Abstract
Reaction of bis(2-picolyl)amine (BPA) with Ni(II) salt yielded [(BPA)NiCl2(H2O)] (NiBPA). The Ni(II) in NiBPA bound to a BPA ligand, two chloride, and one aqua ligands. Because most medications inhibit biological processes by binding to a specific protein, the stopped-flow technique was used to investigate DNA/protein binding in-vitro, and a mechanism was proposed. NiBPA binds to DNA/protein more strongly than BPA via a static quenching mechanism. Using the stopped-flow technique, a mechanism was proposed. BSA interacts with BPA via a fast reversible step followed by a slow irreversible step, whereas NiBPA interacts via two reversible steps. DNA, on the other hand, binds to BPA and NiBPA via the same mechanism through two reversible steps. Although BSA interacts with NiBPA much faster, NiBPA has a much higher affinity for DNA (2077 M) than BSA (30.3 M). Compared to NiBPA, BPA was found to form a more stable BSA complex. When BPA and NiBPA bind to DNA, the Ni(II) center was found to influence the rate but not the mechanism, whereas, for BSA, the Ni(II) center was found to change both the mechanism and the rate. Additionally, NiBPA exhibited significant cytotoxicity and antibacterial activity, which is consistent with the binding constants but not the kinetic stability. This shows that in our situation, biological activity is significantly more influenced by binding constants than by kinetic stability. Due to its selectivity and cytotoxic activity, complex NiBPA is anticipated to be used in medicine.
1. Introduction
Metal complexes with the ability to bind and react with specific DNA locations under physiological conditions are of great interest in the development of anticancer drugs [1]. By attaching to and cleaving DNA, metal complexes inhibit tumor growth [2]. Manufactured nucleases have lower molecular weights and are less reactive under certain conditions than natural DNA-binding enzymes [3,4]. Furthermore, certain transition metal compounds are being studied for therapeutic purposes [5,6]. Nickel complexes have been found to play an important role in the field of metal-based chemotherapeutics due to their biocompatibility [7]. Nickel is also an essential component of life, as it can be found in a variety of enzymes [8]. Nickel (II) complexes, on the other hand, are thought to be one of the most promising anticancer drug alternatives to cisplatin [9,10,11]. The precise mechanism of anticancer activity, on the other hand, is still unknown. In general, clinically approved anticancer treatments are chemicals that either directly damage DNA or indirectly block DNA synthesis by inhibiting nucleic acid precursor production. They have the potential to interfere with the hormonal stimulation of cell growth [12,13]. As a result, considerable efforts are being extended to investigate the interaction of nickel(II) complexes with DNA and their cytotoxic effects [14,15,16,17,18]. Although research has been conducted to better understand the interactions of metal complexes, metal ions, and Schiff base ligands with albumins, it appears that research on nickel(II) Schiff base complexes is limited [19,20,21].
Polypyridyl ligands are very interesting in biological and materials science [22,23,24,25,26,27,28,29]. Tridentate ligands with two identical terminal coordination sites, such as bis(2-picolyl)amine (BPA), are being studied as building blocks in a variety of areas, including chemosensors, catalysts, and biomedicinal applications [30,31,32,33,34,35]. BPA and its secondary nitrogen-substituted derivatives develop stronger complexes with nickel(II), showing the coordination versatility of such ligands and their significance in a variety of biological, catalytic, and magnetic aspects [36,37,38,39,40]. Numerous authors have also explored the use of Ni(II) complexes based on BPA for the cleavage of DNA or RNA [14]. On the other hand, drug interactions at the protein binding level have a significant impact on the apparent distribution volume and elimination rate. As a result, the interactions of metal complexes with serum albumins have received a great deal of attention in the scientific community, with researchers focusing on antitumor, metallopharmaceutical, pharmacokinetics, and structure–activity relationships [41].
BSA is the most extensively researched serum albumin due to its structural similarity to human serum albumin (HSA). This finding emphasizes the importance of studying the interaction behavior of metal complexes with BSA while evaluating their anticancer properties. Our journey with detailed mechanistic studies on DNA/protein binding recently began with the challenge of exploring this type of interaction via related systems while also attempting to answer the question, is the metal center important for binding? [42,43,44,45]. It was discovered that a metal center not only increases ligand affinity to DNA/protein but also alters the binding mechanism. When a ligand and its Ti(IV) complex were mixed with chitosan nanoparticles, the Ti(IV) center was found to slow the affinity but not change the mechanism. These findings suggest that the metal center is not always necessary for binding.
Based on these intriguing findings, and because of the role and activity of nickel and its complexes in biological systems [46], as well as the importance of bis(2-picolyl)amine (BPA) in medicine, we describe the synthesis and characterization of [(BPA)NiCl2(H2O)] (NiBPA). Furthermore, a detailed kinetic investigation of the interaction of BPA and its Ni(II) complex (NiBPA) with ct-DNA/protein has been performed using UV/vis, fluorescence, and the stopped-flow technique, and a mechanism was reported. To evaluate the antibacterial activities of both compounds, Escherichia coli, Salmonella choleraesuis, Staphylococcus aureus, and Proteus mirabilis bacteria strains were chosen. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of the compounds against these bacteria were tested in vitro. Additionally, the cytotoxicity against HepG2 human liver cancer cells is reported, and the results are correlated with the binding constants.
2. Experimental
2.1. Materials and General Methods
All of the chemicals used were of the highest analytical grade. Merck Chemical Company provided the ligand BPA, which was used without further purification. The IR spectra were captured using a Bruker Alpha-Atunated FT-IR Spectrophotometer in the 400–4000 cm−1 range. Shimadzu spectrophotometer UV-240 was used to obtain electronic absorption spectra in a methanolic solution. At room temperature, magnetic moments were measured using Gouy’s method.
2.2. Synthesis of the NiBPA
At 60 °C, a solution of Ni(II)Cl2 (0.13 g, 1.0 mmol) in ethanol (20 mL) was heated, and a solution of BPA (0.2 g, 1.0 mmol) in ethanol (20 mL) was added. The mixture was heated for two hours. After cooling to room temperature, 40 mL of n-hexane was added, and the resulting precipitate was isolated by filtering to give NiBPA: Yield: 75% (0.14 g, 0.4 mmol). Elemental Analysis calculated for C12H15Cl2N3NiO; C, 41.55; H, 4.36; N, 12.11; Found: C, 41.22; H, 4.10; N, 12.03. IR (KBr, cm−1): ν(NH): 3164, 1517. ν(C=N)met: 1678, ν(M-N=C): 721, ν(M–N): 492. Λm = 20 Ω−1mol−1cm2.
2.3. DNA Interaction Studies
Electronic absorption measurements were performed with a constant concentration of ctDNA and increasing concentrations of BPA and NiBPA (Na-phosphate buffer solution, pH 7.2). BPA and NiBPA were dissolved in DMSO and diluted in phosphate buffer (pH 7.2) (DMSO percent by v/v does not exceed 3%). Intrinsic fluorescence was measured with the ct-DNA solution at the excitation wavelength (ex, 330), and emission spectra were recorded at 550–700 nm after aliquot addition of BPA and NiBPA.
2.4. Protein Binding Studies
The excitation and emission wavelengths of BSA at 280 nm were monitored in protein binding studies using BSA (0.5 M) solution prepared in Tris-HCl buffer (pH 7.5), and the solution was stored in a dark place at 4 °C.
