Purification and Epitope Mapping of Jug r 4, a Major Walnut Allergen
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Purification of Jug r 4
3.2. Westsern Blot with Walnut-Allergic Serum IgE, Anti-Ara h 3, and Anti-Jug r 4 Polyclonal Antibodies
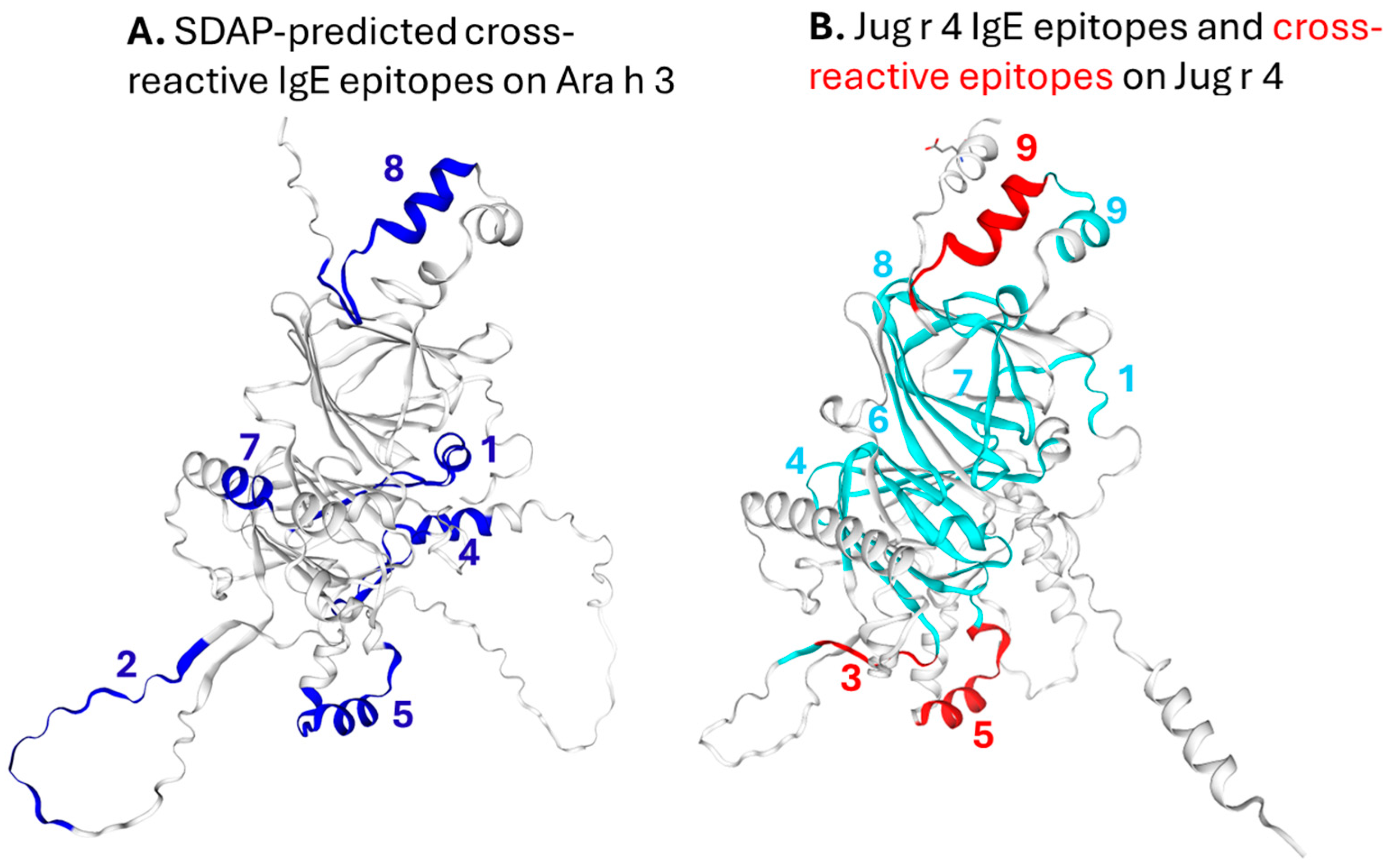
3.3. SDAP-Predicted Epitopes
3.4. IgE Epitopes and Homology Modeling of Ara h 3 and Jug r 4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, R.S.; Warren, C.M.; Smith, B.M.; Jiang, J.; Blumenstock, J.A.; Davis, M.M.; Schleimer, R.P.; Nadeau, K.C. Prevalence and Severity of Food Allergies Among US Adults. JAMA Netw. Open 2019, 2, e185630. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fleischer, D.M.; Conover-Walker, M.K.; Matsui, E.C.; Wood, R.A. The natural history of tree nut allergy. J. Allergy Clin. Immunol. 2005, 116, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, V.; Koplin, J.; Lodge, C.; Tang, M.; Dharmage, S.; Allen, K. The Prevalence of Tree Nut Allergy: A Systematic Review. Curr. Allergy Asthma Rep. 2015, 15, 54. [Google Scholar] [CrossRef] [PubMed]
- Wallowitz, M.; Peterson, W.R.; Uratsu, S.; Comstock, S.S.; Dandekar, A.M.; Teuber, S.S. Jug r 4, a legumin group food allergen from walnut (Juglans regia Cv. Chandler). J. Agric. Food Chem. 2006, 54, 8369–8375. [Google Scholar] [CrossRef] [PubMed]
- Sicherer, S.H.; Furlong, T.J.; Munoz-Furlong, A.; Burks, A.W.; Sampson, H.A. A voluntary registry for peanut and tree nut allergy: Characteristics of the first 5149 registrants. J. Allergy Clin. Immunol. 2001, 108, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Burney, P.G.; Potts, J.; Kummeling, I.; Mills, E.N.; Clausen, M.; Dubakiene, R.; Barreales, L.; Fernandez-Perez, C.; Fernandez-Rivas, M.; Le, T.M.; et al. The prevalence and distribution of food sensitization in European adults. Allergy 2014, 69, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jeong, K.; Jeon, S.A.; Lee, S. Component resolved diagnosis of walnut allergy in young children: Jug r 1 as a major walnut allergen. Asian Pac. J. Allergy Immunol. 2021, 39, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.; Verhoeckx, K.; Knulst, A.; Welsing, P.; de Jong, A.; Gaspari, M.; Ehlers, A.; Verhoeff, P.; Houben, G.; Le, T.M. Co-sensitization between legumes is frequently seen, but variable and not always clinically relevant. Front. Allergy 2023, 4, 1115022. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ballmer-Weber, B.K.; Lidholm, J.; Lange, L.; Pascal, M.; Lang, C.; Gernert, S.; Lozano-Blasco, J.; Gräni, N.; Guillod, C.; Wangorsch, A.; et al. Allergen Recognition Patterns in Walnut Allergy Are Age Dependent and Correlate with the Severity of Allergic Reactions. J. Allergy Clin. Immunol. Pract. 2019, 7, 1560–1567.e6. [Google Scholar] [CrossRef] [PubMed]
