Difficulties in Kinship Analysis for Victims’ Identification in Armed Conflicts
Abstract
1. Introduction
Armed Conflicts and Victim Identification
2. Civil and Family Involvement in the Identification Process
3. The International Commission for Mission Persons—ICMP
4. An Attempt to Harmonize Human Identification—Interpol DVI Protocol
- Biological samples for genetic identification—DNA sources can be medical samples, personal belongings, or relatives of the victim.
- Surgical data—provision of a detailed medical history.
- Dental data—including the facilitation of contact with the last dentist that the victim visited, to obtain an updated dental profile.
- Affiliation data—including the National Identity Document, driver’s wallet, passport, etc.
- Detailed physical description, including scars, moles, malformations, tattoos, etc.
- A detailed description of the clothing he was wearing at the time of his death and his personal effects (piercings, earrings, glasses, rings, etc.)
5. Types of Identification in Armed Conflicts
5.1. Fingerprint Techniques
5.2. Forensic Odontology
5.3. Genetic Identification in Armed Conflicts
- -
- Evaluation of postmortem samples;
- -
- Evaluation and analysis of antemortem samples;
- -
- Family member’s sample(s) selection;
- -
- DNA extraction, quantification, and genotyping;
- -
- Statistical evaluation.
5.3.1. Direct Identification
5.3.2. Indirect Identification
- (a)
- Autosomal markers
- (b)
- X-chromosomal markers
- (c)
- Lineage markers
- (c1)
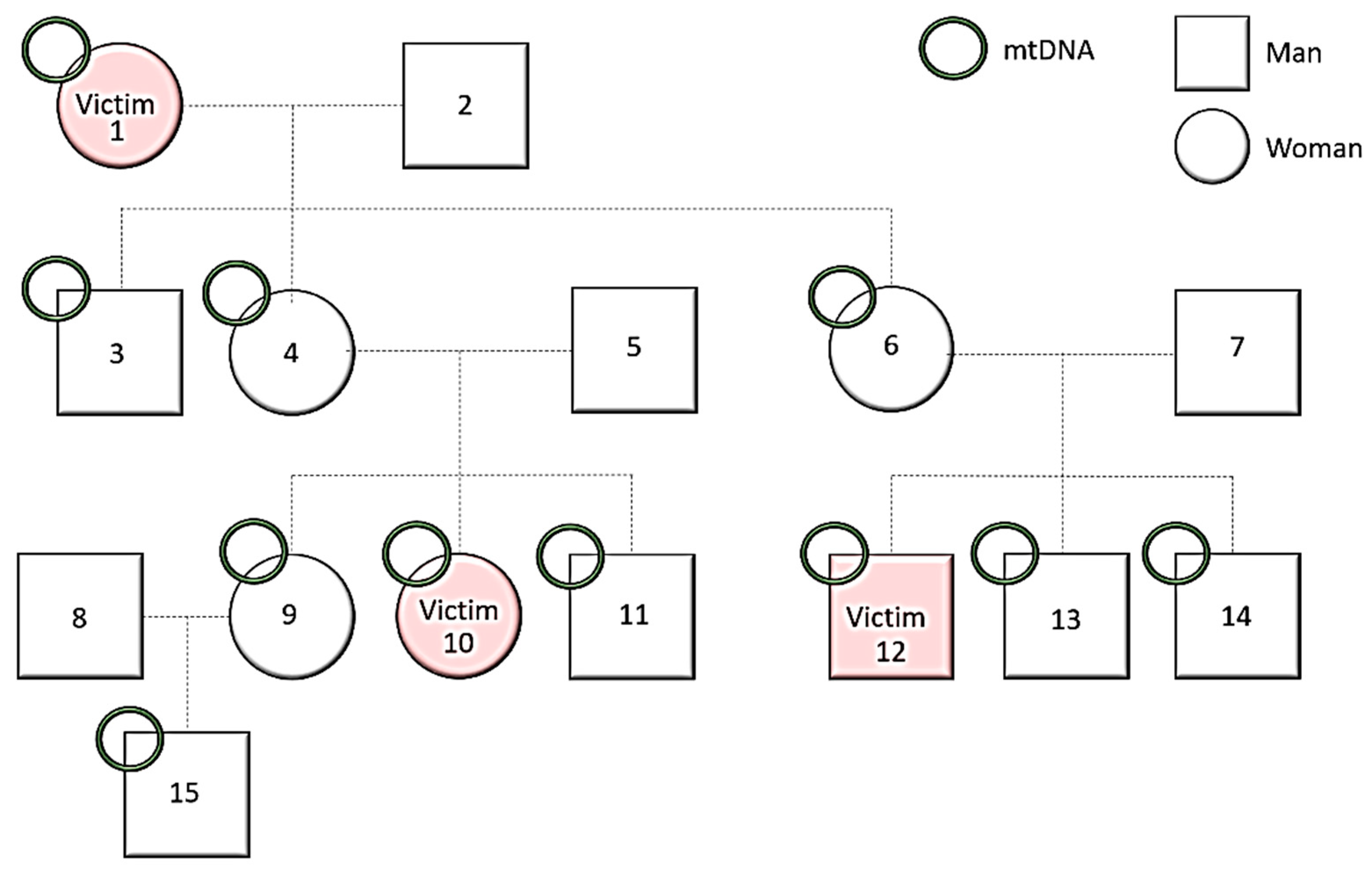
- mtDNA
- (c2)
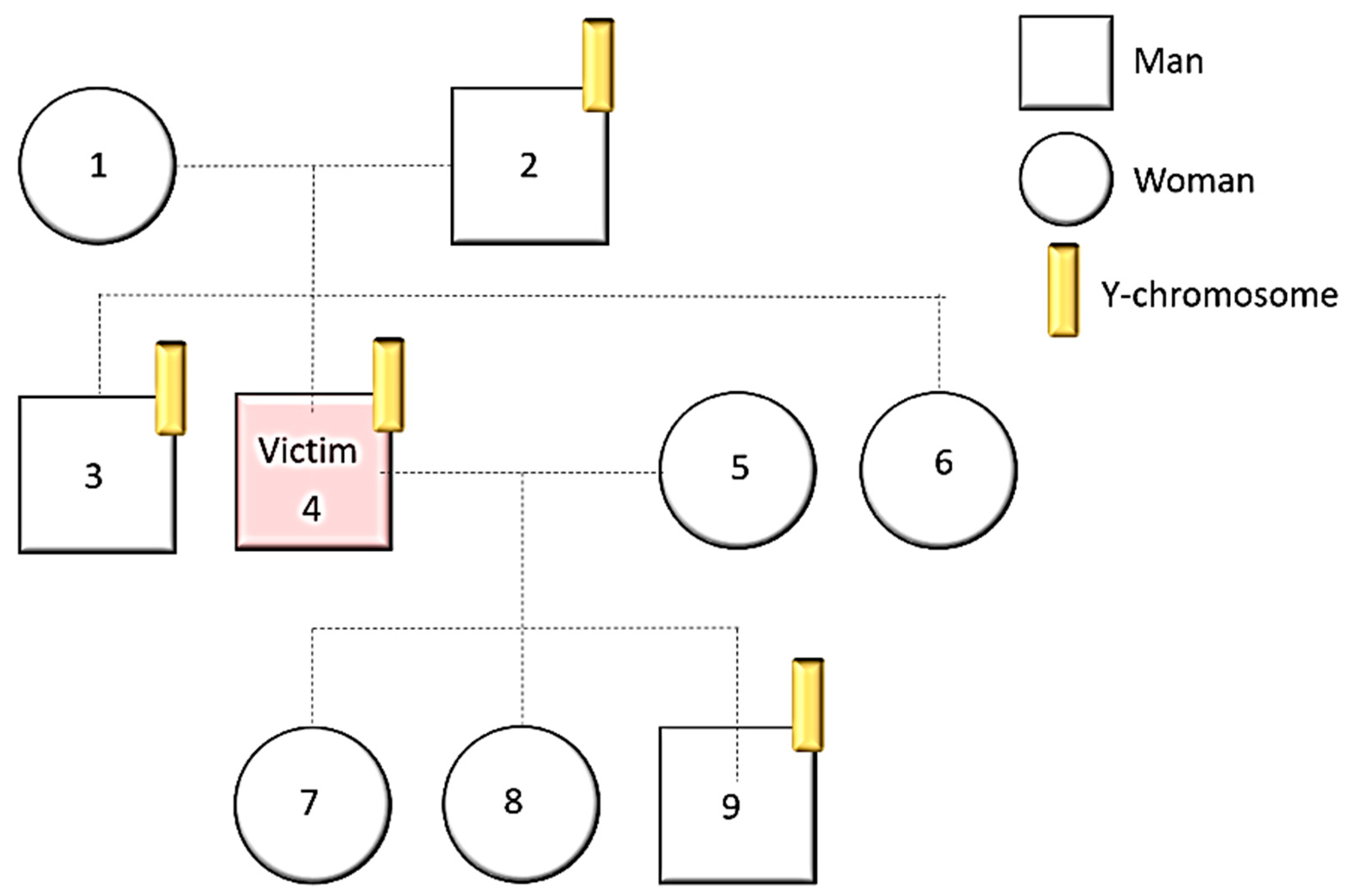
- Y-Chromosome
5.3.3. New Trends in Forensic Genetics
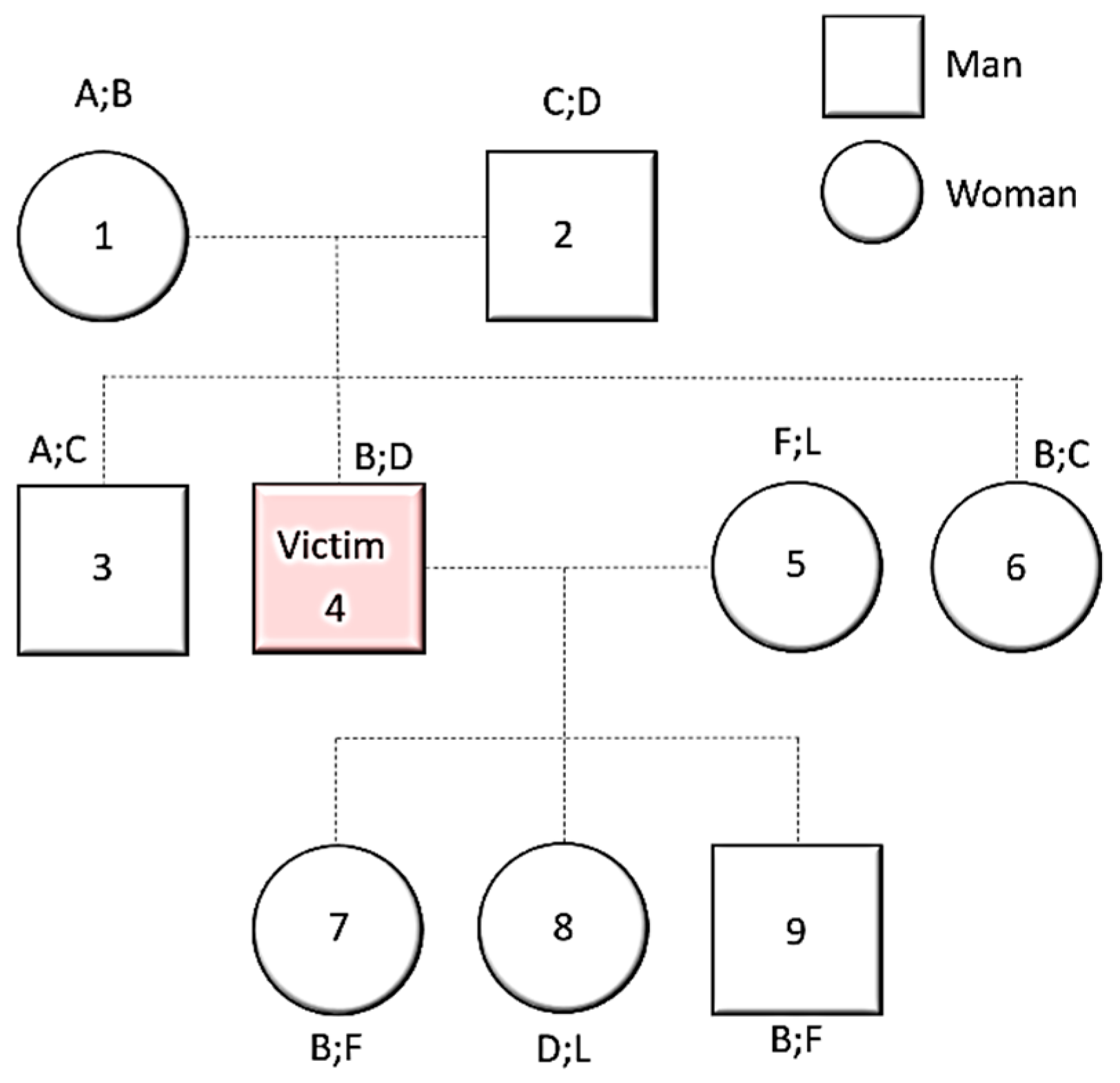
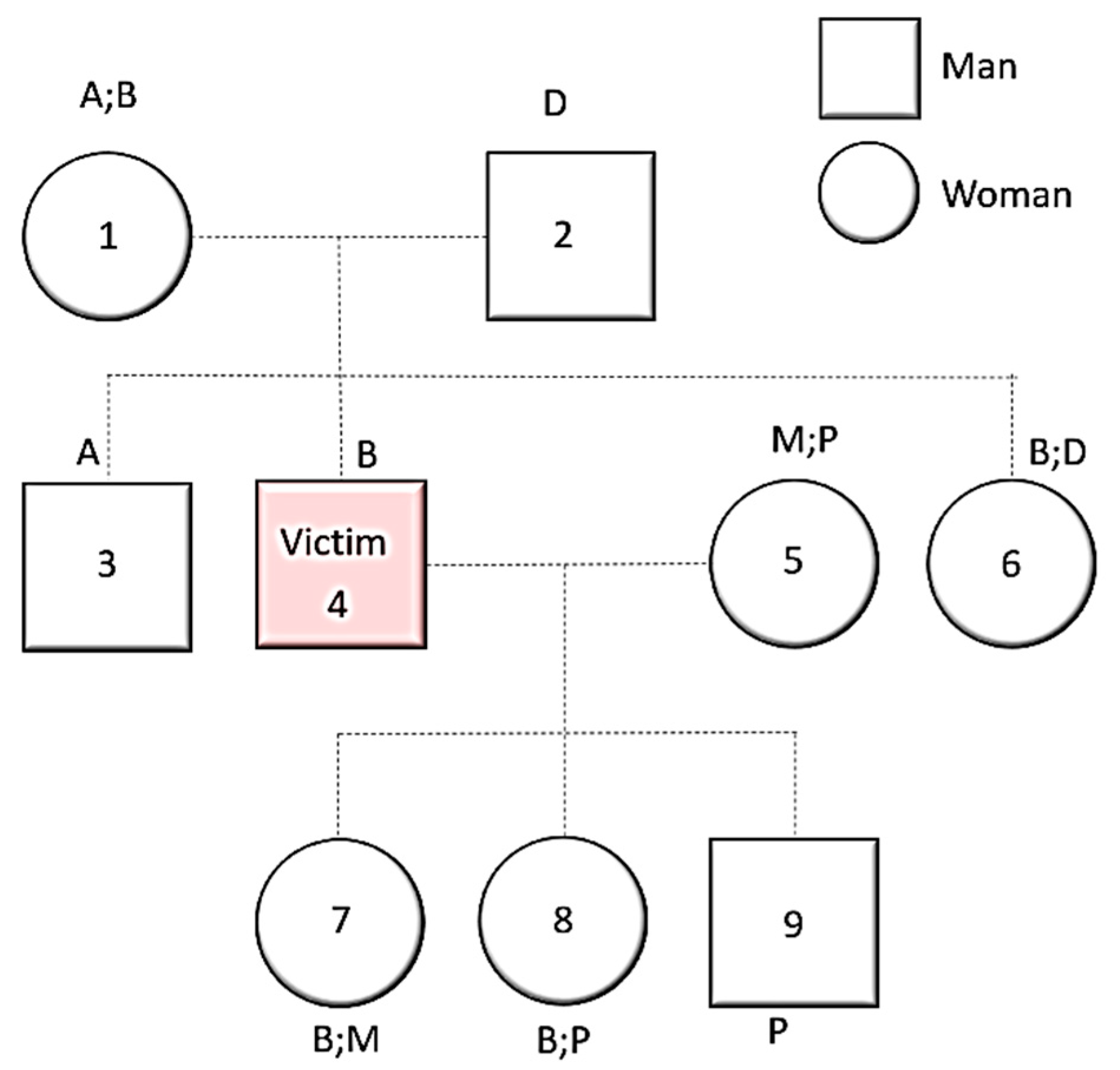
6. Difficulties in Victims’ Genetic Identification in Armed Conflicts
DNA Degradation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adnan, Atif, Allah Rakha, Hayder Lazim, Shahid Nazir, Wedad Saeed Al-Qahtani, Maha Abdullah Alwaili, Sibte Hadi, and Chuan-Chao Wang. 2022. Are Roma People Descended from the Punjab Region of Pakistan: A Y-Chromosomal Perspective. Genes 13: 532. [Google Scholar] [CrossRef]
- Adnan, Atif, Kaidirina Kasimu, Allah Rakha, Guanglin He, Tongya Yang, Chuan-Chao Wang, Jie Lu, and Jin-feng Xuan. 2020. Comprehensive genetic structure analysis of Han population from Dalian City revealed by 20 Y-STRs. Molecular Genetics & Genomic Medicine 8: e1149. [Google Scholar]
- Albarzinji, Balnd M., Farhad M. Abdulkarim, Shaho A. Hussein, Dlshad Rashid, and Hayder Lazim. 2022. Population Genetic Study of 17 Y-STR Loci of the Sorani Kurds in the Province of Sulaymaniyah, Iraq. BMC Genomics 23: 763. [Google Scholar] [PubMed]
