Decoding Brain Signals in a Neuromorphic Framework for a Personalized Adaptive Control of Human Prosthetics
Abstract
1. Introduction
2. Materials and Methods
2.1. Neuromorphic Framework for Adaptive MCD
- A 3D recurrent SNN architecture (3D-SNN), spatially structured and adaptable to an individual 3D brain template, for feature extraction from recorded ECoG brain signals. The SNN is incrementally trained in an unsupervised mode.
- A recurrent Echo State Network (ESN) for decoding of the brain trained in a supervised mode on-line via recursive least squares (RLS) or reinforcement learning (RL) algorithms.
- State (idle, hand or leg movement) (): a vector with dimension corresponding to the number of possible states that was decoded via from the predicted model output.
- Trajectory (left and right hand positions in 3D space) (): a vector of x, y and z coordinates for each hand.
- Satisfaction (): two-dimensional vector that was decoded via from the predicted model output.
- Using STDP rule to continuously adapt the 3D-SNN connectivity in an unsupervised way, based on history of the input signal as well as the state of spiking neurons. This can be used for auto-adaptation of the model, when new data from the subject are continuously entered for unsupervised learning. This will ensure that the model will continuously adapt (auto-adapt) with the subject’s movement improvement in time.
- Using reinforcement learning rule (RL) of the output parameters of the ESN responsible for motor action based on satisfaction prediction.
2.2. Software Implementation
| Algorithm 1 Pseudocode of on-line training algorithm |
| Initialization Initialize ESN module parameters Compose 3D-SNN module using neurons and ECoG positions