2.5. Protein and ctDNA Kinetic Investigation Studies
The kinetics of protein and DNA binding was measured using an Applied KinetAsyst SF-61DX2 stopped-flow instrument. At 25 °C, equal volumes of compounds against protein and DNA solutions were rapidly mixed in the stopped-flow instrument. The binding reactions were studied using a 10-fold excess of the ligand BPA and NiBPA under pseudo-first-order conditions. For each experimental condition, all recorded rate constants were taken into account, from which the mean values of at least three independent kinetic runs were calculated.
2.6. Cell Viability Assay
Methyl thiazolyl tetrazolium (MTT) colorimetry [47] was used to assess the cytotoxicity of BPA and one for human liver cancer (HepG2) cell lines. MTT can be converted into formazan by living cells with an active metabolism, but this ability is lost in dying cells. Therefore, in this study, the MTT assay was used to determine how many viable cells there were. Before the investigated compounds were added, cells were cultured in 96-well flat-bottomed microtiter plates at a density of 1 × 104 cells/well (100 l/well) for 70 to 80 percent confluent cultures, and the cultures were then incubated at 37 °C and 5 percent CO2 for 24 h. The compounds were used in a range of concentrations (31.2, 62.5, 125.0, 250.0, 500.0, and 1000.0 g/mL), and the cells were cultured for 24 h. Then, 10 l of the 12 mM MTT stock solution (5 mg/mL MTT in sterile PBS) was added to each well at the conclusion of the incubation period. After that, the plate was incubated at 37 °C for 4 h. The purple formazan crystal that formed at the bottom of the wells was removed, and 100 l of DMSO was used to dissolve it for 20 min. There was also a negative control that involved adding 10 mL of the MTT stock solution to 100 mL of medium on its own. On an ELISA reader, the absorbance at 570 nm was read (StatFax-2100, Awareness Technology, Inc., USA). The percentage of cells inhibited was calculated using:
where T is the optical density of the test sample, To is the optical density at time zero, and C is the optical density of the control.
3. Results and Discussion
3.1. Synthesis and Structural Characterization
The neutral complex [(BPA)NiCl2(H2O)] (NiBPA) was produced in good yield by treating hydrated NiCl2 with one equivalent of BPA in boiling ethanol at 60 °C. IR and UV-vis spectroscopies, as well as elemental analysis, were used to characterize complex NiBPA. Figure 1 depicts the electronic spectrum of NiBPA, which shows an intense peak at λmax = 284 nm, indicating the inter ligand π→π* transition in the BPA ligand. A less intense band at 477 nm, on the other hand, is attributed to the nickel dπ→π* MLCT transition. At 688 nm, a significant low-energy absorption band assigned for the d–d transition was seen. This may be assigned for the 3A2g(F) to 3T1g(F)(v2) transition and support the octahedral geometry of the nickel complex [19]. The band is observed at around 1603 cm−1 in the IR spectra of NiBPA (Figure 1), corresponding to vibrations of the C=N bond in the pyridine ring, shifted to higher frequencies, compared with free BPA, ν(C=N) = 1583 cm−1, and indicates coordination of nickel (II) to the BPA ligand. The vibration bands at 3050 and 1471 cm−1, ascribed for the ν(N–H) of free BPA, shifted to a higher frequency and appeared at 3238 cm−1 and 1485 cm−1 for NiBPA, respectively. A broad band at 3470 cm−1 was assigned to the coordinated water stretching vibration. Conductivity measurements of NiBPA were carried out in MeOH and DMF solution, and the Λm values are in the range of 22 and 18 Ω−1 mol−1 cm2, respectively. These values indicate that NiBPA is neutral with the participation of two chloride anions in the coordination sphere. The magnetic moment of NiBPA was found to be 2.87 B.M, confirming its paramagnetic character with two unpaired electrons and further confirming the octahedral configuration [48].
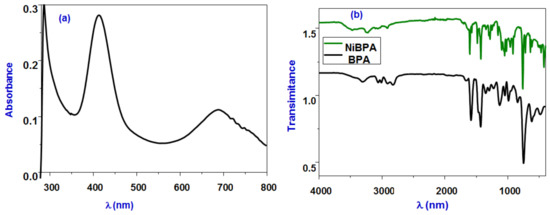
Figure 1.
(a) UV-vis absorption spectrum of NiBPA; (b) FT-IR spectra of BPA and NiBPA.
NiBPA proved to be very difficult to obtain suitable single crystals. A number of structure determinations attempted always resulted in high residual electron density maxima close to the central nickel atoms and showed the presence of numerous solvent molecules. However, the overall connectivity and geometry of NiBPA could be established (Figure 2). A preliminary structure of NiBPA reveals that the single Ni(II) center is tridentate bound to the BPA ligand, with three additional coordination sites occupied by terminally bound two chloride and one aqua ligands. One chloride is trans to the secondary amine, and the other is trans to one of the pyridine moieties. Oxygen from water is trans to the other pyridine moiety, and the nickel center has an approximately octahedral geometry.
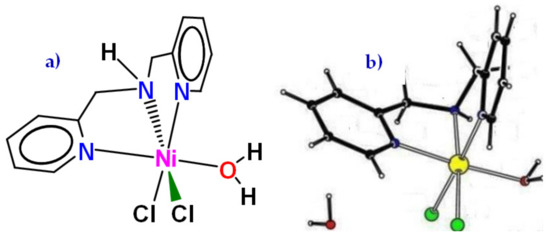
Figure 2.
(a) The structure of [(BPA)NiCl2(H2O)]·H2O (NiBPA·H2O), as well as (b) the overall connectivity and geometry of as obtained from a preliminary X-ray structure determination.
3.2. Protein Binding Studies
Interactions between the most common blood protein, serum albumin, and complexes have sparked a lot of interest recently [49] because of their structural similarity to human serum albumin. To better understand the mechanism of interaction between BPA and its nickel(II) complex NiBPA with BSA, fluorescence quenching studies were carried out. BSA’s fluorescence is due to intrinsic protein characteristics, such as the presence of tryptophan and tyrosine residues. Variations in the emission spectra caused by protein conformational changes, subunit interaction, substrate binding, or denaturation are largely attributed to the tryptophan residue [50,51]. As a result, BSA fluorescence behavior can provide valuable insight into protein structure, dynamics, and folding. Fluorescence titration experiments were performed at room temperature with BSA and different concentrations of BPA and NiBPA in the range 200–600 nm (λex. 280 nm). A slight red shift (40 percent, BPA; 65 percent, NiBPA; Figure 3) reduced the intensity of BSA fluorescence at 343 nm. The presence of the protein’s active site in a hydrophobic environment causes the redshift. It was suggested that BPA or NiBPA and the BSA protein have some sort of interaction [50,51]. The quenching mechanism can be set to either static or dynamic operation. Dynamic quenching occurs when the fluorophore and the quencher come into contact during the excited state’s transient existence, whereas static quenching is caused by the formation of a complex between the quencher and the fluorophore in the ground state. BSA (fluorophore) has a UV-vis absorption spectrum that can be used to investigate the type of quenching quickly and easily. Dynamic quenching does not cause a significant change in the fluorophore’s absorption spectra; however, static quenching does cause fluorophore perturbation [49,50,51]. Figure 4 shows that the intensity of BSA absorption increased with a slight blue shift of about 2 nm, indicating a static interaction between BSA and both BPA and NiBPA. The binding constants for BPA and NiBPA were calculated using Equation (1) and found to be 0.4 × 104 M−1 and 0.82 × 104 M−1, respectively (Table 1). These findings suggest that NiBPA binds to BSA more strongly than the ligand BPA, which is expected given the nickel center’s nature. Furthermore, the findings are in line with previous research [44,52] on related compounds.