- Alonso, Antonio, Pedro Alberto Barrio, Petra Müller, Steffi Köcher, Burkhard Berger, Pablo Martin, Martin Bodner, Sascha Willuweit, Walther Parson, Lutz Roewer, and et al. 2018. Current state-of-art of STR sequencing in forensic genetics. Electrophoresis 39: 2655–68. [Google Scholar] [CrossRef]
- Alonso-Milan, Jesús. 2015. La guerra total en España (1936–1939). Scotts Valley: CreateSpace Independent Publishing Platform (Amazon). [Google Scholar]
- Ambrosio, Isabela Brunelli, Danilo Faustino Braganholi, Larissa Barros Muniz Orlando, Natalia Carolina Andrekenas, Isabel da Mota Pontes, Dayse Aparecida da Silva, Spartaco Astolfi-Filho, Elizeu Fagundes de Carvalho, Regina Maria Barretto Cicarelli, and Leonor Gusmão. 2020. Mutational data and population profiling of 23 Y-STRs in three Brazilian populations. Forensic Science International: Genetics 48: 102348. [Google Scholar]
- Ashirbekov, Yeldar, Arman Abaildayev, Alena Neupokoyeva, Zhaxylyk Sabitov, and Maxat Zhabagin. 2022. Genetic polymorphism of 27 Y-STR loci in Kazakh populations from Northern Kazakhstan. Annals of Human Biology 49: 87–89. [Google Scholar] [CrossRef] [PubMed]
- Ashirbekov, Yeldar, Anastassiya Nogay, Arman Abaildayev, Aigul Zhunussova, Zhaxylyk Sabitov, and Maxat Zhabagin. 2023. Genetic polymorphism of 27 Y-STR loci in Kazakh populations from Eastern Kazakhstan. Annals of Human Biology 49: 87–89. [Google Scholar] [CrossRef]
- Babić Jordamović, Naida, Tamara Kojović, Serkan Dogan, Larisa Bešić, Lana Sali-hefendić, Rijad Konjhodžić, Vedrana Škaro, Petar Projić, Vesna Hadžiavdi5, Adna Ašić, and et al. 2021. Haplogroup Prediction Using Y-Chromosomal Short Tandem Repeats in the General Population of Bosnia and Herzegovina. Frontiers in Genetics 12: 671467. [Google Scholar]
- Baeta, Miriam. 2012. New miniSTRs typing methods help in the identification of ancient skeletal remains from the Spanish civil war. Quaternary International 33: 279–80. [Google Scholar]
- Baeta, Miriam, Carolina Nuñez, Caterina Raffone, Eva Granizo, Leire Palencia-Madrid, Sergio Cardoso, Francisco Etxeberria, Lourdes Herrasti, and Marian M. de Pancorbo. 2019a. Updating data on the genetic identification of bone remains of victims of the Spanish Civil War. Forensic Science International: Genetics Supplement Series 7: 582–84. [Google Scholar] [CrossRef]
- Baeta, Miriam, Sandra García-Rey, Leire Palencia-Madrid, Caterina Raffone, and Marian M. de Pancorbo. 2019b. Forensic application of a mtDNA minisequencing 52plex: Tracing maternal lineages in Spanish Civil War remains. Forensic Science International: Genetics Supplement Series 7: 457–58. [Google Scholar] [CrossRef]
- BBC. 2008. Heavy shelling in Burundi capital. BBC News, April 18. [Google Scholar]
- Bercovitch, Jacob, and Richard Jackson. 1997. International Conflict: A Chronological Encyclopedia of Conflicts and Their Management 1945–1995. Washington, DC: CQ Press. [Google Scholar]