|
- 12th Gen Intel® Core™ i7-12700
- Installed RAM 32 GB
- Base speed 2.10 GHz
- Cores 12
- Logical processors 20
- Initialization includes generation/reading of model parameters and setting-up of two recurrent structures (3D-SNN and ESN): 49 s approximately.
- Single RLS iteration (training on one input/output train data example): 2.5–3.5 s
- Single RL iteration (training on one input/output train data example): 0.55–0.65 s
- Single step model output calculation: 0.58–0.63 s
2.3. Experimental Data
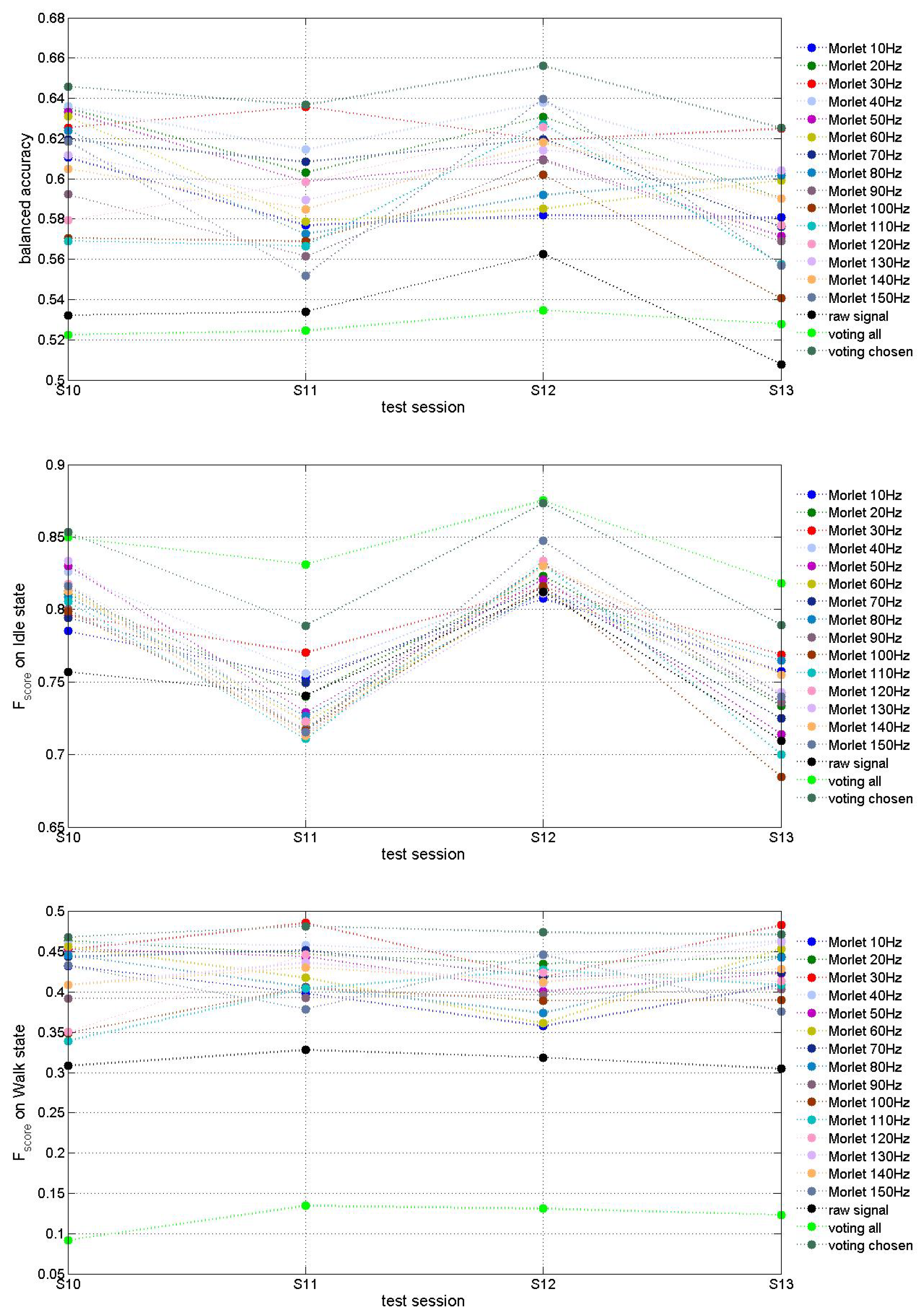
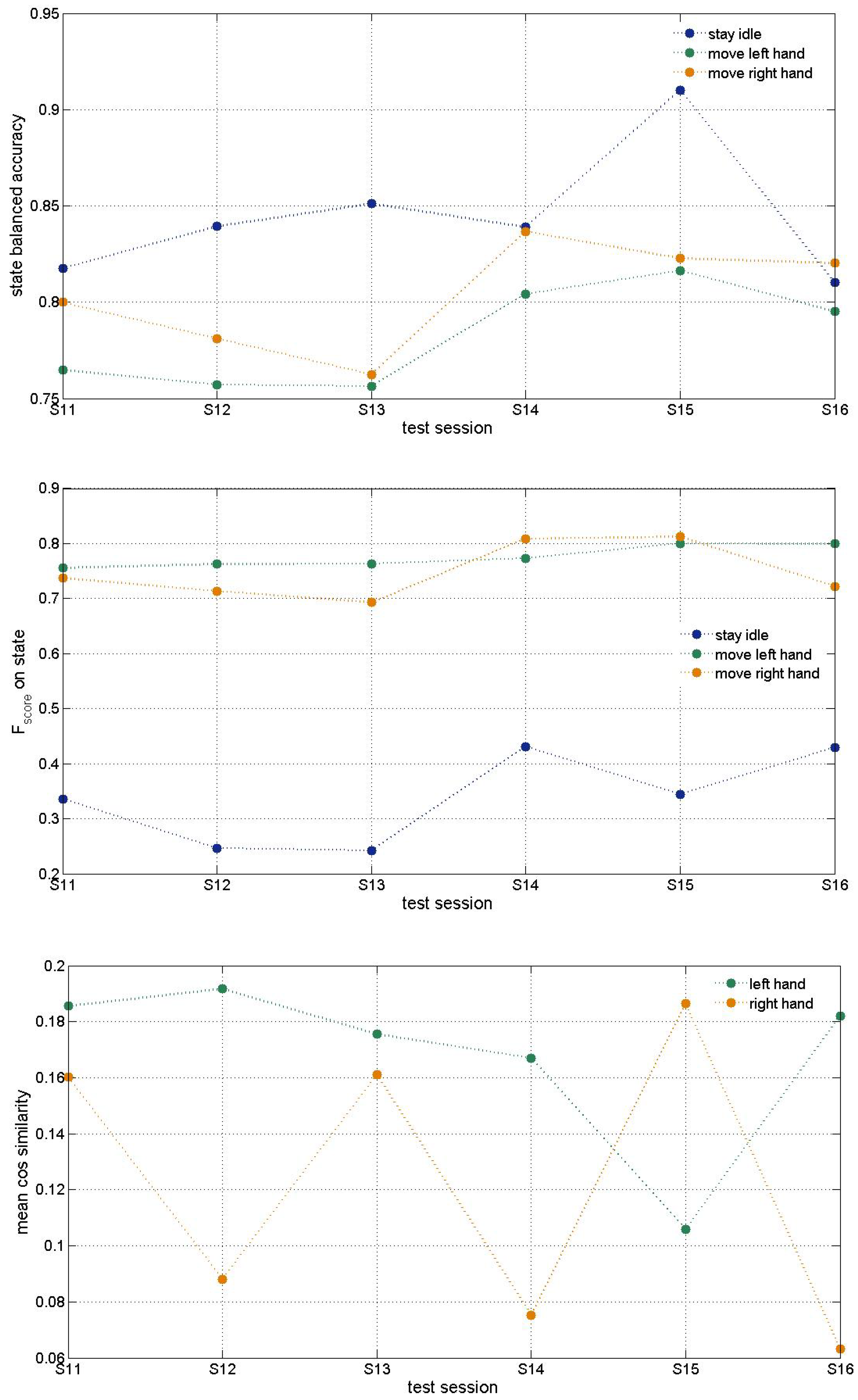
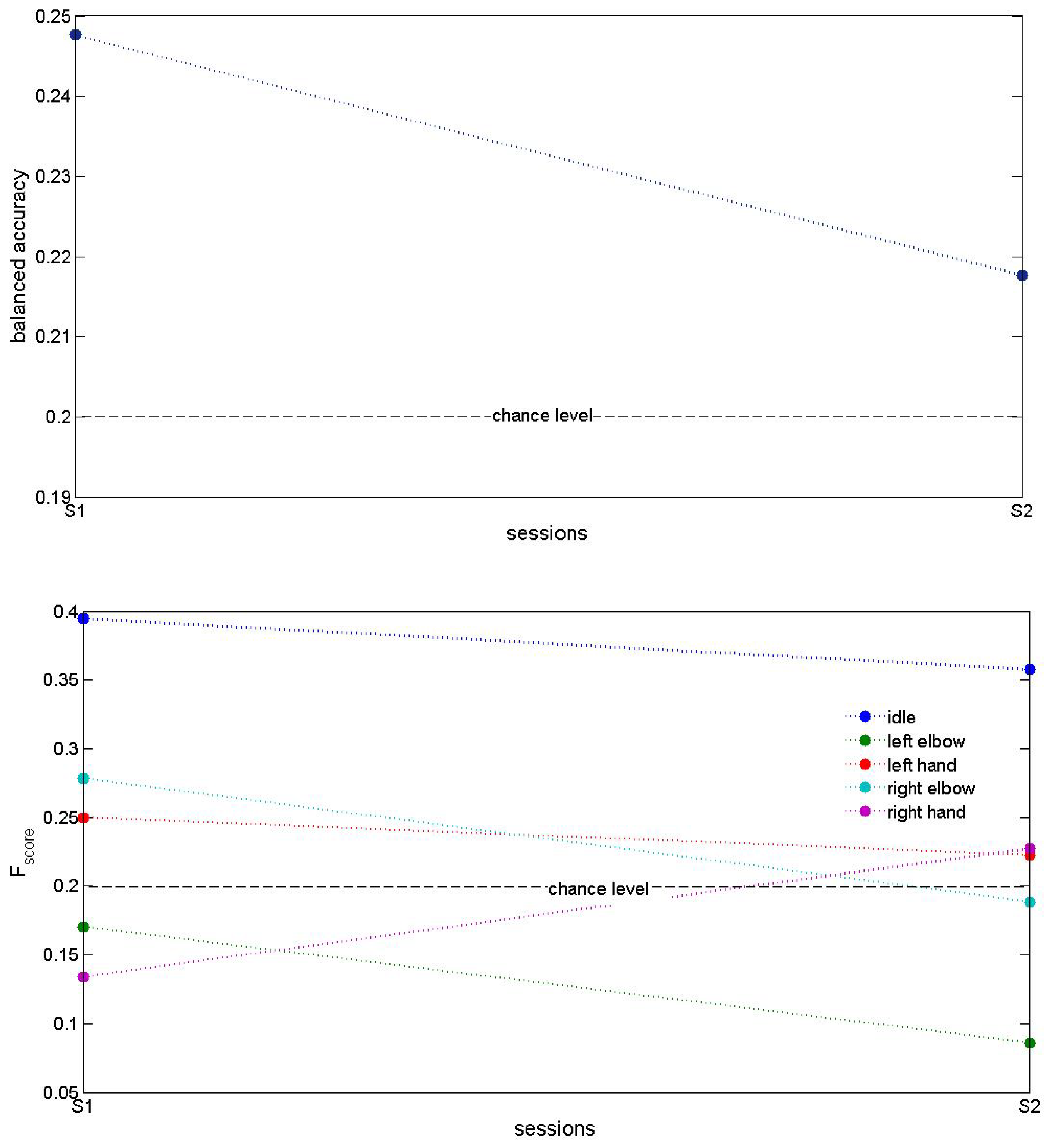
3. Results
- : Shifting up amplitudes of the ECoG signals.
- : Square Morlet transformation [33] of ECoG signals using 15 fundamental frequencies (from 10 to 150 Hz with a step of 10 Hz).
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rudroff, T. Decoding thoughts, encoding ethics: A narrative review of the BCI-AI revolution. Brain Res. 2025, 1850, 149423. [Google Scholar] [CrossRef] [PubMed]
- Sumithra, M.G.; Dhanaraj, R.K.; Milanova, M.; Balusamy, B.; Venkatesan, C. (Eds.) Brain-Computer Interface: Using Deep Learning Applications; Wiley: Hoboken, NJ, USA, 2023. [Google Scholar]
- Awuah, W.A.; Ahluwalia, A.; Darko, K.; Sanker, V.; Tan, J.K.; Pearl, T.O.; Ben-Jaafar, A.; Ranganathan, S.; Aderinto, N.; Mehta, A.; et al. Bridging Minds and Machines: The Recent Advances of Brain-Computer Interfaces in Neurological and Neurosurgical Applications. World Neurosurg. 2024, 189, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Mukherjee, M.; Bakshi, A. A Review of Brain-Computer Interface. In Advances in Medical Physics and Healthcare Engineering; Mukherjee, M., Mandal, J., Bhattacharyya, S., Huck, C., Biswas, S., Eds.; Lecture Notes in Bioengineering; Springer Nature: Berlin/Heidelberg, Germany, 2021; pp. 507–531. [Google Scholar]