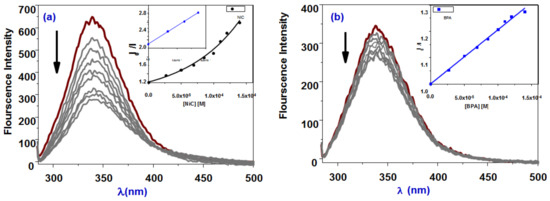
Figure 3.
Fluorescence quenching spectra of BSA in the presence of an increasing concentration of (a) NiBPA and (b) BPA (inset are Stern−Volmer plots).
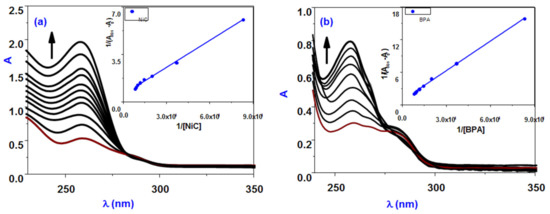
Figure 4.
UV-absorption spectra of BSA in the absence and presence of increasing amounts of (a) NiBPA and (b) BPA in phosphate buffer (inset are Benesi–Hildebrand plots).

Table 1.
Binding constants of BSA to BPA and its nickel complex NiBPA.
Figure 4 shows that at low concentrations, both BPA and NiBPA have linear Stern–Volmer plots, indicating that only dynamic or static quenching is involved in these quenching processes. The KSV values for BPA and NiBPA were found to be 0.21 × 104 and 0.7 × 104 M−1, respectively, indicating that NiBPA has a higher strong binding than the BPA ligand. The Kq value is much higher than the 2 × 1010 M−1s−1 maximum scatters collision quenching constant of quenchers with BSA (Kq = Ksv/τ0), where KSV is the Stern–Volmer constant and τ0 is the average lifetime of the BSA in the absence of the quencher (τ0 = 10 ns). This supports the idea of a static quenching mechanism. Complex NiBPA exhibits an upward curvature of the Stern–Volmer plot at higher concentrations, and this deviation from the linearity of the Stern–Volmer plot should occur when combined quenching (both dynamic and static) occurs, and the fluorophores can be quenched with the same quencher by both collision and complex formation [53,54] according to Equation (1):
where KS and KD are static and dynamic quenching constants, respectively. It is second order in [Q] and thus leads to upward curvy plots of I0/I versus [Q] at higher [Q].
I0/I = (1 + KD [Q]) (1 + Ks[Q]) = 1 + (KD + KS) [Q] + KD KS [Q]2
The quenching constant (Kq) was calculated using the Stern–Volmer and Scatchard equation and plotted as I0/I vs. [Q] to better understand the quenching process (Figure 3). In addition, Scatchard Equation (2) was used to calculate the equilibrium binding constant:
where Kbin is the compound’s BSA binding constant and n is the number of binding sites. From the log (I0 − I)/I vs. log [Q] plot (Figure 5), the binding constant (Kbin) and the number of binding sites (n) have been calculated (Table 1). Table 1 contains the evaluated values of the Kq, Kbin, and n. The estimated value of n (∼1) for BPA and NiBPA strongly supported the existence of a single binding site in BSA. The Kq and Kbin values for these compounds also indicated that NiBPA interacts with BSA more strongly than the BPA under investigation.
log[(I0 − I)/I] = log Kbin + n log [Q]
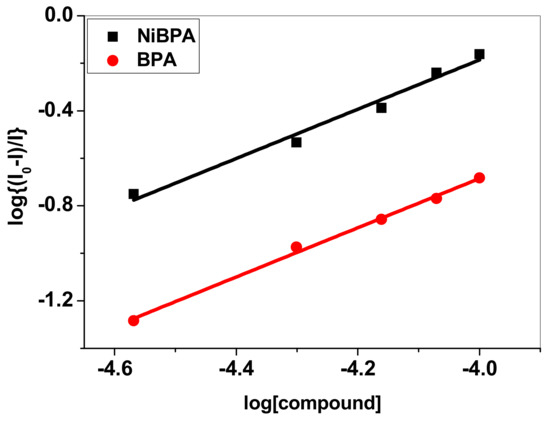
Figure 5.
Plot log (I0 − I)/I vs. log [Q].
3.3. ct-DNA Binding Studies
Absorption titration information was depicted in Figure 6 to understand the mode of interaction of complexes with ct-DNA. Increased addition of a solution of BPA to a solution of ct-DNA causes hyperchromic for the band at 264 nm with a blue shift of 5 nm (259 nm), as shown in the absorption titration spectrum (Figure 6). The band at 278 nm, on the other hand, was hypochromic with a small blue shift of 2 nm (276 nm). The presence of more than two species in the medium was indicated by the isosbestic point in the titration curve at 274 nm. In comparison to BPA, the complex NiBPA exhibited slightly different behavior. When NiBPA was added to a ct-DNA solution, the band at 257 nm showed a hyperchromic shift with a minor redshift. The absorption titration studies revealed a clear interaction between the investigated complexes and ct-DNA. The hyperchromic effect, combined with a redshift, revealed the formation of a ground-state complex between ct-DNA and both BPA and NiBPA, implying the presence of a static quenching mechanism. The hydrogen bonds formed with the –OH and SH groups of BPA and NiBPA are caused by the presence of many accessible hydrogen binding sites in the DNA major and minor grooves. The binding constants for BPA (0.16 × 105 M−1) and NiBPA (0.52 × 105 M−1) are similar to those of well-known groove binding rather than classical intercalation, implying that NiBPA interacts with ct-DNA much more strongly than BPA under investigation.
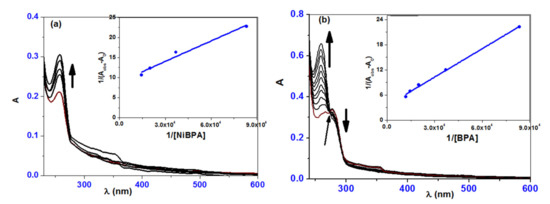
Figure 6.
UV–vis absorption spectral changes recorded for the reaction of different concentrations of (a) NiBPA and (b) BPA with ct-DNA.