- Bini, Carla, Stefania Sarno, Elisabetta Tangorra, Alessandra Iuvaro, Sara De Fanti, Yohannes Ghebremedhin Tseghereda, Susi Pelotti, and Donata Luiselli. 2021. Haplotype data and forensic evaluation of 23 Y-STR and 12 X-STR loci in eight ethnic groups from Eritrea. International Journal of Legal Medicine 135: 449–53. [Google Scholar] [CrossRef]
- Bleay, Stephen M., Ruth S. Croxton, and Marcel De Put. 2018. Fingerprint Development Techniques: Theory and Application, in Developments in Forensic Science. Hoboken: John Wiley & Sons. [Google Scholar]
- Budowle, Bruce, and Angela Van Daal. 2008. Forensically relevant SNP classes. Biotechniques 44: 603–10. [Google Scholar] [CrossRef] [PubMed]
- Calleja, James. 2008. Aged Population, Violence and Nonviolence Toward. In Encyclopedia of Violence, Peace, & Conflict, 2nd ed. Cambridge: Academic Press, pp. 1–14. [Google Scholar]
- Caputo, Mariela, A. Sala, and Daniel Corach. 2019. Demand for larger Y-STR reference databases in ethnic melting-pot countries: Argentina as a test case. International Journal of Legal Medicine 133: 1309–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, Pengyu, Yanyan Han, Guanglin He, Haibo Luo, Tianzhen Gao, Feng Song, Dengfu Wan, Jian Yu, and Yiping Hou. 2018. Genetic diversity and phylogenetic study of the Chinese Gelao ethnic minority via 23 Y-STR loci. International Journal of Legal Medicine 132: 1093–96. [Google Scholar] [CrossRef]
- Claerhout, Sofie, Jennifer Roelens, Michiel Van der Haegen, Paulien Verstraete, Maarten HD Larmuseau, and Ronny Decorte. 2020. Ysurnames? The patrilineal Y-chromosome and surname correlation for DNA kinship research. Forensic Science International: Genetics 44: 102204. [Google Scholar] [CrossRef] [PubMed]
- Clodfelter, Micheal. 2017. Warfare and Armed Conflicts: A Statistical Encyclopedia of Casualty and Other Figures, 1492–2015, 4th ed. Jefferson: McFarland. ISBN 978-0786474707. [Google Scholar]
- Correia, Pedro, and Maria de Fátima Pinheiro. 2013. Perspectivas actuais da Lofoscopia: Aplicação Criminal e civil do Estudo de impressões epidermicas. In Ciencias Forenses ao Serviço da Justiça. Edited by Maria de Fátima Pinheiro. Lisboa: Pactor, pp. 119–22. [Google Scholar]
- Cumings, Bruce. 2011. The Korean War: A History. New York: Modern Library, p. 35. ISBN 9780812978964. [Google Scholar]
- Cunningham, David E. 2011. Barriers to Peace in Civil War. Cambridge: Cambridge University Press. ISBN 9781139499408. [Google Scholar]
- Dixon, Jeffrey, and Meredith Sarkees. 2015. A Guide to Intra-state Wars: An Examination of Civil, Regional, and Intercommunal Wars, 1816–2014. Washington, DC: CQ Press. [Google Scholar]
- Dooley, Kyla Bianca, M. Thabang Madisha, Sonja Strümpher, and Karen Ehlers. 2022. Forensic Genetic Value of 27 Y-STR Loci (Y-Filer® Plus) In The South African Population. Science & Justice 62: 358–64. [Google Scholar]