- Ajiboye, A.B.; Willett, F.R.; Young, D.R.; Memberg, W.D.; Murphy, B.A.; Miller, J.P.; Walter, B.L.; Sweet, J.A.; Hoyen, H.A.; Keith, M.W.; et al. Restoration of reaching and grasping movements through brain-controlled muscle stimulation in a person with tetraplegia: A proof-of-concept demonstration. Lancet 2017, 389, 1821–1830. [Google Scholar] [CrossRef]
- Buttfield, A.; Ferrez, P.W.; Millan, J.R. Towards a robust BCI: Error potentials and online learning. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Eliseyev, A.; Auboiroux, V.; Costecalde, T.; Langar, L.; Charvet, G.; Mestais, C.; Aksenova, T.; Benabid, A.L. Recursive Exponentially Weighted N-way Partial Least Squares Regression with Recursive-Validation of Hyper-Parameters in Brain-Computer Interface Applications. Sci. Rep. 2017, 7, 16281. [Google Scholar] [CrossRef]
- Orsborn, A.L.; Moorman, H.G.; Overduin, S.A.; Shanechi, M.M.; Dimitrov, D.F.; Carmena, J.M. Closed-Loop Decoder Adaptation Shapes Neural Plasticity for Skillful Neuroprosthetic Control. Neuron 2014, 82, 1380–1393. [Google Scholar] [CrossRef]
- Wodlinger, B.; Downey, J.E.; Tyler-Kabara, E.C.; Schwartz, A.B.; Boninger, M.L.; Collinger, J.L. Ten-dimensional anthropomorphic arm control in a human brain-machine interface: Difficulties, solutions, and limitations. J. Neural Eng. 2014, 12, 016011. [Google Scholar] [CrossRef]