3.4. Mechanistic Investigation of Protein/DNA Binding
Because many medicines with lower cellular toxicity have a slow DNA dissociation rate, several factors, such as the strength of binding and the drug’s affinity for DNA, as well as the kinetic stability of DNA-drug adducts, may contribute to antitumor effectiveness. The stopped-flow technique was used to study in-vitro protein/DNA binding to BPA and propose a mechanism in order to obtain such information. The kinetic traces were recorded at 265 nm within 100 s (BPA) and 5 s (NiBPA), respectively, to capture the interaction process with BSA. The absorbance of NiBPA increased sharply in the first 1 s of the reaction with BSA and then gradually decreased over the next 5 s. In contrast, the interaction of BPA with BSA shows a distinct pattern, with a gradual increase in absorption. In the case of ct-DNA interaction, the kinetic traces were recorded at 290 nm, and both BPA and NiBPA followed a similar pattern since kinetic traces for BPA and NiBPA were collected within 100 s, covering the entire interaction process. After a sharp decrease in the first 5 s, the absorbance gradually increased over the next 100 s. Over the concentration range of BPA and NiBPA, the study of kinetic traces performed under pseudo-first-order conditions may be fitted to two kinetic phases (Figure 7) evaluated using the following Equation (3):
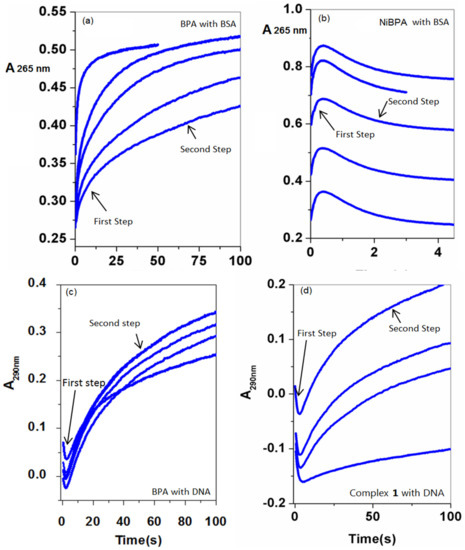
Figure 7.
Kinetic traces of different concentrations of (a) BPA with BSA, (b) NiBPA with BSA, (c) BPA with ct-DNA and (d) NiBPA with ct-DNA.
The results, presented in Table 2, show that BSA binds reversibly to BPA with a second-order association constant of k1 = 12.5 ± 1.1 M−1s−1 and dissociates from the binary complex with a first-order dissociation constant of k−1 = 0.17 ± 0.0 s−1 in the initial phase. Complex NiBPA binds to BSA four times faster (k1 = 53.6 ± 9.0 M−1s−1) and dissociates forty times faster (k−1 = 7.3 ± 0.3 s−1). This means that the binding affinity of BSA to BPA, Ka1, k1/k−1, of 73.5 M−1 is about ten times higher than that of NiBPA (7.3 M−1) and that the equilibrium dissociation constants of BPA, Kd1, k−1/k1, is 13.6 × 10−3 M and about ten times slower than that of NiBPA (136 × 10−3 M). This means that the formed BPA-BSA is far more stable than NiBPA-BSA.

Table 2.
Summary of rate constants and activation parameters for the interaction of BPA and NiBPA to BSA and ct-DNA at 25 °C.
A reversible reaction with a coordination affinity of 0.69 M−1 for NiBPA was detected in the second reaction step, but an irreversible reaction with a K2 = 0.69 M−1 with BPA was observed. The second phase is primarily concerned with the BSA condensation process, in addition to the electrostatic interaction. As shown in Figure 8, the irreversibility of the second reaction step with BPA (k−2 ≈ 0) suggests that the newly formed BPA-BSA complex is kinetically more stable in solution than NiBPA-BSA.
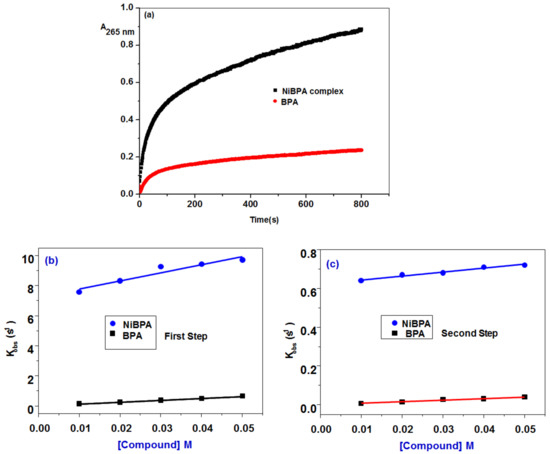
Figure 8.
(a) Kinetic traces of BPA and NiBPA with BSA; variation of kobs vs. [BPA] and [NiBPA] for (b) the first and (c) the second interaction steps to BSA.
The interaction process involves at least two steps, and the stopped-flow records the two kinetic phases as a fast binding (rate constant kobs1) followed by a slow first-order isomerization process (rate constant kobs2). Both kobs1 and kobs2 increased linearly with BPA and NiBPA concentration (Figure 9 and Figure 10) with a slope equal to the second-order association constant, kon [M−1s−1], and an intercept equal to the first-order dissociation constant, koff [s−1], as demonstrated by the equation kobs = koff + kon[Comp]. Both kon and koff were used to calculate the equilibrium association constants Kaff [kon/koff M−1] and the equilibrium dissociation constants Kd [koff/kon M]. A reaction plot is shown in Scheme 1, and all of the data are listed in Table 2.
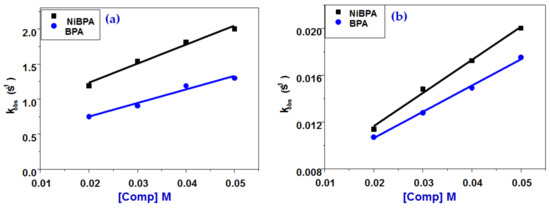
Figure 9.
Variation of kobs vs. [BPA] and [NiBPA] for (a) the first and (b) the second interaction steps to ct-DNA.
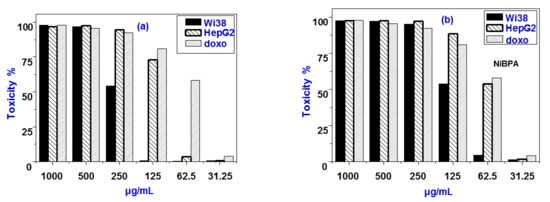
Figure 10.
In vitro cytotoxicity of (a) BPA and (b) NiBPA against different concentrations.
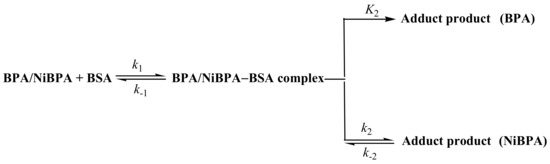
Scheme 1.
Reaction scheme for the binding of BSA to both BPA and NiBPA.
The difference between the free energy, G, of BSA molecules alone in the solution and when bound together is related to the equilibrium dissociation constant, Kd1. Equation (4) [55] gives the binding free energy change Gbind.