- Durney, James. 2011. The Civil War in Kildare. Cork: Mercier Press, p. 159. ISBN 978-1-85635-757-9. [Google Scholar]
- Eckhardt, William, and Ruth Leger Sivard. 1987. World Military and Social Expenditures 1987–1988, 12th ed. Washington, DC: World Priorities. [Google Scholar]
- Emmons, Alexandra. 2015. The Preservation and Persistence of Human DNA in Soil during Cadaver Decomposition. Master’s thesis, University of Tennessee, Knoxville, TN, USA. [Google Scholar]
- Etxeberria, Francisco, Alfredo González-Ruibal, Lourdes Herrasti, Nicholas Márquez-Grant, Laura Muñoz-Encinar, and Jordi Ramos. 2021. Twenty years of forensic archaeology and anthropology of the Spanish Civil War (1936–1939) and Francoist Regime. Forensic Science International: Synergy 3: 100159. [Google Scholar] [CrossRef]
- Fang, Guang-Yao, Zhen-Qing Zhang, Pei-Zhi Tang, Dan-Lu Song, Xing-Kai Zheng, Yi-Jun Zhou, and Meng-Nan Liu. 2021. Forensic and phylogenetic analyses of populations in the Tibetan-Yi corridor using 41 Y-STRs. International Journal of Legal Medicine 135: 783–85. [Google Scholar]
- Fernández-Álvarez, José-Paulino, David Rubio-Melendi, Antxoka Martínez-Velasco, Jamie K. Pringle, and Hector-David Aguilera. 2016. Discovery of a mass grave from the Spanish Civil War using Ground Penetrating Radar and forensic archaeology. Forensic Science International 267: e10–e17. [Google Scholar] [CrossRef]
- Figuero, María Jesús, Peral Diego, and Sánchez José Antonio. 2007. ¿Por qué no se pueden hacer estudios de ADN de restos óseos antiguos en Badajoz? Revista de estudios extremeños 63: 215–31. [Google Scholar]
- Finnish Government Project. 2004. Vuoden 1918 sodan sotasurmat kuolintavan ja osapuolen mukaan. Valtioneuvoston kanslia, Suomi Finland (Finnish Government war casualties project). Suomen sotasurmat 1914–1922, archived from the original on 10 March 2015, retrieved 22 March 2021. Available online: https://www.sotasampo.fi/en/ (accessed on 23 April 2023).
- Forrest, Alex. 2019. Forensic odontology in DVI: Current practice and recent advances. Forensic Sciences Research 4: 316–30. [Google Scholar] [CrossRef] [PubMed]
- Fry, Douglas. 2008. Aggression and Altruism. In Encyclopedia of Violence, Peace, & Conflict, 2nd ed. Edited by Lester R. Kurtz. New York: Academic Press, pp. 14–28. [Google Scholar]
- Gomes, Cláudia. 2020. Investigación de parentesco biológico en muestras críticas-utilidad en casos de investigación histórica, antropológica y/o forense. Ph.D. thesis, University Complutense of Madrid, Madrid, Spain. [Google Scholar]
- Gomes, Cláudia, Alejandro Alonso, Domingo Marquina, Marc Guardià, César López-Matayoshi, Sara Palomo-Díez, Bernardo Perea-Peréz, Juan F. Gibaja, and Eduardo Arroyo-Pardo. 2017. “Inhibiting inhibitors”: Preliminary results of a new “DNA extraction-amplification” disinhibition technique in critical human samples. Forensic Science International: Genetics Supplement Series 6: e197–9. [Google Scholar] [CrossRef]