- Verweij, M.M.; Hagendorens, M.M.; De Knop, K.J.; Bridts, C.H.; De Clerck, L.S.; Stevens, W.J.; Ebo, D.G. Young infants with atopic dermatitis can display sensitization to Cor a 9, an 11S legumin-like seed-storage protein from hazelnut (Corylus avellana). Pediatr. Allergy Immunol. 2011, 22, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, G.N.; Baker, M.G.; Yu, J.; Ford, L.S.; Bencharitiwong, R.; Grishina, G.; Sampson, H.A.; Sicherer, S.; Nowak-Wegrzyn, A. Citrin: A novel food allergen in citrus seeds and citrus-derived pectin that shows cross-reactivity with cashew and pistachio. Ann. Allergy Asthma Immunol. 2023, 131, 759–765.e3. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, L.P.; Wang, R.Q.; Zhu, L.; Chen, F.; Hou, Y.; Ni, K.; Deng, S.; Liu, S.; Ying, W.; et al. Identification, Characterization, Cloning, and Cross-Reactivity of Zan b 2, a Novel Pepper Allergen of 11S Legumin. J. Agric. Food Chem. 2024, 72, 8189–8199. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hurlburt, B.K.; McBride, J.K.; Nesbit, J.B.; Ruan, S.; Maleki, S.J. Purification of Recombinant Peanut Allergen Ara h 1 and Comparison of IgE Binding to the Natural Protein. Foods 2014, 3, 642–657. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Waterhouse, A.M.; Studer, G.; Robin, X.; Bienert, S.; Tauriello, G.; Schwede, T. The structure assessment web server: For proteins, complexes and more. Nucleic Acids Res. 2024, 52, W318–W323. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schein, C.H.; Ivanciuc, O.; Braun, W. Structural Database of Allergenic Proteins (SDAP). In Food Allergy; Maleki, S.J., Burks, A.W., Helm, R.M., Eds.; ASM Press: Washington, DC, USA, 2006; pp. 257–283. [Google Scholar]
- Ivanciuc, O.; Oezguen, N.; Mathura, V.S.; Schein, C.H.; Xu, Y.; Braun, W. Using property based sequence motifs and 3D modeling to determine structure and functional regions of proteins. Curr. Med. Chem. 2004, 11, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Ivanciuc, O.; Schein, C.H.; Braun, W. SDAP: Database and computational tools for allergenic proteins. Nucleic Acids Res. 2003, 31, 359–362. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sharma, E.; Vitte, J. A systematic review of allergen cross-reactivity: Translating basic concepts into clinical relevance. J. Allergy Clin. Immunol. Glob. 2024, 3, 100230. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wallowitz, M.L.; Chen, R.J.Y.; Tzen, J.T.C.; Teuber, S.S. Ses i 6, the sesame 11S globulin, can activate basophils and shows cross-reactivity with walnut in vitro. Clin. Exp. Allergy 2007, 37, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Jacquet, G.; Sordet, C.; Culerrier, R.; Rougé, P. Homology modelling and conformational analysis of IgE-binding epitopes of Ara h 3 and other legumin allergens with a cupin fold from tree nuts. Mol. Immunol. 2007, 44, 3243–3255. [Google Scholar] [CrossRef] [PubMed]
- Blankestijn, M.A.; den Hartog Jager, C.F.; Blom, W.M.; Otten, H.G.; de Jong, G.A.H.; Gaspari, M.; Houben, G.F.; Knulst, A.C.; Verhoeckx, K.C.M. A subset of walnut allergic adults is sensitized to walnut 11S globulin Jug r 4. Clin. Exp. Allergy 2018, 48, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Guo, F.; Chen, Y.W.; Howard, A.; Zhang, Y.Z. Crystal structure of Ara h 3, a major allergen in peanut. Mol. Immunol. 2009, 46, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Robotham, J.M.; Hoffman, G.G.; Teuber, S.S.; Beyer, K.; Sampson, H.A.; Sathe, S.K.; Roux, K.H. Linear IgE-epitope mapping and comparative structural homology modeling of hazelnut and English walnut 11S globulins. Mol. Immunol. 2009, 46, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Teuber, S.; Peterson, W.; Uratsu, S.; Dandekar, A.; Roux, K.; Sathe, S. Identification and cloning of Jug r 4, a major food allergen from English walnut belonging to the legumin group. J. Allergy Clin. Immunol. 2003, 2, S248. [Google Scholar] [CrossRef]
- Restani, P.; Ballabio, C.; Corsini, E.; Fiocchi, A.; Isoardi, P.; Magni, C.; Poiesi, C.; Terracciano, L.; Duranti, M. Identification of the basic subunit of Ara h 3 as the major allergen in a group of children allergic to peanuts. Ann. Allergy Asthma Immunol. 2005, 94, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Rabjohn, P.; Burks, A.W.; Sampson, H.A.; Bannon, G.A. Mutational analysis of the IgE-binding epitopes of the peanut allergen, Ara h 3: A member of the glycinin family of seed-storage proteins. J. Allergy Clin. Immunol. 1999, 103, S101. [Google Scholar]



| Subject | Age | Sex | Peanut | Walnut | Cashew | Almond | Hazelnut | Pecan | Pistachio | Other |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 14 | M | A | x | x | x | sesame, brazil nut, pine nut, garbanzo | |||
| 2. | 19 | F | A | x | x | x | x | brazil nut, pine nut, shellfish, fish | ||
| 3. | 11 | M | x | x | sesame, milk | |||||
| 4. | 8 | ? | A | x | x | x | x | pine nut, brazil nut | ||