While T, R represents the absolute temperature and the universal gas coefficient, respectively. The ΔGbind values, calculated using Equation (4), are −10.6 and −5.1 kJ mol−1 for the first step binding of BSA to BPA and NiBPA, respectively. The negative sign for G indicates spontaneous binding to both BPA and NiBPA, and the greater negative value of BPA compared to NiBPA indicates that binding with BPA is very favorable. Both compounds interact with ct-DNA through a similar mechanism that is reversible in both reaction steps (Figure 9 and Scheme 2). The initial reaction step is thought to be the complexation and production of binary complexes. BPA binds to ct-DNA with a second-order association constant of k1 = 27.0 ± 2.5 M−1s−1 and dissociates from the binary complex with a first-order dissociation constant of k−1 = 0.69 ± 0.1 s−1, whereas 1 binds to ct-DNA with (k1 = 19.3 ± 2.3 M−1s−1) and dissociates from the binary complex with (k−1 = 0.36 ± 0.1 s−1). This means that the DNA binding affinity of 1, Ka1, k1/k−1 is slightly higher (53.6 M−1) than that of BPA (40.0 M−1) and that the equilibrium dissociation constant of NiBPA, Kd1, k−1/k1 is ×10−3 M, which is slightly slower (25.6 × 10−3 M). Because of the presence of a Ni(II) center, NiBPA has a higher affinity and a lower dissociation constant than BPA, indicating that the formed 1–DNA is much more stable than BPA–DNA. Gbind values of −9.1 kJ mol−1 (BPA) and −9.7 kJ mol−1 (NiBPA) indicate that this step is spontaneous for both compounds. The data from the second reaction step, which included internal DNA rearrangement as well as electrostatic interaction and isomerization reaction, followed the same pattern as the first; a reversible reaction was observed for both BPA and NiBPA. The DNA affinity for BPA (Ka2 = k2/k−2) is 47.5 M−1, which is slightly higher than NiBPA (39 M−1), and the equilibrium dissociation constant for BDA (Kd2 = k−2/k2) is 2.1 × 10−2 M, which is slightly slower than NiBPA (2.6 × 10−3 M). Gbind values for this phase are −9.4 kJ mol−1 (BPA) and −9.0 kJ mol−1 (NiBPA), indicating that this step is spontaneous for both compounds.
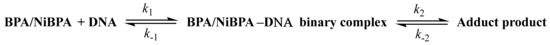
Scheme 2.
Reaction scheme for the reversible binding of ct-DNA to BPA and NiBPA.
Equation (5) shows how to calculate the overall equilibrium dissociation constant, Kd, from the individual equilibrium dissociation constants, Kd1 and Kd2, as well as the overall association constants, Ka.
The data in Table 2 show that the DNA binding affinity of NiBPA (2077 M) is higher than that of BPA (1899). The overall reaction Gbind values are −18.7 kJ mol−1 (BPA) and −18.9 (NiBPA) kJ mol−1, indicating that the reaction is spontaneous for both compounds.
3.5. Cytotoxic Activity
The interesting findings from BSA binding studies inspired us to use the MTT assay to test all compounds’ anticancer activity. In vitro cytotoxic activity of BPA and NiBPA were tested against human liver cancer (HepG2) and normal liver (Wi38) cell lines at various concentrations by MTT assay. The percentages of cell viability in the presence of BPA and NiBPA with different concentration levels (31.2, 62.5, 125.0, 250.0, 500.0, and 1000.0 μg/mL) are depicted in Figure 10. The cell viability was found to decrease with increasing compound concentrations for both liver cancer (HepG2) and normal (THLE2) cell lines. However, the cancer cell growth exhibited by BPA and NiBPA can be measured by the IC50 values and were found to be 100.0 ± 1.0 and 75.0 ± 0.5 μg/mL, respectively. From this data, one can conclude the following. Firstly, both complexes were cytotoxic active, but the activity was less than that for the normal drug (doxo, IC50 = 52.29 ± 1.01 μg/mL). Secondly, the Ni(II) complex NiBPA exhibits higher cytotoxicity compared to BPA. The complexation of BPA to nickel center in complex NiBPA may be the main factor for the better cytotoxicity of NiBPA. The cytotoxic selectivity of both compounds was investigated by measuring their effect on the cell viability of normal liver (Wi38) cell lines. In the case of the normal cells, the IC50 values suppressed by BPA and its Ni(II) complex NiBPA are 237.2 ± 1.3 and 120.0 ± 0.5 μg/mL, respectively. The IC50 values of the normal cells are much higher than that of the cancer cells, indicating that both compounds can be effective and selective toward cancer cells. Thus, complex NiBPA is the most active, indicating that nickel ion coordination was crucial for cytotoxicity and that complexation improved the anticancer activity. The positive charge in NiBPA due to the neutral BPA ligand may increase cytotoxicity. Additionally, these findings are in agreement with the affinities and binding constants of every compound that was studied for interaction with BSA.
3.6. Antibacterial Activity
Several studies have shown that organic compound coordination with a metallic element results in significant changes in the biological activity of both the organic ligand and the metal. As a result, BPA and its nickel complex NiBPA were tested in vitro for antibacterial activity against Gram-positive (S. aureus and S epidermidis) and Gram-negative (P. mirabilis and E. coli) bacteria. Figure 10 depicts the antibacterial function of BPA and NiBPA as the diameter of the growth-inhibition region in millimeters. Table 3 shows the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of complexes required to inhibit bacterial growth. The MBC is identified by determining the lowest concentration of antibacterial agents that reduces the viability of the initial bacterial inoculum by 99.9%. The MIC or MBC strain was chosen based on the value agreed upon on two or more occasions. The antibiotic tobramycin (TOB) was used to control the situation. The inhibition zone, as well as the MIC and MBC data, leads to the following conclusions. To begin, NiBPA exhibited strong antibacterial activity, and its antibacterial activity was much greater than that of BPA, but it was less than that of the standard drug (TOB). Second, NiBPA demonstrated greater antimicrobial activity against Gram-negative bacteria than Gram-positive bacteria. This is explained by the metal ion’s polarity and cationic charge, as NiBPA has a cationic property with two positive charges. Gram-negative bacteria were more sensitive to NiBPA than Gram-positive bacteria due to differences in cell surface characteristics; Gram-negative bacteria had a higher negative charge on their cell surface than Gram-positive bacteria [56,57]. The interaction between Gram-negative bacteria and NiBPA was stronger than that of Gram-positive bacteria due to the higher negative charge on the cell surface, despite the presence of an outer membrane in Gram-negative bacteria (see Figure 11).

Table 3.
MIC and MBC results of BPA and NiBPA positive control, Tobramycin antibiotic (TOB).
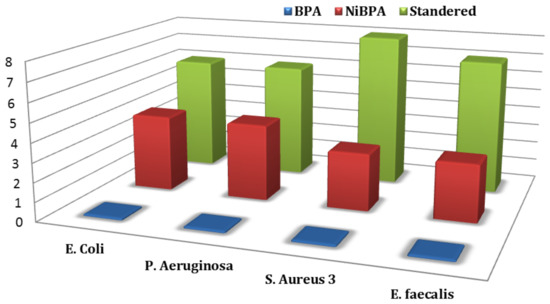
Figure 11.
Mean inhibition zone of BPA, NiBPA, and standard against Gram-negative (E. coli and P. aeruginosa) and Gram-positive (S. aureus and E. faecalis) bacteria (mm) compared to ‘Standard’.
