance of convolutional neural networks for the classification of brain tumors using magnetic resonance imaging. Heliyon 2024, 10, e25468. [Google Scholar] [CrossRef]
- Gayathri, P.; Dhavileswarapu, A.; Ibrahim, S.; Paul, R.; Gupta, R. Exploring the potential of VGG-16 architecture for accurate brain tumor detection using deep learning. J. Comput. Mech. Manag. 2023, 2, 13–22. [Google Scholar] [CrossRef]
- Hapsari, P.A.; Dewinda, J.R.; Cucun, V.A.; Nurul, Z.F.; Joan, S.; Anggraini, D.S.; Peter, M.A.; IKetut, E.P.; Mauridhi, H.P. Brain tumor classification in MRI images using en-CNN. Int. J. Intell. Eng. Syst. 2021, 14, 437–451. [Google Scholar]
- Younis, A.; Qiang, L.; Nyatega, C.O.; Adamu, M.J.; Kawuwa, H.B. Brain tumor analysis using deep learning and VGG-16 ensembling learning approaches. Appl. Sci. 2022, 12, 7282. [Google Scholar] [CrossRef]
- Gu, H.; Zhang, X.; Di Russo, P.; Zhao, X.; Xu, T. The current state of radiomics for meningiomas: Promises and challenges. Front. Oncol. 2020, 10, 567736. [Google Scholar] [CrossRef]
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging imaging and genomics. Abdom Radiol. 2019, 44, 1960–1984. [Google Scholar] [CrossRef]
- Court, L.E.; Fave, X.; Mackin, D.; Lee, J.; Yang, J.; Zhang, L. Computational resources for radiomics. Trans. Cancer Res. 2016, 5, 340–348. [Google Scholar] [CrossRef]
- Bang, J.-I.; Ha, S.; Kang, S.-B.; Lee, K.-W.; Lee, H.-S.; Kim, J.-S.; Oh, H.-K.; Lee, H.-Y.; Kim, S.E. Prediction of neoadjuvant radiation chemotherapy response and survival using pretreatment [(18)F]FDG PET/CT scans in locally advanced rectal cancer. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Warfield, S.K.; Zou, K.H.; Wells, W.M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 2004, 23, 903–921. [Google Scholar] [CrossRef] [PubMed]
- Alippi, C.; Disabato, S.; Roveri, M. Moving convolutional neural networks to embedded systems: The AlexNet and VGG-16 case. In Proceedings of the 2018 17th ACM/IEEE International Conference on Information Processing in Sensor Networks (IPSN), Porto, Portugal, 11–13 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 212–223. [Google Scholar]
- Gui, Y.; Zhang, J. Research Progress of Artificial Intelligence in the Grading and Classification of Meningiomas. Acad. Radiol. 2024, 31, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
| Histological Type | Histological Malignancy Grade |
|---|---|
| Meningothelial meningioma | 1/2 |
| Fibrous meningioma | 1/2 |
| Transitional meningioma | 1/2 |
| Psammomatous meningioma | 1/2 |
| Angiomatous meningioma | 1/2 |
| Microcystic meningioma | 1/2 |
| Secretory meningioma | 1/2 |
| Lymphoplasmacyte-rich meningioma | 1/2 |
| Atypical meningioma (including brain infiltrative meningiomas) | 2 |
| Chordoid meningioma | 2 |
| Clear cell meningioma | 2 |
| Anaplastic (malignant) meningioma | 3 |
| Genetic Alteration | Clinicopathological Significance |
|---|---|
| NF2 mutation | Convexity meningiomas, fibrous, and transitional subtypes, more often CNS WHO grade 2/3 |
| TRAF7 mutations | Secretory subtype |
| TERT promotor mutation | CNS WHO grade 3 |
| SMARCE1 mutation | Clear cell subtype |
| BAP1 mutation | Rhabdoid and papillary subtypes |
| CDKNA2A/B loss | CNS WHO grade 3 |
| H3K27me3 loss | Increased risk of recurrence |
| DNA methylation profiling | Methylation classes associated with increased risk of recurrence |
| Model | Accuracy |
|---|---|
| VGG-16 (our model) | 99% |
| EasyDL | 96.6% |
| GoogLeNet | 92.54% |
| GrayNet | 95% |
| ImageNet | 91% |
| CNN | 96% |
| Multivariable Regression and Neural Network | 95% |
| Test Case No. | Prediction Result |
|---|---|
| 1 | Meningioma |
| 2 | Meningioma |
| 3 | Meningioma |
| 4 | Meningioma |
| 5 | Meningioma |
| 6 | Meningioma |
| 7 | Normal |
| 8 | Normal |
| 9 | Normal |
| 10 | Normal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, D.; Sarkar, C.; Das, B. Real-Time Detection of Meningiomas by Image Segmentation: A Very Deep Transfer Learning Convolutional Neural Network Approach. Tomography 2025, 11, 50. https://doi.org/10.3390/tomography11050050
Das D, Sarkar C, Das B. Real-Time Detection of Meningiomas by Image Segmentation: A Very Deep Transfer Learning Convolutional Neural Network Approach. Tomography. 2025; 11(5):50. https://doi.org/10.3390/tomography11050050
Chicago/Turabian StyleDas, Debasmita, Chayna Sarkar, and Biswadeep Das. 2025. "Real-Time Detection of Meningiomas by Image Segmentation: A Very Deep Transfer Learning Convolutional Neural Network Approach" Tomography 11, no. 5: 50. https://doi.org/10.3390/tomography11050050
APA StyleDas, D., Sarkar, C., & Das, B. (2025). Real-Time Detection of Meningiomas by Image Segmentation: A Very Deep Transfer Learning Convolutional Neural Network Approach. Tomography, 11(5), 50. https://doi.org/10.3390/tomography11050050