- Gomes, Cláudia, and Eduardo Arroyo-Pardo. 2022. Usefulness of the X-Chromosome on Forensic Science. In Handbook of DNA Profiling. Edited by Hirak Ranjan Dash, Pankaj Shrivastava and José Antonio Lorente. Singapore: Springer Nature Singapore, pp. 455–77. [Google Scholar]
- Gomes, Cláudia, José David Quintero-Brito, Jesús Martínez-Gómez, Rui Pereira, Carlos Baeza-Richer, Mercedes Aler Gay, Laura Díez-Juárez, Sara Palomo-Díez, Ana María López-Parra, Elena Labajo-González, and et al. 2020. Spanish allele and haplotype database for 32 X-chromosome Insertion-Deletion polymorphisms. Forensic Science International: Genetics 46: 102262. [Google Scholar] [CrossRef]
- Gomes, Cláudia, María Fondevila, Concepción Magaña-Loarte, Juan Fernández-Jiménez, José Fernández-Serrano, Sara Palomo-Díez, C. Baeza-Richer, A. M. López-Parra, and E. Arroyo-Pardo. 2019a. An unusual kinship case from the Spanish Civil War (1936–1939): Ancient versus degraded sample’s investigation. Forensic Science International: Genetics Supplement Series 7: 690–91. [Google Scholar] [CrossRef]
- Gomes, Cláudia, Marta Magalhães, Cíntia Alves, António Amorim, Nádia Pinto, and Leonor Gusmão. 2012. Comparative evaluation of alternative batteries of genetic markers to complement autosomal STRs in kinship investigations: Autosomal indels vs. X-chromosome STRs. International Journal of Legal Medicine 126: 917–21. [Google Scholar] [CrossRef] [PubMed]
- Gomes, Cláudia, Sara Palomo-Díez, Carlos Baeza-Richer, Ana María López-Parra, Ivon Cuscó, Elena Garcia-Arumí, Eduardo Tizzano, Andrea Fernández-Vilela, Diego López-Onaindia, Ares Vidal Aixalà, and et al. 2019b. X-InDels efficacy evaluation in a critical samples paternity case: A Spanish Civil War case from the memorial of the camposines (Tarragona, Spain). Forensic Science International: Genetics Supplement Series 7: 494–95. [Google Scholar] [CrossRef]
- Gomes, Iva, Nádia Pinto, Sofia Antão-Sousa, Verónica Gomes, Leonor Gusmão, and António Amorim. 2020. Twenty Years Later: A Comprehensive Review of the X Chromosome Use in Forensic Genetics. Frontiers in Genetics 11: 926. [Google Scholar]
- González, Angéla, Catherine Cannet, Vincent Zvénigorosky, Annie Geraut, Guillaume Koch, Tania Delabarde, Bertrand Ludes, Jean-Sébastien Raul, and Christine Keyser. 2020. The petrous bone: Ideal substrate in legal medicine? Orensic Science International: Genetics 47: 102305. [Google Scholar] [CrossRef]
- Hagelstein, Roman. 2007. Where and When Does Violence Pay Off? The Algerian Civil War. Available online: https://www.researchgate.net/publication/228470456_Where_and_When_Does_Violence_Pay_Off_The_Algerian_Civil_War (accessed on 21 April 2023).