4. Conclusions
New Ni(II) complexes containing bispicolylamine have been described and characterized in this study. Because almost all pharmaceuticals work by interfering with biological functions by binding to a specific protein or DNA, in vitro DNA/protein binding of BPA has been studied mechanistically and compared to its Ni(II) complex NiBPA. In comparison to BPA, complex NiBPA binds strongly to DNA/protein via a static quenching mechanism. A mechanism for the interaction was proposed using stopped-flow techniques. BSA interacts with BPA and NiBPA via two different mechanisms. BPA’s interaction is characterized by fast reversible followed by slow irreversible steps, whereas NiBPA’s interaction is characterized by two reversible steps. The binding parameters for the first step of BPA (Ka1 = 73.5 M−1, Kd1 = 13.6 × 10−3 M, and ΔG1 = −10.6 kJ mol−1) and NiBPA (Ka1 = 7.3 M−1, Kd1 = 136 × 10−3 M, ΔG1 = −5.1 kJ mol−1) and for the second step of BPA (K2 = 0.82 M−1) and 1 (Ka1 = 3.2 M−1, Kd1 = 31.2×10−2 M, ΔG1 = −8.3 kJ mol−1) were determined. Overall binding parameters for NiBPA (Ka = 30.3 M−1, Kd = 34.0 M, ΔG0 = −8.4 kJ mol−1). BPA and its Ni(II) complex NiBPA bind to DNA in a similar way, with two reversible steps: fast second-order binding followed by slow first-order isomerization. The binding parameters for the first step of BPA (Ka1 = 40 M−1, Kd1 = 25.6 × 10−3 M, and ΔG1 = −9.1 kJ mol−1) and NiBPA (Ka1 = 53.6 M−1, Kd1 = 19.0 × 10−3 M, ΔG1 = −9.7 kJ mol−1) and for the second step of BPA (Ka2 = 47.5 M−1, Kd1 = 2.1 × 10−2 M, and G1 = −9.4 kJ mol−1) and NiBPA (Ka1 = 39 M−1, Kd1 = 2.6×10−2 M, ΔG1 = −9.0 kJ mol−1) were determined. Overall binding parameters for BPA (Ka = 1899 M−1, Kd = 52.7 × 10−3 M, ΔG0 = −18.7 kJ mol−1) and NiBPA (Ka = 2077 M−1, Kd = 48.2 × 10−3 M, ΔG0 = −18.9 kJ mol−1) were also determined and showed that the relative reactivity is approximately (BPA)/(NiBPA) = 9/10. The significantly negative G values support a spontaneous binding reaction to both BPA and its Ni(II) complex NiBPA. The following points can be summarized by comparing the results of the interactions of BPA and NiBPA with DNA with those of BSA. BSA interacts with BPA and NiBPA via two different mechanisms: (i) A fast reversible followed by slow irreversible steps characterized the interaction of BPA, whereas two reversible steps are observed with NiBPA.; (ii) BPA and NiBPA interact with DNA via a similar mechanism via two reversible steps; (iii) Complex NiBPA was found to have much higher affinity to DNA (2077 M) than BSA (30.3 M), despite the fact that BSA interacts with this, which is explained by the fact that the fast association of NiBPA with BSA is followed by a fast dissociation reaction; (iv) BPA formed a more stable BSA complex than its Ni(II) complex. This is also understandable because they both bind via different mechanisms, with the second binding step for BPA being irreversible and the second binding step for NiBPA being reversible; (v) Interestingly, when BPA and NiBPA bind to both DNA, the Ni(II) center was found to affect the rate but not the mechanism. In the case of BSA interaction with BPA and its Ni(II) complex, the Ni(II) center was found to change both the mechanism and the rate. Additionally, NiBPA exhibited significant cytotoxicity against the human liver cancer HepG2 cell as well as antibacterial activity, which is consistent with the binding constants but not the kinetic stability. This shows that in our situation, cytotoxic and antibacterial activities are significantly more influenced by binding constants than by kinetic stability. Due to its selectivity and cytotoxic activity, the complex NiBPA is anticipated to be used in medicine.
Author Contributions
Conceptualization, S.Y.S. and M.M.I.; methodology, E.R.; software, F.I.E.; validation, E.R.; formal analysis, F.I.E.; investigation, E.R. and F.I.E.; resources, I.M.E.-M.; data curation, S.Y.S. and F.I.E.; writing—original draft preparation, E.R. and F.I.E.; writing—review and editing, S.Y.S. and R.v.E.; visualization, S.Y.S.; supervision, S.Y.S., A.E.-M.M.R. and I.M.E.-M.; project administration S.Y.S.; funding acquisition, M.M.I. and R.v.E. All authors have read and agreed to the published version of the manuscript.
Funding
Taif University Researchers Supporting Project (TURSP-2020/05), Taif University, Taif, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this research paper.
References
- Kramer, R. Dimeric versus polymeric coordination in copper(II) cationic complexes with bis(chelating) oxime and amide ligands Coord. Chem. Rev. 1999, 182, 243. [Google Scholar]
- Usman, M.; Zaki, M.; Khan, R.A.; Alsalm, A.; Ahmad, M.; Tabassum, S. Coumarin centered copper(ii) complex with appended-imidazole as cancer chemotherapeutic agents against lung cancer: Molecular insight via DFT-based vibrational analysis. RSC Adv. 2017, 7, 36056. [Google Scholar] [CrossRef]
- Cowan, J.A. Metal Activation of Enzymes in Nucleic Acid Biochemistry. Chem. Rev. 1998, 98, 1067. [Google Scholar] [CrossRef] [PubMed]
- Sreedhara, A.; Cowan, J.A. Catalytic hydrolysis of DNA by metal ions and complexes. J. Biol. Inorg. Chem. 2001, 6, 337. [Google Scholar] [CrossRef] [PubMed]
- Bales, B.C.; Kodama, T.; Weledji, Y.N.; Pitie, M.; Meunier, B.; Greenberg, M.M. Water-soluble CuII and MnII bis-phenanthroline octanedioate complexes with unprecedented nano and picomolar in vitro complexes with unprecedented nano and picomolar in vitro cytotoxicity as promising leads for chemotherapeutic drug cytotoxicity as promising leads for chemotherapeutic drug development. Nucleic Acids Res. 2005, 33, 5371. [Google Scholar]
- Fernandes, G.L.; Parrilha, J.A.; Lessa, L.J.M.; Santiago, M.M.; Kanashiro, F.; Boniolo, F.S.; Bortoluzzi, A.J.; Vugman, N.V.; Herbst, M.H.; Horn, A. Catalase vs. Peroxidase Activity of a Manganese (II) Compound: Identification of a Mn(III)-(μ-O)2-Mn(IV) Reaction Intermediate by Electrospray Ionization Mass Spectrometry and Electron Paramagnetic Resonance Spectroscopy. Inorg. Chim. Acta 2006, 359, 3167. [Google Scholar] [CrossRef]