| 5. | 31 | F | x | x | x | x | x | pine nut | ||
| 6. | 21 | F | A | x | x | x | x | x | x | macadamia, egg, milk |
| 7. | 24 | M | x | x | x | x | x | brazil nut | ||
| 8. | 11 | F | x | x | x | x | x | x | x | macadamia nut, milk, egg, brazil, pine |
| 9. | 21 | F | LT | LT | x | |||||
| 10. | 25 | F | LT | LT | LT | LT | LT | ne | x | brazil nut |
| 11. | 63 | F | LT | LT | x | x | x | LT | ne | brazil nut, potato, egg |
| 12. | n | F | x | x | sunflower | |||||
| 13. | 38 | M | x | x | x | x | coconut | |||
| 14. | 42 | F | x | x | ne | ne | ne | x | macadamia, brazil nut, | |
| 15. | 16 | M | x | x | x | x | x | x | x | crab, lobster, flax, soy |
| 16. | 35 | F | x | x | x | x | x | x | x | soy |
| 17. | 25 | F | x | x | x | x | x | x | x | poppy, coconut, fruits, shellfish |
| 18. | 26 | M | x | x | x | x | x | x | x | pine nut, crab, lobster |
| 19. | 25 | x | x | x | ne | ne | x | |||
| 20. | 46 | M | x | x | x | x | x | x | x | brazil, pine, macadamia, shellfish, fish |
| 21. | 25 | F | x | x | x | ne | x | ne | x | pea, soy, sesame |
| 22. | 9 | M | x | x | x | |||||
| 23. | 22 | M | x | |||||||
| 24. | 25 | F | A | soy | ||||||
| 25. | 13 | M | A | x | sesame | |||||
| 26. | 15 | M | x | x | x | outgrew egg | ||||
| 27. | 10 | M | x | |||||||
| 28. | 35 | x | ||||||||
| 29. | 19 | x | ||||||||
| 30. | 16 | M | x | soy | ||||||
| 31. | ch | Chicken serum negative control | ||||||||
| 32. | 23 | F | x | x | ||||||
| 33. | 9 | M | x |
| Allergen | NCBI | PD Sequence Similarity Index | Start Residue | Matching Region | End Residue |
|---|---|---|---|---|---|
| Ara h 3 Epitope 1 | |||||
| Ara h 3 | O82580 | 0.00 | 28 | GYIETWNPNNQEFECAGVAL | 47 |
| Jug r 4.0101 | Q2TPW5 | 2.94 | 55 | GVIESWDPNNQQFQCAGVAV | 74 |
| Ara h 3 Epitope 2—Cross-reactive with Jug r 4 Epitope 3 | |||||
| Ara h 3 | 3703107 | 0.00 | 91 | HYEEPHTQGRRSQSQR | 106 |
| Jug r 4.0101 | Q2TPW5 | 7.77 | 115 | TFEESQRQSQQGQSRE | 130 |
| Ara h 3 Epitope 3 QQQRDSHQKVHFDEGD—No low PD match to Jug r 4 | |||||
| Ara h 3 Epitope 4—Cross-reactive with Jug r 4 Epitope 5 | |||||
| Ara h 3 | O82580 | 0.00 | 170 | PRRFNLAGNTEQEFLRYQQQSRQ | 192 |
| Jug r 4.0101 | Q2TPW5 | 8.82 | 185 | PRNFYLAGNPDDEFRPQGQQEYE | 207 |
| Ara h 3 Epitope 5—Cross-reactive with previously identified Jug r 4 epitope | |||||
| Ara h 3 | 3703107 | 0.00 | 242 | IFSGFTPEFLEQAFQVD | 258 |
| Jug r 4.0101 | Q2TPW5 | 5.13 | 233 | VFSGFDADFLADAFNVD | 249 |
| Ara h 3 Epitope 6 VTVRGGLRILSPDRK—No low PD match to Jug r 4 | |||||