- Hakim, Hashom Mohd, Hussein Omar Khan, Siti Afifah Ismail, Japareng Lalung, Abban Edward Kofi, Bryan Raveen Nelson, Mohd Tajuddin Abdullah, Geoffrey Keith Chambers, and Hisham Atan Edinur. 2020. Population data for 23 Y chromosome STR loci using the Powerplex® Y23 STR kit for the Kedayan population in Malaysia. International Journal of Legal Medicine 134: 1335–37. [Google Scholar] [CrossRef]
- Harbom, Lotta, and Peter Wallensteen. 2007. Armed Conflict, 1989—2006. Journal of Peace Research 44: 623–34. [Google Scholar] [CrossRef]
- Herrasti, Lourdes, Nicholas Márquez-Grant, and Francisco Etxeberria. 2021. Spanish Civil War: The recovery and identification of combatants. Forensic Science International 320: 110706. [Google Scholar] [CrossRef] [PubMed]
- Hilton, Stanley. 1982. A Guerra Civil Brasileira. Rio de Janeiro: Nova Fronteira. [Google Scholar]
- Holquist, Peter. 2002. Making War, Forging Revolution: Russia’s Continuum of Crisis, 1914–1921. Cambridge: Harvard University Press. ISBN 0-674-00907-X. [Google Scholar]
- ICMP—International Commission on Missing Persons. 2022. About Us, What We Do, Where We Work. Available online: https://www.icmp.int/ (accessed on 12 June 2022).
- INTERPOL. 2018. Identificación de Víctimas de Catástrofes (IVC). Lyon: Interpol. [Google Scholar]
- Jankova, Renata, Maria Seidel, Alja Videtič Paska, Sascha Willuweit, and Lutz Roewer. 2019. Y-chromosome diversity of the three major ethno-linguistic groups in the Republic of North Macedonia. Forensic Science International: Genetics 42: 165–70. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, Christopher. 2000. The Rise and Fall of Albania’s Pyramid Schemes. Finance & Development: A Quarterly Magazine of the IMF. Washington, DC: International Monetary Fund. [Google Scholar]
- Jayakrishnan, Jijin Mekkadath, Jagat Reddy, and R. B. Vinod Kumar. 2021. Role of forensic odontology and anthropology in the identification of human remains. Journal of Oral and Maxillofacial Pathology 25: 543–47. [Google Scholar] [CrossRef] [PubMed]
- Jones, Howard. 1989. A New Kind of War. Oxford: Oxford University Press,. [Google Scholar]
- Kidd, Kenneth K., Andrew J. Pakstis, William C. Speed, Elena L. Grigorenko, Sylvester LB Kajuna, Nganyirwa J. Karoma, Selemani Kungulilo, Jong-Jin Kim, Ru-Band Lu, Adekunle Odunsi, and et al. 2006. Developing a SNP panel for forensic identification of individuals. Forensic Science International 164: 20–32. [Google Scholar] [CrossRef] [PubMed]
- Labajo, Elena, and Bernardo Perea. 2022. Odontología Forense: El papel del odontólogo en la investigación criminal. In Manual para el estudio de las Ciencias Forenses. Madrid: Tébar Flores Editorial, pp. 51–75. [Google Scholar]
- Le, Kien, and My Nguyen. 2020. Armed conflict and birth weight. Economics & Human Biology 39: 100921. [Google Scholar]
- Lee, Dong Gyu, Su Jin Kim, Woo-Cheol Cho, Yoonjung Cho, Ji Hwan Park, Jinmyung Lee, and Ju Yeon Jung. 2023. Analysis of mutation rates and haplotypes of 23 Y-chromosomal STRs in Korean father–son pairs. Forensic Science International: Genetics 65: 102875. [Google Scholar] [CrossRef]
- Lehne, Inge, and Johnson Lonnie. 1985. Vienna: The Past in the Present. Wien: Österreichischer Bundesverlag Gesellschaft. ISBN 3-215-05758-1. [Google Scholar]
- Lennard, Chris. 2019. Fingerprint Techniques. In Encyclopedia of Analytical Science, 3rd ed. Edited by Paul Worsfold, Alan Townshend, Colin F. Poole and Manuel Miró. New York: Academic Press, pp. 38–47. [Google Scholar]
- Lewy, Guenter. 1980. America in Vietnam. Oxford: Oxford University Press, pp. 450–53. ISBN 9780199874231. [Google Scholar]
- Lomperis, Timothy. 1996. From People’s War to People’s Rule. Chapel Hill: University of North Carolina Press. [Google Scholar]
- Luque, Juan Antonio. 2019. Estudio de las relaciones de parentesco. In Genética Forense- Del laboratorio a los Tribunales. Edited by Manuel C. Crespillo Márquez and Pedro Barrio Caballero. Madrid: Díaz de Santos, pp. 351–81. [Google Scholar]
- Marshall, Charla, Kimberly Sturk-Andreaggi, Joseph D. Ring, Cassandra R. Taylor, Suzanne Barritt-Ross, Walther Parson, and Timothy P. McMahon. 2019. Advancing mitochondrial genome data inter-pretation in missing persons casework. Forensic Science International: Genetics Supplement Series 7: 721–23. [Google Scholar]
- Mienkerd, Sirirat, Anillada Nettakul, and Worawee Waiyawuth. 2019. The using of massively parallel sequencing of mitochondrial DNA to assist the missing person identification: Human remains in the wild. Forensic Science International: Genetics Supplement Series 7: 716–17. [Google Scholar] [CrossRef]
- Mihajlovic, Milica, Vanja Tanasic, Milica Keckarevic Markovic, Miljana Kecmanovic, and Dusan Keckarevic. 2022. Distribution of Y-chromosome haplogroups in Serbian population groups originating from historically and geographically significant distinct parts of the Balkan Peninsula. Forensic Science International: Genetics 61: 102767. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, Faraz, Arishiya T. Fairozekhan, Subraya Bhat, and Ritesh G. Menezes. 2022. Forensic Odontology. In StatPearls. Tampa: StatPearls Publishing. [Google Scholar]