- Benabid, A.L.; Costecalde, T.; Eliseyev, A.; Charvet, G.; Verney, A.; Karakas, S.; Foerster, M.; Lambert, A.; Morinière, B.; Abroug, N.; et al. An exoskeleton controlled by an epidural wireless brain–machine interface in a tetraplegic patient: A proof-of-concept demonstration. Lancet Neurol. 2019, 18, 1112–1122. [Google Scholar] [CrossRef]
- Lorach, H.; Galvez, A.; Spagnolo, V.; Martel, F.; Karakas, S.; Intering, N.; Vat, M.; Faivre, O.; Harte, C.; Komi, S.; et al. Walking naturally after spinal cord injury using a brain–spine interface. Nature 2023, 618, 126–133. [Google Scholar] [CrossRef]
- Moly, A.; Costecalde, T.; Martel, F.; Martin, M.; Larzabal, C.; Karakas, S.; Verney, A.; Charvet, G.; Chabardes, S.; Benabid, A.L.; et al. An adaptive closed-loop ECoG decoder for long-term and stable bimanual control of an exoskeleton by a tetraplegic. J. Neural Eng. 2022, 19, 026021. [Google Scholar] [CrossRef] [PubMed]
- Furber, S.B.; Galluppi, F.; Temple, S.; Plana, L.A. The SpiNNaker Project. Proc. IEEE 2014, 102, 652–665. [Google Scholar] [CrossRef]
- Davies, M.; Srinivasa, N.; Lin, T.-H.; Chinya, G.; Joshi, P.; Lines, A.; Wild, A.; Wang, H.; Mathaikutty, D. Loihi: A Neuromorphic Manycore Processor with On-Chip Learning. IEEE Micro. 2018, 38, 82–99. [Google Scholar] [CrossRef]
- Kasabov, N. Time-Space, Spiking Neural Networks and Brain-Inspired Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2018; 750p. [Google Scholar]
- Costa, F.; Schaft, E.V.; Huiskamp, G.; Aarnoutse, E.J.; van’t Klooster, M.A.; Krayenbühl, N.; Ramantani, G.; Zijlmans, M.; Indiveri, G.; Sarnthein, J. Robust compression and detection of epileptiform patterns in ECoG using a real-time spiking neural network hardware framework. Nat. Commun. 2024, 15, 3255. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-H.; Jeong, J. An electrocorticographic decoder for arm movement for brain–machine interface using an echo state network and Gaussian readout. Appl. Soft Comput. 2022, 117, 108393. [Google Scholar] [CrossRef]