- Marzano, M.; Pellei, F.; Tisato, F.; Santini, C. Copper complexes as anticancer agents. Anticancer. Agents Med. Chem. 2009, 9, 185. [Google Scholar] [CrossRef]
- Mobley, H.L.; Hausinger, R.P. Microbial ureases: Significance, regulation, and molecular characterization. Microbiol. Rev. 1989, 53, 85. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hille, A.; Frias, C.; Kater, B.; Bonitzki, B.; Wölfl, S.; Scheffler, H.; Prokop, A.; Gust, R. [Fe(III)(salophene)Cl], a potent iron salophene complex overcomes multiple drug resistance in lymphoma and leukemia cells. J. Med. Chem. 2010, 53, 6064. [Google Scholar] [CrossRef]
- Pradeepa, S.M.; Naik, H.S.B.; Kumar, B.V.; Priyadarsini, K.I. Cobalt(II), Nickel(II) and Copper(II) complexes of a tetradentate Schiff base as photosensitizers: Quantum yield of 1O2 generation and its promising role in the antitumor activity. Spectrochim. Acta 2013, 101, 132. [Google Scholar] [CrossRef]
- Sathisha, M.P.; Shetti, U.N.; Revankar, V.K.; Pai, K.S.R. Synthesis and antitumor studies on novel Co(II), Ni(II) and Cu(II) metal complexes of bis(3-acetylcoumarin)thiocarbohydrazone. Eur. J. Med. Chem. 2008, 43, 2338. [Google Scholar] [CrossRef]
- Foye, W.O. Cancer Chemotherapeutic Agents; American Chemical Society: Washington, DC, USA, 1995; p. 369. [Google Scholar]
- Yin, H.D.; Liu, H.; Hong, M. Synthesis, structural characterization and DNA-binding properties of organotin(IV) complexes based on Schiff base ligands derived from 2-hydroxy-1-naphthaldy and 3- or 4-aminobenzoic acid. J. Organomet. Chem. 2012, 713, 11. [Google Scholar] [CrossRef]
- Sathyadevi, P.; Krishnamoorthy, P.; Butorac, R.R.; Cowley, A.H.; Bhuvanesh, N.S.P.; Dharmaraj, N. Potentially cytotoxic new copper(II) hydrazone complexes: Synthesis, crystal structure and biological properties. Dalton Trans. 2011, 40, 9690. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jin, Y.; Lewis, M.A.; Gokhale, N.H.; Long, E.C.; Cowan, J.A. DNA cleavage behavior of a new p-xylyl spaced bis-Cu(BPA)Cl2 complex: The steric effect of a bulky p-xylyl-derived spacer. J. Am. Chem. Soc. 2007, 129, 8353. [Google Scholar] [CrossRef]
- Bisceglie, F.; Baldini, M.; Belicchi-Ferrari, M.; Buluggiu, E.; Careri, M.; Pelosi, G.; Pinelli, S.; Tarasconi, P. Metal complexes of retinoid derivatives with antiproliferative activity: Synthesis, characterization, and DNA interaction studies. Eur. J. Med. Chem. 2007, 42, 627. [Google Scholar] [CrossRef]
- Barone, G.; Cambino, N.; Ruggirello, A.; Silvestri, A.; Terenzi, A.; Liveri, V.T. Spectroscopic study of the interaction of Ni(II)-5-triethyl ammonium methyl salicylidene ortho-phenylendiiminate. Chem. -A Eur. J. 2009, 88, 3233. [Google Scholar]
- Kirin, S.I.; Happel, C.M.; Hrubanova, S.; Weyhermüller, T.; Klein, C.; Metzler-Nolte, N. Synthesis, structure and comparison of the DNA cleavage ability of metal complexes M(II)L with the N-(2-ethoxyethanol-bis(2-picolyl)amine ligand L (M = Co, Ni, Cu and Zn). Dalton Trans. 2004, 1201. [Google Scholar] [CrossRef]
- Stamler, J.S.; Loscalzo, D.J.S.J. Nitric oxide directly activates calcium-dependent potassium channels in vascular smooth muscle. Science 1992, 258, 1898. [Google Scholar] [CrossRef]
- Shrivastava, H.Y.; Kanthimathi, M.; Niar, B.U. DNA Cleavage by a Chromium(III) Complex. Biochem. Biophys. Res. Commun. 1999, 265, 311. [Google Scholar] [CrossRef]
- Raja, D.S.; Bhuvanesh, N.S.P.; Natarajan, K. Cubane core/1D polynuclear copper(II) complexes: Structure, protein/DNA binding, and magnetic properties. Inorg. Chem. 2011, 50, 12852. [Google Scholar]
- Wende, C.; Lüdtke, C.; Kulak, N. Copper Complexes of N-Donor Ligands as Artificial Nucleases. Eur. J. Inorg. Chem. 2014, 2014, 2597. [Google Scholar] [CrossRef]
- Ma, D.-L.; He, H.-Z.; Leung, K.-H.; Chan, D.S.-H.; Leung, C.-H. Bioactive Luminescent Transition-Metal Complexes for Biomedical Applications. Angew. Chem. Int. Ed. 2013, 52, 7666. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.R.; Thomas, J.A. Ruthenium(ii) polypyridyl complexes and DNA—from structural probes to cellular imaging and therapeutics. Chem. Soc. Rev. 2012, 41, 3179. [Google Scholar] [CrossRef] [PubMed]
- Salassa, L. Polypyridyl Metal Complexes with Biological Activity. Eur. J. Inorg. Chem. 2011, 2011, 4931. [Google Scholar] [CrossRef]
- Schatzschneider, U. Photoactivated Biological Activity of Transition-Metal Complexes. Eur. J. Inorg. Chem. 2010, 2010, 1451. [Google Scholar] [CrossRef]
- Hancock, R.D. The pyridyl group in ligand design for selective metal ion complexation and sensing. Chem. Soc. Rev. 2013, 42, 1500. [Google Scholar] [CrossRef]
- Vos, J.G.; Kelly, J.M. Ruthenium polypyridyl chemistry; from basic research to applications and back again. Dalton Trans 2006, 41, 4869. [Google Scholar] [CrossRef]
- Alstrum-Acevedo, J.H.; Brennaman, M.K.; Meyer, T.J. Chemical Approaches to Artificial Photosynthesis. 2. Inorg. Chem. 2005, 44, 6802. [Google Scholar] [CrossRef]
- Mortezaei, S.; Caterineu, N.; Canary, J.W. A Redox-Reconfigurable, Ambidextrous Asymmetric Catalyst. J. Am. Chem. Soc. 2012, 134, 8504. [Google Scholar] [CrossRef]
- Sundaravel, K.; Suresh, E.; Saminathan, K.; Palaniandavar, M. Iron(III) complexes of N2O and N3O donor ligands as functional models for catechol dioxygenase enzymes: Ether oxygen coordination tunes the regioselectivity and reactivity. Dalton Trans. 2011, 40, 8092. [Google Scholar] [CrossRef]
- Frezza, M.; Hindo, S.S.; Tomco, D.; Allard, M.M.; Cui, Q.C.; Heeg, M.J.; Chen, D.; Dou, Q.P.; Verani, N.C. Comparative Activities of Nickel(II) and Zinc(II) Complexes of Asymmetric [NN′O] Ligands as 26S Proteasome Inhibitors. Inorg. Chem. 2009, 48, 5928. [Google Scholar] [CrossRef]
- Carney, M.J.; Robertson, N.J.; Halfen, J.A.; Zakharov, L.N.; Rheingold, A.L. Octahedral Chromium(III) Complexes Supported by Bis(2-pyridylmethyl)amines: Ligand Influence on Coordination Geometry and Ethylene Polymerization Activity. Organometallics 2004, 23, 6184. [Google Scholar] [CrossRef]