- Neyra-Rivera, Carlos David, Andres Ticona Arenas, Edgardo Delgado Ramos, Margarita Rosa Eugenia Velasquez Reinoso, Omar Alberto Caceres Rey, and Bruce Budowle. 2021. Population data of 27 Y-chromosome STRS in Aymara population from Peru. Australian Journal of Forensic Sciences 54: 596–610. [Google Scholar] [CrossRef]
- O’Balance, Edgar. 1966. The Greek Civil War: 1944–1949. New York: Praeger. [Google Scholar]
- OCHA Services. 2012. 17,800 People Died during Conflict Period, Says Ministry of Peace. New York: OCHA—ONU. [Google Scholar]
- Pääbo, Svante, Hendrik Poinar, David Serre, Viviane Jaenicke-Després, Juliane Hebler, Nadin Rohland, Melanie Kuch, Johannes Krause, Linda Vigilant, and Michael Hofreiter. 2004. Genetic analyses from ancient DNA. Annual Review of Genetics 38: 645–79. [Google Scholar] [CrossRef]
- Pajnič, Irena. 2021. Identification of a Slovenian prewar elite couple killed in the Second World War. Forensic Science International 327: 110994. [Google Scholar] [CrossRef]
- Pajnič, Irena Zupanič, Jezerka Inkret, Tomaž Zupanc, and Eva Podovšovnik. 2021. Comparison of nuclear DNA yield and STR typing success in Second World War petrous bones and metacarpals III. Forensic Science International: Genetics 55: 102578. [Google Scholar] [CrossRef]
- Pajnič, Irena Zupanič, Marcel Obal, and Tomaž Zupanc. 2020. Identifying victims of the largest Second World War family massacre in Slovenia. Forensic Science International 306: 110056. [Google Scholar] [CrossRef]
- Palomo-Díez, Sara, and Ana María López-Parra. 2022. Utility and Applications of Lineage Markers: Mitochondrial DNA and Y Chromosome. In Handbook of DNA Profiling. Edited by Hirak Ranjan Dash, Pankaj Shrivastava and José Antonio Lorente. Singapore: Springer Nature Singapore, pp. 423–54. [Google Scholar]
- Palomo-Díez, Sara, Cláudia Gomes, Ana María López-Parra, Carlos Baeza-Richer, Ivón Cuscó, C. Raffone, E. García-Arumí, Núria Montes, Diego López Onaindia, Assumpció Malgosa Morera, and et al. 2019. Genetic identification of Spanish civil war victims. The state of the art in Catalonia (Northeastern Spain). Forensic Science International: Genetics Supplement Series 7: 419–21. [Google Scholar] [CrossRef]
- Palomo-Díez, Sara, Cláudia Gomes, Ana María López-Parra, Ivon Cuscó, Eduardo Tizzano, María Eulalia Subirá, Andrea Fernández Vilela, Núria Montes, Diego López Onaindia, Assumpció Malgosa Morera, and et al. 2021. Estado actual de la identificación de víctimas de la Guerra Civil Española y de la posguerra en Cataluña. Revista d’Arqueologia de Ponent 31: 273–86. [Google Scholar]
- Pamjav, Horolma, Ábel Fóthi, Dániel Dudás, Attila Tapasztó, Virág Krizsik, and Erzsébet Fóthi. 2022. The paternal genetic legacy of Hungarian-speaking Rétköz (Hungary) and Váh valley (Slovakia) populations. Frontiers in Genetics 13: 977517. [Google Scholar] [CrossRef] [PubMed]
- Pereira, Vânia, and Leonor Gusmão. 2019. Análisis de Marcadores STR de Cromosoma Y. in Genética Forense—Del laboratorio a los tribunals. Edited by Manuel Crespillo and Pedro Barrio. Madrid: Díaz de Santos. [Google Scholar]
- Pettersson, Therése, and Peter Wallensteen. 2015. Armed conflicts, 1946–2014. Journal of Peace Research 52: 536–50. [Google Scholar] [CrossRef]
- Pilli, Elena, Stefania Vai, Marina Grazia Caruso, Giancarlo D’Errico, Andrea Berti, and David Caramelli. 2018. Neither femur nor tooth: Petrous bone for identifying archaeological bone samples via forensic approach. Forensic Science International 283: 144–49. [Google Scholar] [CrossRef]
- Pinto, Nádia, Leonor Gusmão, and António Amorim. 2010. Likelihood ratios in kinship analysis: Contrasting kinship classes, not genealogies. Forensic science international. Genetics 43: 21. [Google Scholar]
- Pinto, Nádia, Leonor Gusmão, and António Amorim. 2011. X-chromosome markers in kinship testing: A generalisation of the IBD approach identifying situations where their contribution is crucial. Forensic Science International: Genetics 5: 27–32. [Google Scholar] [CrossRef]
- Pinto, Nádia, Pedro V. Silva, and António Amorim. 2012. A general method to assess the utility of the X-chromosomal markers in kinship testing. Forensic Science International: Genetics 6: 198–207. [Google Scholar] [CrossRef] [PubMed]
- Resource Information Center. 2000. Angola: Current Political and Human Rights Conditions in Angola. Washington, DC: United States Bureau of Citizenship and Immigration Services. [Google Scholar]
- Ríos, Luis. 2012. Identificación en fosas comunes de la guerra civil: Limitaciones y posibilidades a partir del caso de Burgos. Boletín Gallego de Medicina Legal e Forense 12: 125–42. [Google Scholar]
- Simkin, John. 2012. Spanish Civil War. The Spanish Civil War Encyclopedia (Ser. Spanish Civil War). Brighton and Hove: University of Sussex, Spartacus Educational E-Books. [Google Scholar]
- Singh, Risha, Srinivas Goli, and Abhra Singh. 2022. Armed conflicts and girl child marriages: A global evidence. Children and Youth Services Review 137: 106458. [Google Scholar] [CrossRef]
- Strathern, Andrew J., and Pamela J. Stewart. 2008. Anthropology of Violence and Conflict, Overview. In Encyclopedia of Violence, Peace, & Conflict, 2nd ed. Edited by Lester Kurtz. New York: Academic Press, pp. 75–86. [Google Scholar]
- SWGDAM. n.d. Interpretation Guidelines for Y Chromosome STR Testing. Available online: https://www.swgdam.org/publications (accessed on 19 April 2023).