- Koprinkova-Hristova, P.; Penkov, D.; Nedelcheva, S.; Yordanov, S.; Kasabov, N. On-line Learning, Classification and Interpretation of Brain Signals using 3D SNN and ESN. In Proceedings of the International Joint Conference on Neural Networks, Gold Coast, Australia, 18–23 June 2023. art. no. 23518355. [Google Scholar]
- Blumberg, J.; Rickert, J.; Waldert, S.; Schulze-Bonhage, A.; Aertsen, A.; Mehring, C. Adaptive Classification for Brain Computer Interfaces. In Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; pp. 2536–2539. [Google Scholar]
- Gürel, T.; Mehring, C. Unsupervised adaptation of brain machine interface decoders. Front. Neurosci. 2012, 6, 164. [Google Scholar] [CrossRef]
- Llera, A.; Gómez, V.; Kappen, H.J. Adaptive Classification on Brain-Computer Interfaces Using Reinforcement Signals. Neural Comput. 2012, 24, 2900–2923. [Google Scholar] [CrossRef] [PubMed]
- Rouanne, V.; Śliwowski, M.; Costecalde, T.; Benabid, A.; Aksenova, T. Detection of Error Correlates in the Motor Cortex in a Long Term Clinical Trial of ECoG based Brain Computer Interface. In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021)–Volume 4: BIOSIGNALS, Online, 11–13 February 2021; pp. 26–34. [Google Scholar]
- Spüler, M.; Walter, A.; Ramos-Murguialday, A.; Naros, G.; Birbaumer, N.; Gharabaghi, A.; Rosenstiel, W.; Bogdan, M. Decoding of motor intentions from epidural ECoG recordings in severely paralyzed chronic stroke patients. J. Neural Eng. 2014, 11, 066008. [Google Scholar] [CrossRef] [PubMed]