- Andres, P.R.; Schubert, U.S. New Functional Polymers and Materials Based on 2,2:6,2-Terpyridine Metal Complexes. Adv. Mater. 2004, 16, 1043. [Google Scholar] [CrossRef]
- Ojida, A.; Mito-oka, Y.; Sada, K.; Hamachi, I. Molecular Recognition and Fluorescence Sensing of Monophosphorylated Peptides in Aqueous Solution by Bis(zinc(II)−dipicolylamine)-Based Artificial Receptors. J. Am. Chem. Soc. 2004, 126, 2454. [Google Scholar] [CrossRef]
- Goetzke, L.; Gloe, K.; Jolliffe, K.A.; Lindoy, L.F.; Heine, A.; Doert, T.; Joeger, A.; Gloe, K. Nickel(II) and zinc(II) complexes of N-substituted di(2-picolyl)amine derivatives: Synthetic and structural studies. Polyhedron 2011, 30, 708–714. [Google Scholar] [CrossRef]
- Nagataki, T.; Ishii, K.; Tachi, Y.; Itoh, S. Ligand effects on NiII-catalysed alkane-hydroxylation with m-CPBA. Dalton Trans. 2007, 1120. [Google Scholar] [CrossRef]
- Wikstrom, J.P.; Nazarenko, A.Y.; Reiff, W.M.; Rybak-Akomova, E.V. Synthesis and characterization of tetrakis(μ-hydroxo)tetrakis(2,2′-dipicolylamine)tetranickel perchlorate, a nickel-hydroxy cubane complex. Inorg. Chim. Acta 2007, 360, 3733. [Google Scholar] [CrossRef]
- Sarker, S.; Mondal, A.; el Fallah, M.S.; Ribas, J.; Stoeckli-Evans, H.; Rajak, K.K. Synthesis, structure and magnetic properties of two end-on double azido bridged nickel(II) dinuclear entities incorporating N,N,N-coordinating tridentate reduced Schiff base ligands. Polyhedron 2006, 25, 25. [Google Scholar] [CrossRef][Green Version]
- Škalamera, Đ.; Sanders, E.; Vianello, R.; Maršavelski, A.; Pevec, A.; Turel, I.; Kirin, S.I. Synthesis and characterization of ML and ML2 metal complexes with amino acid substituted bis(2-picolyl)amine ligands. Dalton Trans. 2016, 45, 2845. [Google Scholar] [CrossRef]
- Esposito, B.P.; Najjar, R. DNA/Protein binding, antioxidative, antimicrobial studies of metal complexes with anthranilic acid Schiff base. Coord. Chem. Rev. 2002, 232, 137. [Google Scholar]
- Shaban, N.Z.; Yehia, S.A.; Shoueir, K.R.; Saleh, S.R.; Awad, D.; Shaban, S.Y. Design, DNA binding, and kinetic studies, antibacterial and cytotoxic activities of stable dithiophenolato titanium(IV)-chitosan Nanocomposite. J. Mol. Liq. 2019, 287, 111002. [Google Scholar] [CrossRef]
- Shaban, N.Z.; Aboelsaad, A.M.; Shoueir, K.R.; Abdulmalek, S.A.; Awad, D.; Shaban, S.Y. Chitosan-based dithiophenolato nanoparticles: Preparation, mechanistic information of DNA binding, antibacterial and cytotoxic activities. J. Mol. Liq. 2020, 318, 114252. [Google Scholar] [CrossRef]
- Elshami, F.I.; Ramadan, A.; Ibrahim, M.M.; Elmehasseb, I.M.; Al-Juaid, S.; Shaban, S.Y. Metformin Containing Nickel (II) Complexes: Synthesis, Structural Characterization, Binding and Kinetic Interactions with BSA, Antibacterial and in-vitro Cytotoxicity Studies. Appl. Organometal. Chem. 2019, 34, 5437. [Google Scholar] [CrossRef]
- Shereef, H.A.; Shaban, S.Y.; Moemen, Y.S.; El-Khouly, M.E.; El-Nahas, A.M. Biophysicochemical studies of a ruthenium (II) nitrosyl thioether-thiolate complex binding to BSA: Mechanistic information, molecular docking, and relationship to antibacterial and cytotoxic activities. Appl. Organomet. Chem. 2022, 34, e6583. [Google Scholar] [CrossRef]
- Ibrahim, M.M.; Ramadan, A.M.; Mersal, G.A.M.; El-Shazly, S.A. Synthesis, superoxide dismutase, nuclease, and anticancer activities of copper(II) complexes incorporating bis(2-picolyl)amine with different counter anions. J. Mol. Struct. 2011, 998, 1–10. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Liu, W.; Migdisov, A.; Williams-Jones, A. The stability of aqueous nickel(II) chloride complexes in hydrothermal solutions: Results of UV-Visible spectroscopic experiments. Geochim. Cosmochim. Acta 2012, 94, 276–290. [Google Scholar] [CrossRef]
- Ramachandran, E.; Raja, D.S.; Bhuvanesh, N.S.P.; Natarajan, K. Self-assembled Cu(ii) and Ni(ii) metallamacrocycles formed from 3,3,3′,3′-tetrabenzyl-1,1′-aroylbis(thiourea) ligands: DNA and protein binding studies, and cytotoxicity of trinuclear complexes. Dalton Trans. 2012, 41, 13308. [Google Scholar] [CrossRef]
- Raja, D.S.; Paramaguru, G.; Bhuvanesh, N.S.P.; Reibenspies, J.H.; Renganathan, R.; Natarajan, K. Effect of terminal N-substitution in oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicar-benzones on the mode of coordination structure.interaction of protein, radical scavenging and cytotoxic activity of copper (II) complexes. Dalton Trans. 2011, 40, 4548. [Google Scholar] [CrossRef]
- Raja, D.S.; Bhuvanesh, N.S.P.; Natarajan, K. Synthesis, Characterization and Biological Evaluation of Fe(III) and Cu(II) Complexes with 2,4-Dinitrophenyl hydrazine and Thiocyanate Ions. Eur. J. Med. Chem. 2011, 46, 4584. [Google Scholar]
- Karami, K.; Parsianrad, F.; Alinaghi, M.; Amirghofran, Z. In vitro cytotoxicity studies of palladacyclic complexes containing the symmetric diphosphine bridging ligand: Studies of their interactions with DNA and BSA. J. Inorg. Chim. Acta 2017, 467, 46. [Google Scholar] [CrossRef]
- Gok, E.; Ozturk, C.; Akbay, N. Interaction of oxovanadium(IV)–salphen complexes with bovine serum albumin and their cytotoxicity against cancer. J. Fluoresc. 2008, 18, 781. [Google Scholar]
- Papadopoulou, A.; Green, R.J.; Frazier, R.A. Interaction of flavonoids with bovine serum albumin: A fluorescence quenching study. J. Agric. Food Chem. 2005, 53, 158. [Google Scholar] [CrossRef]
- Williams, M.; Daviter, T. Protein-Ligand Interactions. Biophys. Physicobiol. 2016, 13, 85. [Google Scholar]
- Chung, Y.-C.; Su, Y.P.; Chen, C.-C.; Jia, G.; Wang, H.L.; Wu, J.G.; Lin, J.G. Relationship between antibacterial activity of chitosan and surface characteristics of cell wall. Acta Pharmacol. Sin. 2004, 25, 932. [Google Scholar]
- Xue, Z.-X.; Yang, G.-P.; Zhang, Z.-P.; He, B.-L. Application of chitosan microspheres as carriers of LH-RH analogue TX46 React. Funct. Polym. 2006, 66, 893. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).