- Tätte, Kai, Ene Metspalu, Helen Post, Leire Palencia-Madrid, Javier Rodríguez Luis, Maere Reidla, Erika Tamm, Anne-Mai Ilumäe, Marian M. de Pancorbo, Ralph Garcia-Bertrand, and et al. 2022. Genetic characterization of populations in the Marquesas Archipelago in the context of the Austronesian expansion. Scientific Reports 12: 5312. [Google Scholar] [CrossRef] [PubMed]
- Theune, Claudia. 2022. Archeology of War and Armed Conflict. In Encyclopedia of Violence, Peace, & Conflict, 3rd ed. Edited by Lester Kurtz. New York: Academic Press, pp. 552–61. [Google Scholar]
- Thomas, Hugh. 1977. The Spanish Civil War. London: Penguin Books. [Google Scholar]
- Toscanini, Ulises. 2019. Historia y evolución de la genética forense. Grupos de Trabajo de estandarización científica. In Genética Forense- Del laboratorio a los Tribunales. Edited by Manuel C. Crespillo Márquez and Pedro Barrio Caballero. Madrid: Díaz de Santos, pp. 1–21. [Google Scholar]
- Truth Commission: Chile 90. n.d. Report of the Chilean National Commission on Truth and Reconciliation. Available online: https://www.usip.org/publications/1990/05/truth-commission-chile-90 (accessed on 23 April 2023).
- U.S. Committee for Refugees and Immigrants. 2001. Sudan: Nearly 2 Million Dead as a Result of the World’s Longest-Running Civil War. Arlington: U.S. Committee for Refugees and Immigrants. [Google Scholar]
- Vullo, Carlos María. 2019. Identificación de desaparecidos en gran escala: Grandes catástrofes (DVI) y desapariciones en contextos de crises Humanitarias (MPI). In Genética Forense- Del laboratorio a los Tribunales. Edited by Manuel C. Crespillo Márquez and Pedro Barrio Caballero. Madrid: Díaz de Santos, pp. 405–24. [Google Scholar]
- Ziętkiewicz, Ewa, Magdalena Witt, Patrycja Daca, Jadwiga Żebracka-Gala, Mariusz Goniewicz, Barbara Jarząb, and Michał Witt. 2012. Current genetic methodologies in the identification of disaster victims and in forensic analysis. Journal of Applied Genetics 53: 41–60. [Google Scholar] [CrossRef]
| Armed Conflict | Date | Estimated Number of Deaths |
|---|---|---|
| Finnish civil war | 1918 | Over 33,000 (Finnish Government Project 2004) |
| Russian civil war | 1917–1921 | 7,000,000–12,000,000 (Holquist 2002) |
| Irish civil war | 1922–1923 | 300–400 (Durney 2011) |
| Austrian civil war | 1934 | 100–1000 (Lehne and Lonnie 1985) |
| Spanish civil war | 1936–1939 | Over 300,000 (Thomas 1977; Simkin 2012; Alonso-Milan 2015; Clodfelter 2017) |
| Greek civil war | 1945–1949 | Over 150,000 (O’Balance 1966; Jones 1989; Lomperis 1996; Bercovitch and Jackson 1997) |
| Albanian civil war | 1997 | 2000–3800 (Jarvis 2000) |
| Korean civil war | 1950–1953 | 2,000,000–3,000,000 (Lewy 1980; Cumings 2011) |
| Nepalese civil war | 1996–2006 | 12,000 (OCHA Services 2012) |
| Angola civil war | 1961–1974 | 500,000–800,000 (Resource Information Center 2000) |
| Algeria civil war | 1954–1962 | 150,000 (Hagelstein 2007) |
| Burundi civil war | 1993–2005 | 300,000 (BBC 2010) |
| Rwanda civil war | 1994 | 500,000–800,000 (Cunningham 2011) |
| Second Sudanese Civil War | 2013–2020 | 1,000,000–2,500,000 (U.S. Committee for Refugees and Immigrants 2001) |
| Cuban Revolution | 1956–1959 | Over 3000 (Dixon and Sarkees 2015) |
| Chaco War (Paraguay–Bolivia,) | 1932–1935 | 70,000 (Eckhardt and Sivard 1987) |
| Brazil–Constitutionalist Revolution | 1964–1985 | 2500–3000 (Hilton 1982) |
| Chile | 1973–1990 | Over 3000 (Truth Commission: Chile 90 n.d.) |
| Genealogical Relationship | Allele Sharing Partitions | ||
|---|---|---|---|
| k2 | k1 | k0 | |
| Parent–Child | 0 | 1 | 0 |
| Full siblings | 1/4 | 1/2 | 1/4 |
| Half siblings | 0 | 1/2 | 1/2 |
| Avuncular | 0 | 1/2 | 1/2 |
| Grandparent–Grandchild | 0 | 1/2 | 1/2 |
| Double first-cousins | 1/16 | 3/8 | 9/16 |
| First Cousins | 0 | 1/4 | 3/4 |
| Double half-cousins | 1/64 | 7/32 | 49/64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manera-Scliar, G.; Hernández, S.; Martín-López, M.; Gomes, C. Difficulties in Kinship Analysis for Victims’ Identification in Armed Conflicts. Genealogy 2023, 7, 31. https://doi.org/10.3390/genealogy7020031
Manera-Scliar G, Hernández S, Martín-López M, Gomes C. Difficulties in Kinship Analysis for Victims’ Identification in Armed Conflicts. Genealogy. 2023; 7(2):31. https://doi.org/10.3390/genealogy7020031
Chicago/Turabian StyleManera-Scliar, Gabriel, Santiago Hernández, Miguel Martín-López, and Cláudia Gomes. 2023. "Difficulties in Kinship Analysis for Victims’ Identification in Armed Conflicts" Genealogy 7, no. 2: 31. https://doi.org/10.3390/genealogy7020031
APA StyleManera-Scliar, G., Hernández, S., Martín-López, M., & Gomes, C. (2023). Difficulties in Kinship Analysis for Victims’ Identification in Armed Conflicts. Genealogy, 7(2), 31. https://doi.org/10.3390/genealogy7020031