| Ara h 3 Epitope 7—Similar peptide found in Jug r 4, but with low likelihood of being cross-reactive | |||||
| Ara h 3 | O82580 | 0.00 | 300 | DEDEYEYDEEDRRRGRGSR | 318 |
| Jug r 4.0101 | Q2TPW5 | 9.40 | 294 | RERERESESERRQSRRGGR | 312 |
| Ara h 3 Epitope 8—Similar peptide found in Jug r 4 Epitope 9, but with low likelihood of being cross-reactive | |||||
| Ara h 3 | O82580 | 0.00 | 480 | REQARQLKNNNPFKFFVPP | 498 |
| Jug r 4.0101 | Q2TPW5 | 9.87 | 473 | REDARRLKFNRQESTLVRS | 491 |
| Jug r 4 Epitope 1 CQLNRLDALEPTNRI—Similar peptide found in Ara h 3, but no IgE binding detected on microarray | |||||
| Jug r 4 Epitope 3—Cross-reactive with Ara h 3 Epitope 2 | |||||
| Jug r 4.0101 | Q2TPW5 | 0.00 | 111 | GCPETFEESQRQSQQGQSR | 129 |
| Ara h 3 | 3703107 | 6.54 | 87 | GCPRHYEEPHTQGRRSQSQ | 105 |
| Jug r 4 Epitope 4—Cross-reactive with peptide found in Ara h 3 | |||||
| Jug r 4.0101 | Q2TPW5 | 0.00 | 149 | AFPAGVAHWSYNDGSNP | 165 |
| Ara h 3 | 3703107 | 7.55 | 137 | AVPTGVAFWLYNDHDTD | 153 |
| Jug r 4 Epitope 6 ISTVNSHTLPVLRWL—Similar peptide found in Ara h 3, but with low likelihood of being cross-reactive | |||||
| Jug r 4 Epitope 9—Microarray-confirmed cross reactivity with Ara h 3 Epitope 8 despite elevated PD score. | |||||
| Jug r 4.0101 | Q2TPW5 | 0.00 | 465 | LATAFQIPREDARRLKFNRQESTLVR | 490 |
| Ara h 3 | 3703107 | 10.36 | 475 | VANSYGLQREQARQLKNNNPFKFFVP | 500 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gipson, S.A.Y.; Nesbit, J.B.; Swientoniewski, L.T.; Rogers, S.I.; Mustafa, S.S.; Dreskin, S.C.; Teuber, S.S.; Cheng, H.; Maleki, S.J. Purification and Epitope Mapping of Jug r 4, a Major Walnut Allergen. Allergies 2025, 5, 8. https://doi.org/10.3390/allergies5010008
Gipson SAY, Nesbit JB, Swientoniewski LT, Rogers SI, Mustafa SS, Dreskin SC, Teuber SS, Cheng H, Maleki SJ. Purification and Epitope Mapping of Jug r 4, a Major Walnut Allergen. Allergies. 2025; 5(1):8. https://doi.org/10.3390/allergies5010008
Chicago/Turabian StyleGipson, Stephen A. Y., Jacqueline B. Nesbit, Lauren T. Swientoniewski, Stephen I. Rogers, S. Shahzad Mustafa, Stephen C. Dreskin, Suzanne S. Teuber, Hsiaopo Cheng, and Soheila J. Maleki. 2025. "Purification and Epitope Mapping of Jug r 4, a Major Walnut Allergen" Allergies 5, no. 1: 8. https://doi.org/10.3390/allergies5010008
APA StyleGipson, S. A. Y., Nesbit, J. B., Swientoniewski, L. T., Rogers, S. I., Mustafa, S. S., Dreskin, S. C., Teuber, S. S., Cheng, H., & Maleki, S. J. (2025). Purification and Epitope Mapping of Jug r 4, a Major Walnut Allergen. Allergies, 5(1), 8. https://doi.org/10.3390/allergies5010008