- Zeyl, T.J.; Chau, T. A case study of linear classifiers adapted using imperfect labels derived from human event-related potentials. Pattern Recognit. Lett. 2014, 37, 54–62. [Google Scholar] [CrossRef]
- Kasabov, N.K. NeuCube: A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Netw. 2014, 52, 62–76. [Google Scholar] [CrossRef]
- Gütig, R.; Aharonov, R.; Rotter, S.; Sompolinsky, H. Learning input correlations through nonlinear temporally asymmetric hebbian plasticity. J. Neurosci. 2003, 23, 3697–3714. [Google Scholar] [CrossRef]
- Jaeger, H. Tutorial on Training Recurrent Neural Networks, Covering BPPT, RTRL, EKF and the “Echo State Network” Approach; GMD Report 159, German National Research Center for Information Technology; GMD-Forschungszentrum Informationstechnik: Bonn, Germany, 2002. [Google Scholar]
- Rouanne, V. Adaptation of Discrete and Continuous Intracranial Brain-Computer Interfaces Using Neural Correlates of Task Performance Decoded Continuously From the Sensorimotor Cortex of a Tetraplegic. Ph.D. Thesis, Université Grenoble Alpes, Saint-Martin-d’Hères, France, 2022. [Google Scholar]
- Rouanne, V.; Costecalde, T.; Benabid, A.L.; Aksenova, T. Unsupervised adaptation of an ECoG based brain-computer interface using neural correlates of task performance. Sci. Rep. 2022, 12, 21316. [Google Scholar] [CrossRef] [PubMed]
- Andrew, B.; Richard, S.S. Reinforcement Learning: An Introduction, 2nd ed.; The MIT Press: Cambridge, MA, USA; London, UK, 2018. [Google Scholar]
- Spreizer, S.; Mitchell, J.; Gutzen, R.; Lober, M.; Linssen, C.; Trensch, G.; Jordan, J.; Plesser, H.E.; Kurth, A.; Vennemo, S.B.; et al. 2022. NEST 3.3 (3.3). Windows. Zenodo. Available online: https://zenodo.org/records/6368024 (accessed on 9 March 2025).
- Mestais, C.S.; Charvet, G.; Sauter-Starace, F.; Foerster, M.; Ratel, D.; Benabid, A.L. WIMAGINE: Wireless 64-channel ECoG recording implant for long term clinical applications. IEEE Trans. Neural Syst. Rehabil. Eng. 2015, 23, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Morlet, J. Sampling Theory and Wave Propagation. In Issues in Acoustic Signal—Image Processing and Recognition; NATO ASI Series; Chen, C.H., Ed.; Springer: Berlin/Heidelberg, Germany, 1983; Volume 1. [Google Scholar]




| Hyperparameter | Value |
|---|---|
| 3D-SNN | |
| membrane potential threshold | −65.0 mV |
| refractory time | 0 ms |
| STDP learning rate | 0.001 |
| ESN | |
| leaking rate a | 0.5 |
| number of reservoir neurons | 15,000 |
| sparsity of reservoir connectivity matrix | 0.5 |
| Model | Mean Balanced Accuracy | STD of Balanced Accuracy | Mean on Idle | STD of on Idle | Mean on Walk | STD of on Walk |
|---|---|---|---|---|---|---|
| Raw signal | 0.534 | 0.022 | 0.755 | 0.043 | 0.315 | 0.011 |
| Morlet 10 Hz | 0.587 | 0.016 | 0.776 | 0.026 | 0.399 | 0.031 |
| Morlet 20 Hz | 0.615 | 0.022 | 0.777 | 0.047 | 0.447 | 0.012 |
| Morlet 30 Hz | 0.626 | 0.007 | 0.787 | 0.022 | 0.460 | 0.032 |
| Morlet 40 Hz | 0.623 | 0.017 | 0.785 | 0.045 | 0.457 | 0.008 |
| Morlet 50 Hz | 0.603 | 0.026 | 0.773 | 0.060 | 0.431 | 0.024 |
| Morlet 60 Hz | 0.599 | 0.023 | 0.772 | 0.047 | 0.422 | 0.044 |
| Morlet 70 Hz | 0.606 | 0.021 | 0.771 | 0.041 | 0.435 | 0.015 |
| Morlet 80 Hz | 0.598 | 0.021 | 0.778 | 0.040 | 0.417 | 0.034 |
| Morlet 90 Hz | 0.583 | 0.022 | 0.771 | 0.053 | 0.396 | 0.005 |
| Morlet 100 Hz | 0.571 | 0.025 | 0.754 | 0.063 | 0.383 | 0.024 |
| Morlet 110 Hz | 0.580 | 0.032 | 0.762 | 0.066 | 0.395 | 0.038 |
| Morlet 120 Hz | 0.595 | 0.022 | 0.779 | 0.055 | 0.409 | 0.041 |
| Morlet 130 Hz | 0.605 | 0.011 | 0.776 | 0.056 | 0.430 | 0.025 |
| Morlet 140 Hz | 0.600 | 0.015 | 0.778 | 0.054 | 0.420 | 0.011 |
| Morlet 150 Hz | 0.592 | 0.044 | 0.780 | 0.062 | 0.409 | 0.036 |
| Voting all | 0.527 | 0.005 | 0.844 | 0.025 | 0.120 | 0.019 |
| Voting chosen | 0.641 | 0.013 | 0.826 | 0.044 | 0.474 | 0.006 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rusev, G.; Yordanov, S.; Nedelcheva, S.; Banderov, A.; Sauter-Starace, F.; Koprinkova-Hristova, P.; Kasabov, N. Decoding Brain Signals in a Neuromorphic Framework for a Personalized Adaptive Control of Human Prosthetics. Biomimetics 2025, 10, 183. https://doi.org/10.3390/biomimetics10030183
Rusev G, Yordanov S, Nedelcheva S, Banderov A, Sauter-Starace F, Koprinkova-Hristova P, Kasabov N. Decoding Brain Signals in a Neuromorphic Framework for a Personalized Adaptive Control of Human Prosthetics. Biomimetics. 2025; 10(3):183. https://doi.org/10.3390/biomimetics10030183
Chicago/Turabian StyleRusev, Georgi, Svetlozar Yordanov, Simona Nedelcheva, Alexander Banderov, Fabien Sauter-Starace, Petia Koprinkova-Hristova, and Nikola Kasabov. 2025. "Decoding Brain Signals in a Neuromorphic Framework for a Personalized Adaptive Control of Human Prosthetics" Biomimetics 10, no. 3: 183. https://doi.org/10.3390/biomimetics10030183
APA StyleRusev, G., Yordanov, S., Nedelcheva, S., Banderov, A., Sauter-Starace, F., Koprinkova-Hristova, P., & Kasabov, N. (2025). Decoding Brain Signals in a Neuromorphic Framework for a Personalized Adaptive Control of Human Prosthetics. Biomimetics, 10(3), 183. https://doi.org/10.3390/biomimetics10030183








